Abstract
Mixed cultures represent better alternatives to ferment organic waste and dark fermentation products in anerobic conditions because the microbial associations contribute to electron transfer mechanisms and combine metabolic possibilities. The understanding of the microbial interactions in natural and synthetic consortia and the strategies to improve the performance of the processes by bioaugmentation provide insight into the physiology and ecology of the mixed cultures used for biotechnological purposes. Here, synthetic microbial communities were built from three hydrogen (bioH2) and poly-hydroxy-alkanoates (PHA) producers, Clostridium pasteurianum, Rhodopseudomonas palustris and Syntrophomonas wolfei, and a photoheterotrophic mixed consortium C4, and their performance was evaluated during photofermentation. Higher hydrogen volumetric production rates (H2VPR) were determined with the consortia (28–40 mL/Lh) as compared with individual strains (20–27 mL/Lh). The designed consortia reached the highest bioH2 and PHA productions of 44.3 mmol and 50.46% and produced both metabolites simultaneously using dark fermentation effluents composed of a mixture of lactic, butyric, acetic, and propionic acids. When the mixed culture C4 was bioaugmented with S. wolfei, the bioH2 and PHA production reached 32 mmol and 50%, respectively. Overall, the consumption of organic acids was above 50%, which accounted up to 55% of total chemical oxygen demand (COD) removed. Increased bioH2 was observed in the condition when S. wolfei was added as the bioaugmentation agent, reaching up to 562 mL of H2 produced per gram of COD. The enhanced production of bioH2 and PHA can be explained by the metabolic interaction between the three selected strains, which likely include thermodynamic equilibrium, the assimilation of organic acids via beta-oxidation, and the production of bioH2 using a proton driving force derived from reduced menaquinone or via electron bifurcation.
1. Introduction
Biological hydrogen (bioH2) is a clean biofuel produced during dark fermentation (DF) and photofermentation (PF). BioH2 production during microbial fermentation is a way to dissipate excess reductants (mainly by regenerating NAD+ from NADH) as a gas [1]. During DF, bioH2 is produced with organic acids (OAs) and alcohols, leading to low bioH2 yields. To improve that yield, coupling DF and PF processes allow for the assimilation of the OAs produced during the DF in the PF to produce more bioH2 from the substrates [2,3] toward closed production cycles. The sequential DF-PF process for bioH2 has been previously studied [3,4,5,6,7,8]; however, the role of the microbial interaction using consortiums, particularly in the PF processes, are still not clear.
While bioH2 production is still not commercially viable, its coproduction with other products in a biorefinery context should contribute to increase its viability, in particular as the world moves to a circular economy. Bioplastics, in particular those derived from polyhydroxyalkanoates (PHAs) have attracted attention as replacement to petroleum-based plastics. Their production faces similar problems in terms of cost, as compared to traditional plastics, and naturally, their biological production from cheap sources has been addressed [9]. Among the bacteria capable of producing PHAs, several can also produce hydrogen, and the “competition” for reducing equivalents between bioH2 and PHAs has been widely documented [8,10,11]; however, their simultaneous production might be advantageous to reducing their cost. Thulasidharan et al. [12] reviewed their simultaneous production and noted that, as suggested by Wang et al. [13], as PHA production increases, the conversion of pyruvate to Acetyl-CoA increases, which increases the rate of formate production and thus the rate of hydrogen formation, making their simultaneous production appealing. Moreover, ensuring the long-term sustainability of bioH2 and PHA production requires the optimization of their yields, since the cost of producing bioH2 has been estimated to be between USD 10 GJ−1 and USD 20 GJ−1, which is not competitive with gasoline (USD 0.33 GJ−1) [14]. Similarly, the cost of PHA of microbial origin (68 USD/kg, Goodfellow®) is approximately 20–80% higher than that of fossil-based synthetic plastics [15,16]. Simultaneous PHA accumulation can contribute to abatement of the costs of bioH2 production [17]. In this context, the production of biofuels and bioplastics from cheap substrates like organic residues and wastewater is a challenge to address to make it a competitive process. Still, organic waste and DF products are challenging to ferment in anaerobic conditions, especially using single strains. Additionally, the use of pure cultures in long-term processes at a large scale represents a considerable challenge because of the sterile conditions.
Mixed cultures represent better alternatives for processing complex substrates because they combine the metabolic possibilities of several microorganisms [18]. Clostridium pasteurianum and Rhodopseudomonas palustris exhibit a solid potential to produce bioH2 and different polymers when grown as pure cultures. Different species belonging to the genus Clostridium have been categorized as typical acids and bioH2 producers, fermenting carbohydrates to lactate (HLac), acetate (HAc), butyrate (HBu), bioH2, carbon dioxide and organic solvents. Similarly, photoheterotrophic purple nonsulfur bacteria (PNSB), such as R. palustris, constitute an attractive option for various biotechnological purposes because of their bioH2 yields and the diversity of valuable products they can generate, including biodegradable polymers, poly-hydroxy-alkanoates (PHAs) and their copolymers (PHB-PHV) [19], carotenoids and pigments [20]. Although the individual capabilities of C. pasteurianum and R. palustris have been demonstrated, their role in consortia has yet to be elucidated. Microbial consortia may establish electron transfer mechanisms that permit additional consumption of the complex substrates derived from DF. These and other possible interactions are essential at the fundamental and practical levels because they provide insight into the physiology and ecology of the mixed cultures and define their potential biotechnological utility [21,22].
Some researchers have extensively studied bioH2 production in R. palustris and nifA- mutants and developed metabolic models [8,10,23]. The coculture of R. palustris and C. pasteurianum has also been previously studied [24,25]. However, there is still a lack of information on the interactions established between these two microorganisms that allow for higher bioH2 production. Moreover, knowledge concerning the population dynamics, final products that could be obtained as a function of the culture conditions, and the predominant microbes in photoheterotrophic mixed cultures, is also scarce. Thus, a better understanding of the mechanisms that define final metabolite production is needed. In PNSB, it is well known that there is competition for reducing equivalents among biomass production, CO2 fixation, and bioH2 and PHA production, given that these compounds are electron sinks [8,10,11]. The transcriptional levels of the enzymes involved in a photoheterotrophic microbial consortium were studied using real-time quantitative PCR (RT-QPCR) [26]. Data showed that bioH2 production was associated with high transcription levels of the nouF gene (NADH ubiquinone dehydrogenase) and low expression of the uptake hydrogenase. Since the transcription of nouF observed in the photoheterotrophic culture is not related to R. palustris, this suggests that other microorganisms in the photoheterotrophic consortium influenced the metabolic activity or modified the regulation of the reducing equivalents (redox balance), thereby promoting bioH2 production.
Based on the understanding of the microbial interactions in natural and synthetic consortia and the strategies to improve the performance of bioprocesses, bioaugmentation is a suitable approach that allows for enhancing the transformation of substrates to products by increasing the concentration of one of the microbial populations or the addition of a different microorganism [21].
Here, we aim to describe the interactions between the microbial population in a natural photoheterotrophic consortium (“NPC-C4”) by evaluating the individual behavior of the strains R. palustris ATCC 1007 and C. pasteurianum ATCC 6013 to explain the metabolic mechanisms involved during bioH2 and PHA production. These microorganisms were selected based on a previous molecular characterization that identified them as the main microbial population in the consortium [19,26]. Additionally, the kinetic behavior of “NPC-C4” was evaluated when it was bioaugmented with Syntrophomonas wolfei ATCC BAA-1933 (“BNPC-C4”) to determine if it was possible to simultaneously obtain bioH2, PHA and biomass. In addition, a “designed consortium” containing C. pasteurianum, R. palustris and S. wolfei was evaluated to determine its ability to handle the complex composition (high concentrations of HLac and HBu) of DFE and produce PHA and bioH2 simultaneously. Using DFE as feedstock opens the possibilities of obtaining different biotechnological products and simultaneous elimination of residues.
2. Materials and Methods
A two-staged process was performed. Overall, organic wastes were used as inoculum for dark fermentation (DF). The effluent was then filtered and used as a carbon source for the photofermentation (PF). The following diagram (Figure 1) illustrates the process:
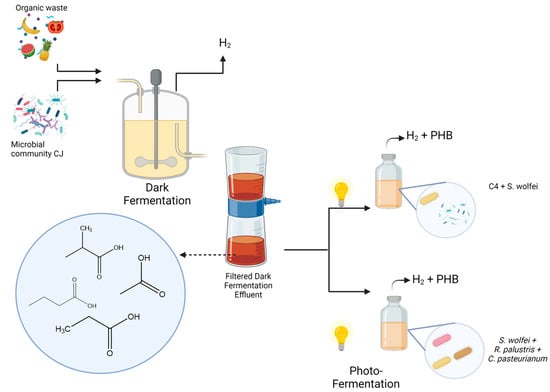
Figure 1.
Fermentation strategy. During the dark fermentation stage, organic wastes were used to produce H2 and organic acids. The effluent was then filtered and used as substrate for the photofermentation in serum bottles. Created with BioRender.com.
2.1. Inoculums for Dark- and Photofermentation Processes
2.1.1. Dark Fermentation with a Lactose-Adapted Consortium
The DF inoculum was previously adapted with lactose and FVW, as described by [27]. The FVWs were half diluted (1:1) with a mineral medium containing (g/L): (NH4)2SO4 3, KH2PO4 0.6, MgSO4 1.5, K2HPO4 2.4, CaSO4 0.15, FeSO4 0.03. The pH was adjusted and kept at 5.5 ± 0.2 during fermentation. Cultivation was performed in serological bottles of 120 mL of total volume filled with the FVW feedstock (50 mL), sparged with N2 gas for 10 min to assure anaerobic conditions and incubated at 35 °C and 160 rpm for 24 h. The HLac-adapted inoculum was subsequently used for the PF.
2.1.2. R. palustris ATCC 17001 and the NPC-C4 Growth Stage
R. palustris ATCC 17001 and “NPC-C4” were grown in sterile complete media that contained (g/L) KH2PO4 0.0495, MgSO4.7H2O 0.2, CaCl2.2H2O 0.075, (NH4)2SO4 1, and Na2HPO4. H2O 0.1295, EDTA 0.02, FeSO4 0.012, yeast extract 0.5 and HAc 1.0 as substrates. The pH of the mineral medium was adjusted to 6.8 ± 0.2. Serological bottles were filled with 70 mL of mineral medium, sparged with argon for 10 min, and inoculated and incubated at 35 °C and 160 rpm.
2.1.3. C. pasteurianum ATCC 6013
The growth media recommended by ATCC for C. pasteurianum was composed of (g/L) peptone 10, meat yeast extract 10, extract 3, dextrose 5, NaCl 5, starch soluble 1, L-cysteine 0.5 and HAc 3 g, with a pH of 6.8 ± 0.2. The bottles were sparged with N2 for 5 min, inoculated and then incubated at 35 °C and 160 rpm.
2.1.4. S. wolfei ATCC BAA 1933
The pure culture of S. wolfei was grown in a defined medium containing (in g/L): NaHCO3, 8.4; solution A, 10 mL; solution B, 2 mL; yeast extract, 2; tryptone, 2; trace solution, 2 mL; reducing agent, 10 mL. Solution A contained (g/L): NH4Cl, 100; MgCl2·6H2O, 1000; CaCl2, 40. Solution B contained (g/L): K2HPO4∙3 H2O, 200. The trace solution contained (in g/L) EDTANa2, 0.05; MgSO4∙7 H2O, 3; MnSO4∙H2O, 0.5; NaCl, 1; FeSO4∙7 H2O, 0.1; Co(NO3)2∙6 H2O, 0.1; CaCl2, 0.1; ZnSO4∙7 H2O, 3; CuSO4∙5 H2O, 0.5; Na2MoO4∙2 H2O, 1; and NiCl4∙6 H2O, 0.02. The reducing agent consisted of 30 g/L L-cysteine. The medium was sparged with a gas mixture of 20% CO2 and 80% N2 for approximately 30 min. Each solution was prepared separately, sterilized, and kept refrigerated at −4 °C for periods no longer than 6 months. S. wolfei was initially grown with 8 g/L crotonic acid as a substrate.
2.2. Experimental Conditions
2.2.1. Dark Fermentation Process
The DF inoculum described above was used to inoculate a 5 L reactor (Winpact One fermentation systems, FS-06, Major Science) filled with the diluted FVW mixture. The medium was sparged with N2 gas for 30 min after inoculation to assure anaerobic conditions [27]. The pH and temperature were automatically controlled at 6.8 ± 0.2 and 30 °C, respectively. The DF process lasted 96 h, and liquid and gas samples were regularly withdrawn to determine the concentration of the OAs and the bioH2 production during the process. The DFEs were used as substrates for PF.
2.2.2. Photofermentation Process
After an initial growth stage, DFEs were used to evaluate the bioH2 and PHA production by the individual strains, the “NPC-C4”, the bioaugmented natural photoheterotrophic consortium (NPC-C4 + S. wolfei; “BNPC-C4”), and the “designed consortium” (R. palustris, C. pasteurianum and S. wolfei). This last “designed consortium” and the “BNPC-C4” were inoculated with a 1:1-mass proportion of each strain.
DFEs were centrifuged at 3600 rpm for 30 min and sterilized for 1 h at 15 psi and 121 °C to be used as the substrate for the PF process. The DFE was diluted 1:2 with the mineral medium described above for R. palustris and “NPC-C4”, but without HAc, (NH4)2SO4, and yeast extract at pH 6.8 ± 0.2.
A total of 180 mL of diluted DFEs was added to modified 250 mL Wheaton bottles and inoculated with 20 mL of C. pasteurianum, R. palustris, S. wolfei and “NPC-C4”. The bottles were sparged with argon gas and incubated at 35 °C. The experiments were performed in duplicate.
The PF process lasted 166 h, and gas and liquid samples were taken every 4 h to quantify the bioH2 (0.25 mL), biomass (1 mL), and OAs. At the middle and end of the cultivation period, 50 mL was recovered to analyze the PHA accumulation.
2.3. Chemical Analysis
2.3.1. Biomass (Protein) Quantification
The biomass concentration in liquid samples was determined by the Bradford method [28] using bovine serum albumin as a standard solution, following the provider’s instructions.
2.3.2. bioH2 Gas Production
Gaseous samples were taken every 4 h from the headspace of each experimental system. The bioH2 was measured in gas samples by a TCD-GC as previously reported [26].
2.3.3. Nitrogen Quantification
The nitrogen content was quantified as total nitrogen Kjeldahl (TNK) with a HACH kit, following the provider’s instructions.
2.3.4. OA Quantification
Liquid samples were centrifuged, and the supernatant was filtered (0.2 µm) and analyzed by high-pressure liquid chromatography (HPLC) (Perkin Elmer®, Waltham, MA, USA) as previously reported [19].
2.3.5. PHA Accumulation
Extraction
Liquid samples (50 mL) were taken at the middle and end of the cultivation to extract the PHA content from the previously centrifuged biomass by the Soxhlet method. The extracting solvent was chloroform at 61 °C; the refluxes were 4 h long, and 8 refluxes were performed. After PHA extraction, the solvent was recovered by a distillation process, and the residue, considered PHA, was weighed and kept at ambient temperature for further characterization [19].
Biopolymer Characterization
Hydrogen nuclear magnetic resonance (HNMR) spectroscopy was used to characterize the chemical structure of the PHA. The NMR equipment was a Varian Mercury operating at 300 MHz. To prepare the samples for the characterization analysis, the PHA was dissolved in deuterated chloroform (CDCl3) [1% (w/v)]. The relative intensities of the 3HB and 3HV methyl resonance of the PHA were analyzed from 0.9 to 4.2 ppm [19].
3. Results
The initial OA compositions of the DFEs were 24 g/L, 0.87 g/L, 5.6 g/L and 0.68 g/L for HLac, HAc, HBu, and HPr, respectively. The DFE was diluted twofold to be used as feedstock for the PF process.
3.1. Individual Strain Cultures
Figure 2 shows the growth kinetics of C. pasteurianum, R. palustris and S. wolfei cultures grown in the DFEs.
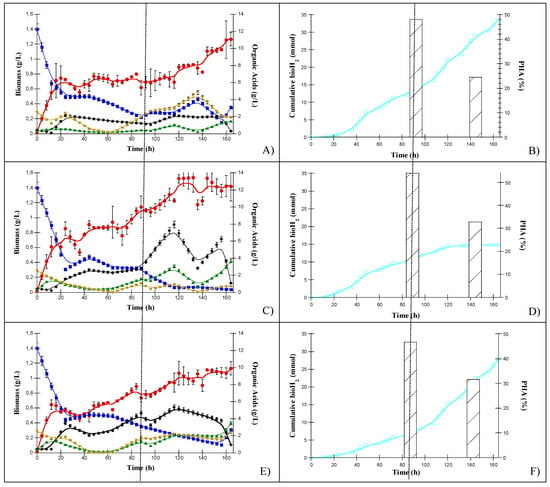
Figure 2.
Individual strain patterns of organic acid consumption: HLac (■), HAc (◆), HPr (▲), HBu (▼), biomass (λ), bioH2 (□) and PHA production (bars) obtained by C. pasteurianum in the upper panel (A,B), R. palustris in the middle panel (C,D), and S. wolfei in the bottom panel (E,F).
3.1.1. C. Pasteurian
The fermentation process follows three phases. The initial stage showed a fast growth rate (µ = 0.25 h−1), where HLac and HBut were used as substrates and HAc was the main product (Figure 2A). This period lasted approximately 44 h, reaching up to 0.87 g/L biomass with a low bioH2 production (cumulative bioH2 10 mmol) (Figure 2B). Then, a relatively constant phase of biomass and bioH2 production was maintained up to 110 h. A second increment in biomass and bioH2 was determined after 110 h of culture (1.43 g/L, µ = 0.0125 h−1). Higher biomass (Figure 2A) and bioH2 (Figure 2B) production were observed at the third stage of the cultivation process, and these were associated with the simultaneous HLac and HAc consumption from 50 h to 90 h of culture. The highest cumulative bioH2 production was 33 mmoL, corresponding to a bioH2 yield (H2Y) of 158.8 mL/g per total OA consumed (114.6 mL/g DQO).
During the first 12 h of cultivation, 52, 53 and 77% of HLAc, HBu and HAc were consumed, respectively, as shown in Figure 2A. After the first 24 h, HLac consumption reached a plateau at approximately 5 g/L until 50 h, when it was constantly consumed to a final concentration of 2 g/L at 88 h. By 137 h, the HLac concentration increased slightly. HBu was initially consumed for 12 h. Then, its concentration slightly increased at 24 h. It was exhausted between 24 and 50 h. At 60 h, the HBu concentration increased and reached a maximum of approximately 5 g/L at 137 h before being reassimilated. HAc was also initially consumed, but after 15 h, its concentration increased to up to 2 g/L at 25 h of cultivation. By 50 h, it was slowly consumed together with HLac until 90 h. This OA assimilation led to HBu and bioH2 production. The initial HPr concentration was consumed from 24 h to 48 h, and then increased to reach a concentration of 1 g/L at 116 h.
As seen in Figure 2B, C. pasteurianum produced bioH2 and PHA, and the cumulative PHA production was 48% and 24% at 88 h and 164 h, respectively. This PHA production was associated with the lowest OA concentration.
A linear correlation was found between the initial growth and OA consumption from the beginning of cultivation until 20 h. The low initial bioH2 production from 24 to 48 h was correlated with the consumption of HAc, HBu and HPr, with HBu being the more consumed substrate (with a yield of 0.067 mol/mol). Similarly, a positive correlation was determined during the second period of bioH2 production between 50 and 140 h, when HLac was consumed together with HAc, and HBu and HPr were produced. From 140 h onward, the increase in biomass was related to the consumption of HLac and HBut.
3.1.2. R. palustris
The growth kinetics of R. palustris are shown in Figure 2C. The growth and substrate consumption patterns were different from those determined for C. pasteurianum. R. palustris consumed 9.5 g/L HLac, 1 g/L HBu and 0.2 g/L HAc in 24 h, and the initial HLac consumption was higher than that of C. pasterianum. From the beginning of the process until 60 h, HBu was steadily consumed, while the HLac concentration remained almost constant from 48 to 90 h. HLac was depleted at 120 h. R. palustris initially assimilated Hac and then produced it, and the concentration remained at 2 g/L until 90 h. HPr was produced at the beginning and end of the fermentation process. At the first maximum, it reached a concentration of 3.5 g/L, and then it was consumed following a similar pattern to that of HBu. Thus, HPr showed an oscillating behavior during the culture because R. palustris can assimilate HLac for synthesis of HAc and HPr.
The biomass concentration of R. palustris increased to reach a plateau at a concentration of 0.9 g/L after 48 h (µ = 0.165 h−1). Then, it increased gradually, reaching 1.4 g/L at 120 h (µ = 0.005 h−1) (Figure 2D). An initial low cumulative bioH2 production (8 mmol) was observed at approximately 44 h (Figure 2D). After that, the bioH2 production remained constant up to 90 h, attaining a final cumulative bioH2 production of 15 mmol (H2Y = 81.9 mL/g OA consumed; 45.8 mL/g DQO). The bioH2 production was half of that determined for C. pasteurianum. R. palustris produced a PHA percentage of 54 after 88 h, which could explain the lower H2Y, as PHA and bioH2 are both electron acceptors. However, the PHA concentration decreased to 32% after 164 h (Figure 2D).
A linear correlation was found between the initial growth and bioH2 production and HLac and HBu consumption (0–44 h). Similarly, a second bioH2 production occurred when consuming HLac and HBu with concurrent production of HAc (90–120 h); later, HAc was also used as substrate. The OA most used as substrate was HLac. For both C. pasteurianum and R. palustris, the particular combinations of OAs that were substrates or products changed throughout the cultivation.
3.1.3. S. wolfei
The results for S. wolfei cultures are presented in Figure 2E,F. Similar to C. pasteurianum, two growth and bioH2 production stages were observed. During the first stage, S. wolfei consumed 8 g/L HLac in 24 h, along with 0.64 g/L HBu within the first 12 h. HBu was no longer consumed for the next 12 h but the HBu was totally consumed from 24 h until 44 h. Associated with HBu consumption, an increase in HAc was observed for most of the cultivation, reaching a concentration up to 5 g/L at 90 h. HPr also increased at the initial, intermediate, and final stages of the process, but there were two intercalate periods when it was consumed. HLac was steadily consumed from 70 h and was almost exhausted by 136 h. Then, it was produced at the final stage of cultivation (164 h), reaching a concentration of 2.7 g/L. An initial biomass production of 0.64 g/L was determined in the first 12 h (µ = 0.29 h−1); it decreased and then gradually increased again to a final concentration of 1.13 g/L after 164 h (Figure 2F).
The bioH2 production (Figure 2F) also showed two production stages. It started after 40 h, attaining a low cumulative bioH2 production (approximately 4–5 mmol) that increased slowly until 96 h and subsequently increased rapidly to reach a cumulative bioH2 production of 28 mmol (94.4 mL/g total OA consumed; 167.8 mL/g DQO) at the end of the second stage of production. The higher bioH2 production was correlated with increased biomass concentration (Figure 2F).
S. wolfei accumulated lower PHA concentrations than R. palustris and C. pasteurianum, reaching 46% and 31% at 88 and 164 h, respectively (Figure 2F).
3.2. Consortiums, Bioaugmented and Designed Cultures
3.2.1. Natural Photoheterotrophic Consortium “NPC-C4”
The kinetic profiles of “NPC-C4” are depicted in Figure 3. “NPC-C4” showed a faster growth rate during the first 48 h (µ = 0.339 h−1) and reached 1.2 to 1.4 g/L of biomass. The biomass concentration oscillated between these values until 120 h, when the consortium grew slower at a specific growth rate of µ = 0.0024 h−1.
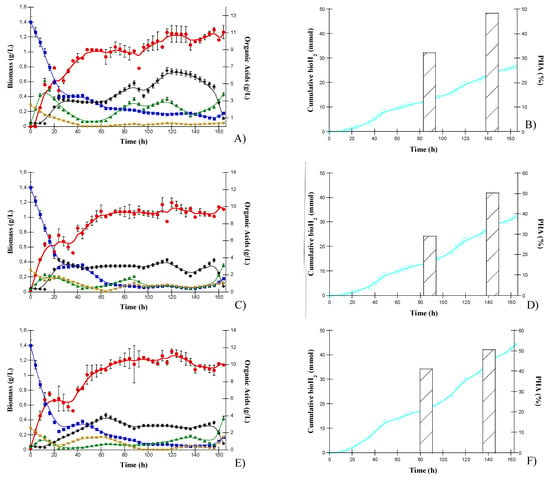
Figure 3.
Consortium culture patterns of organic acid consumption: HLac (■), HAc (◆), HPr (▲), HBu (▼), biomass (λ), bioH2 (□) and PHA production (bars) obtained by NPC-C4 in the upper panel (A,B), BNPC-C4 in the middle panel (C,D), and the designed consortium (C. pasteurianum, R. palustris and S. w) in the lower panel (E,F).
The HLac consumption rate was slower than that of the pure strain cultures. HBu was depleted by 40 h. HPr was produced in the process up to 120 h and was then reassimilated by the cells. HAc was produced during the growth stage and then assimilated alongside HLac and HBu; the production and consumption subsequently oscillated. The initial consumption of HLac, HBu and HPro resulted in biomass production, reaching the highest concentration of 1.3 g/L at 50 h of culture. The HAc concentration increased to 6 g/L after 112 h and was subsequently consumed, which correlates with the biomass increase observed from 112 to 164 h. The faster HAc consumption observed at the end of the culture agreed with the results obtained for R. palustris, which showed complete assimilation of this microorganism in the culture as an individual strain.
The data show less variability in the periods when the OAs were substrates or products. HLac and HBu were consumed for a longer period, even though HBu was exhausted at 44 h and then produced and consumed again. HLac was produced in the first half of the cultivation and then consumed, and the role of HPr changed from substrate to product several times. The highest increase in bioH2 was observed when HLac was the substrate and HBu, HLac and HPr were produced.
3.2.2. Bioaugmented Natgural Photoheterotrophic Consortium with S. wolfei “BNPC-C4”
The results obtained for “BNPC-C4” are presented in Figure 3C,D. The biomass production behavior was similar to that determined for “NPC-C4”; the biomass remained relatively constant after 60 h, with a maximum of 1.31 g/L (µ = 0.0083 h−1). The growth rate and biomass concentration obtained for “NPC-C4” and “BNPC-C4” were very similar (Figure 3B,D, respectively). Nevertheless, the cumulative bioH2 production was higher in “BNPC-C4”, 32 mmol, equivalent to 161.8 mL/g total OAs consumed, and 106.7 mL/g COD, which constitutes an increase of 19%, presumably because of the metabolic activity of S. wolfei (Figure 3D). Considering that bioH2 production showed a two-stage dynamic, it is plausible that the first bioH2 production was driven by the activity of C. pasteurianum and R. palustris. The second production could be the result of the activity of C. pasteurianum and S. wolfei.
Before 24 h of culture, HLac and HBu were almost wholly consumed, 82 and 98%, respectively. After 24 h, HLac was steadily consumed until reaching a concentration of 1.5 g/L (90%) at 148 h. During the first 24 h, HAc was not consumed; after this period, its concentration increased to 5 g/L at 40 h, higher than in the other cultures. This could be explained by its high production from HBu consumption by S. wolfei, followed by its consumption by C. pasteurianum and R. palustris. The role of OAs during this culture was similar to that of the natural consortium “NPC-C4”. HLac and HBu were initially consumed for an extended period, and HBu was exhausted at 64 h and then produced. There were several changes in the consumption and production of HPr. The highest bioH2 production was observed when HLac was consumed. However, this consumption was marginal and, as no other OA was consumed in that period, it is not clear how bioH2 originated; bioH2 production could possibly occur via the consumption of some reserve, such as PHA.
3.2.3. Designed Consortium
Finally, Figure 3E,F show the substrate and product patterns obtained with the “designed consortium” grown in the DFE. The designed culture had a similar metabolic activity to that of the bioaugmented consortium “BNPC-C4” regarding the biomass, bioH2 and PHA production. Before 24 h of cultivation, HLac and HBu were consumed, up to 82.0 and 98%, respectively. After 24 h, HLac was consumed, reaching a concentration of 1.5 g/L at 148 h (Figure 3E). The biomass growth was similar to those of “NPC-C4” and the bioaugmented “BNPC-C4”; that is, the biomass remained constant after 60 h and reached a maximum value of 1.31 g/L (Figure 3F).
The highest cumulative bioH2 production was 45 mmol, equivalent to 195.4 mL/g total OAs consumed, and was obtained with the “designed consortium”. These results suggest a positive interaction between the three microorganisms regarding bioH2 production. Moreover, the artificial elimination of other strains contained in the natural consortium did not negatively affect the production of secondary metabolites.
The OA patterns were similar to both the natural and the bioaugmented consortium, which suggests that some interactions are established. However, the HBu was not exhausted in this case until 96 h, suggesting the possible absence of some consuming microorganisms. Most notably, higher bioH2 production was observed when HBu, HAc, HLac and HPr were all consumed. In the other cultures, this period was characterized by HLac consumption, but with the designed consortium, all the OAs were consumed simultaneously, a desirable feature in the context of DF effluents.
4. Discussion
The growth kinetics show that the pure cultures present a two-growth pattern characterized by two exponential phases, each separated by a lag phase. Such growth could be attributed to the co-consumption of different substrates [29]. This pattern was less apparent in the microbial consortium, suggesting that the simultaneous assimilation of substrates by the members of the microbial population follows a less defined pattern.
C. pasteurianum showed the highest bioH2 production compared to the other individual strains, while R. palustris exhibited the lowest production at the end of the culture period (Table 1). The bioH2 production profile of C. pasteurianum and S. wolfei alone showed similar behavior during the two production periods. On the other hand, R. palustris showed a single period of production (Figure 2D). The bioH2 volumetric production rates (H2VPR) obtained in all the systems are presented in Table 1 and the profiles can be found in the Figure S1. The higher H2VPR obtained during the first stage was similar for C. pasteurianum and R palustris, approximately 20 mL/Lh; however, during the second stage (~110 h), the H2VPR of C. palustris increased to reach a final value of 27 mL/Lh, while the H2VPR of P. palustris decreased. In general, the H2PVR of the consortiums was higher than those of the individual strains; the H2VPRs determined for “NPC-C4” and “BNPC-C4” were similar during the first period (a maximum of 28 mL/Lh), but after 110 h, the H2VPR of “BNPC-C4” increased slightly, reaching a value similar to that measured for C. pasteurainum. The highest H2VPR (40 mL/Lh) was determined with the “designed consortium”. Lo et al. [6] reported the effect of the type and concentration of OAs as individual substrates on the biomass and bioH2 produced by R. palustris WP3-5. The highest bioH2 production rate was obtained at an HAc concentration of 4.0 g/L, reaching a value of 19.8 mL/L h. This was highger than those observed when HLac and HBu were used as substrates, 17.0 mL/L h and 11.67 mL/L h at concentrations of 5.0 g/L and 2.0 g/L, respectively. The results obtained with R. palustris in the present work are consistent with the data presented by the above authors. Regarding the overall yield of bioH2, the highest values were those of S. wolfei and the designed consortium, suggesting that S. wolfei increases the yield; however, its addition to NPC-C4 did not have a significant impact. Apparently, the complexity of the consortium mitigated its impact, which did not happen in the simpler, designed consortium.

Table 1.
Summary and comparison of OA consumption, biomass production, and bioH2 and PHA production.
Despite the extensive work developed for the study of C. pasteurianum and R. palustris [30,31,32,33,34], it is still not clear how these microorganisms grow and produce bioH2 from a mixture of OAs (HLac, HAc, HLac and HBu). This aspect is of particular interest for the use of effluents of organic residues after DF or the use of wastewater. Sarma et al. [35] found that HAc and HLac can be consumed by C. pasteurianum to obtain HBu (2 g/L) and bioH2 (2 mmol) as the main byproducts. In addition, the conversion of malate and HLac to bioH2 by photosynthetic bacteria has been documented [36]. However, it is still unknown whether R. palustris can assimilate HLac as a sole carbon source or if it is only possible in mixed-substrate conditions. Govindaraju et al. [29] demonstrated that R. palustris utilized HLac with other organic acids simultaneously.
When grown in pure cultures, C. pasteurianum, R. palustris and S. wolfei consumed 57, 63 and 39%, respectively, of the total OAs. Based on the OA consumption in the DFE and bioH2 production patterns observed in this work, it can be suggested that the production of bioH2 in C. pasteurianum implies the consumption of HLac and HAc, with the simultaneous generation of HBu. While this behavior is typical of chain-elongation microorganisms [37,38], the observed growth and product kinetics provide further evidence of complex mechanisms. These involve thermodynamic equilibrium, the assimilation of OAs via beta-oxidation [39], and the production of bioH2 using a proton driving force derived from reduced menaquinone [40] or via electron bifurcation [41].
The presented results show that HLac was the better substrate for all the evaluated microorganisms. HLac oxidation is a thermodynamically attractive process compared to that of other OAs and alcohols. The assimilation of HLac is likely the result of the bifurcation reaction catalyzed by a stable complex formed between a FAD-dependent lactate dehydrogenase LDH (GlcD domain) and an electron transfer flavoprotein (EtfA/B) [42]:
HLac + Fd2− + 2NAD+ → pyruvate + Fd + 2NADH
Previous reports suggest that the mechanism is present in members of the Clostridiales, Halanaerobiales, Fusobacteriales, Thermotogales, and Thermoanaerobacteriales [42], which coincides with these groups and the consortia, and supports the selection of C. pasteurianum.
The OA consumption behavior determined for S. wolfei is consistent with other reports in the literature. Beaty and McInerney [43] evaluated different concentrations of HLac (0–12 g/L) for the growth of S. wolfei and found that concentrations up to 1.12 g/L reduced the growth rate and increased the lag phase of the culture (0–350 h). Nevertheless, S. wolfei does not lose its metabolic ability to produce HAc and bioH2; this last byproduct is detected when the concentration of HLac is approximately 2.5 g/L, as shown in the results of the present study at 100 h (Figure 2E). A recent study on anaerobic HLac oxidation [44,45] summarized the ΔG0′ values/reaction for the oxidation of HAc, HBu, Hpr, ethanol, and HLac. The authors suggest that HLac oxidation favorably drives the microbial communities’ performance toward producing HAc that, in the absence of methanogens, leads to increased bioH2 production. As per the consumption of HBu, syntropy is the most likely process occurring during the early stages of the cultures, as lower concentrations of bioH2 and HAc are required to keep the reaction favorable toward the products [46].
The OA assimilation patterns could indicate differences in metabolic pathways. For example, during the process developed with “NPC-C4”, the cumulative HPr concentration was three times larger than the concentrations in the individual cultures (1 g/L for C. pasteurianum and R. palustris). Therefore, it is reasonable to conclude that this is the result of the simultaneous growth of R. palustris, C. pasteurianum and the other microorganisms comprising “NPC-C4”. Additionally, HPr is involved in the PHA production pathways, as discussed in previous works [19]. Therefore, the higher PHA production obtained with “NPC-C4” could explain why its bioH2 production was not the sum of the individual strains.
When analyzing the microbial consortia, the total OA removal percentages were similar to those found for the individual strains, between 53–59%. NPC-C4 showed a similar OA assimilation to that determined for R. palustris. During fermentation, HLac and HBut were simultaneously assimilated. However, the higher bioH2 production obtained with this consortium compared with the one determined for the single culture of R. palustris suggests the metabolic activity of other members of the microbial population. “BNPC-C4” showed a similar pattern to “NPC-C4” but higher HAc production, which suggests the consumption of HBu to produce HAc, as determined by the individual growth of S. wolfei. Finally, the “design consortium” showed a complex metabolic interaction in which OAs were constantly produced and consumed, resulting in the highest bioH2 production.
The PHA production obtained with R. palustris was the highest, attaining 54% at 88 h and 31% at 164 h. The other two strains showed similar PHA accumulation at the same culture times (C. pasteurianum, 48.80% and 24.49%; S. wolfei, 46.51% and 31.61%). The biopolymer percentages obtained with R. palustris agree with the data provided in the literature [37] using R. palustris (49% CDW), and our previous work where 44% of PHA was determined with a photoheterotrophic mixed consortia [19] using HAc and HBu as substrates. PHA accumulation in S. wolfei has been reported by other authors [43,47], with a production of approximately 39.5%; the individual results obtained in this work were higher than those reported. The synthesis of PHA in S. wolfei is mediated by HBu degradation toward the production of 3-hydroxybutyryl CoA, which also enables the production of bioH2 from the electrons generated by the dehydrogenation of butyryl CoA [48].
Comparing the results obtained in the present work (Table 1) with the literature, it can be noticed that the overall H2 yields obtained at the two-stage DF-PF process is in the range of those previously reported by [49] where starch was used as feedstock (16.1 mmolH2/gCOD), by [24] using starch/glucose (8.3 mmolH2/gCOD), and by [50] obtained using cassava starch (15.2 mmolH2/gCOD), but lower than those obtained in other works for sucrose (26 mmolH2/gCOD) and for cassava and food waste (28.1 mmolH2/gCOD) as substrates [4,51]. The values determined here are also lower than the higher H2 yields obtained for another photo-heterotrophic consortium C2 previously evaluated in a similar scheme of a coupled process of DF-PF, which reached 24.7 and 21.7 mmolH2/gCOD (equivalent to 695.4 and 793.7 mLH2/gCOD, respectively) [2]. The cumulative bioH2 production obtained in the present work is similar to those determined in our previous study however the initial TOAs concentration were twice that used previously [2], this influences the consumed COD and in turn the H2 yields. Differences in H2 yields could be explained by (a) the simultaneous high PHA production, (b) changes in the proportion of the main microbial population (Clostridium and Rhodopseudomonas) in the natural photoheterotrophic consortiums C2 and C4 depending on the culture conditions, as reported previously [2,19,26]. Several previous studies for bioH2 production using two-stage systems have been conducted using DF effluents that mainly contain acetate and butyrate. It is important to highlight that in this work it was possible to obtain similar overall H2 yield by using DF effluents with higher lactate concentration. PHA yields were comparable among the cultures, the higher one being that of R. palustris at 88 h, however, at 134 h the value for the designed consortium was only slightly lower. Considering the H2VPR and the PHA yield at 134 h, the results suggest that the designed consortium is better suited for the production of both H2 and PHA from reduced substrates than the individual cultures and the natural consortia, which should add to the economic viability of both products.
The experiments highlight the interaction between the different microbial populations and give some insight into the processes driving the biomass, bioH2, and PHA production in each case, considering that competition exists for reducing equivalents during the process and these products constitute electron sinks. At this stage, further analysis must be implemented to confirm the metabolic processes that occur within each strain in the mixed culture and describe how they contribute to the production of bioH2 and PHA. Based on our results and the metabolic potential of each strain annotated in their genome, we can suggest the key metabolic process involved (Figure 4). The individual strains rely on the use of organic acids for growth and production of bioH2 and PHB, which is only possible with the use of an external energy source or a proton diving force. While R. palustris uses photons to generate ATP and reducing power, S. wolfei and C. pasteurianus are likely to use the beta-oxidation pathway [28] coupled with electron bifurcation [30]. These mechanisms are likely to be performed using nitrogenases (Nfn) and hydrogenases (Hyd) complexes [52]. The combination of all three mechanisms in the mixed cultures likely explain the increased availability of NADH/NADPH that can be diverted toward H2 and PHB. The results suggest that S. wolfei enhances the assimilation of the organic acids, leading to increased bioH2 and PHB production.
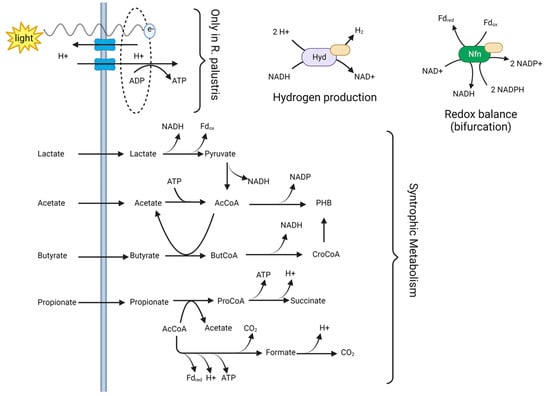
Figure 4.
Proposed metabolic mechanisms involved in bioH2 and PHB production in mixed cultures bioaugmented with S. wolfei. Acetyl CoA, AcCoA. Butiryl CoA, ButCoA. Propionyl Co A, ProCoA. Crotonyl CoA, CroCoA. Reduced ferredoxin, Fdred. Oxidised ferredoxin, Fdox. Created with BioRender.com.
5. Conclusions
The main contribution of this work constitutes a demonstration of the feasibility of the simultaneous production of bioH2 and PHA using DFE. The selected experimental conditions resulted in the simultaneous production of bioH2 and PHA accumulation using DFE as a carbon source with high concentrations of OAs. Both consortia, “BNPC-C4” and the “designed consortium”, showed higher PHA accumulation. The designed consortium was the only consortium that maintained PHA accumulation (50%) throughout the entire culture period while producing the highest cumulative bioH2 concentration (45 mmol). The initial cumulative PHA production obtained by the individual strains was consumed at the final stage of the cultivation time. Conversely, all the consortia maintained PHA production throughout the process. Moreover, the higher biomass and substrate consumption is indicative of an improvement in the efficiency of using DFE as a substrate by the cooperative action of the microbial populations in the consortiums. The OA substrate consumption patterns showed that HBu assimilation was promoted by the presence of HLac in the feedstock mixture. Overall, the results suggest that the microbial communities use synergistic mechanisms, such as beta-oxidation, proton driving force derived from reduced menaquinone and electron bifurcation, which makes the observed production of bioH2 a favorable process. Nevertheless, a more in-depth systematic analysis is required to precisely describe the metabolic processes that drive the interactions between the microbial populations. The results attained in this study motivate the design of new microbial consortia considering their metabolic capacities to produce more complex byproducts with industrial interest.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation8110644/s1, Figure S1. Comparison between bioH2 volumetric production rates (H2VPR) and PHA production obtained by pure cultures: (A) H2VPR of C. pasteurianum ( ), H2VPR of R. palustris (
), H2VPR of R. palustris ( ) and H2VPR of S. wolfei (
) and H2VPR of S. wolfei ( ); (C) PHA C. pasteurianum (blue solid column), R. palustris (pink column), S. wolfei (crosse black column); and by the microbial consortiums: (B) H2VPR of “NPC-C4” (
); (C) PHA C. pasteurianum (blue solid column), R. palustris (pink column), S. wolfei (crosse black column); and by the microbial consortiums: (B) H2VPR of “NPC-C4” ( ), H2VPR of “BNPC-C4” (
), H2VPR of “BNPC-C4” ( ) and H2VPR of the “design consortium” (
) and H2VPR of the “design consortium” ( ); (D) PHA “NPC-C4” (solid golden column), “BNPC-C4” (brown column), “design consortium” (cross green column).
); (D) PHA “NPC-C4” (solid golden column), “BNPC-C4” (brown column), “design consortium” (cross green column).
 ), H2VPR of R. palustris (
), H2VPR of R. palustris ( ) and H2VPR of S. wolfei (
) and H2VPR of S. wolfei ( ); (C) PHA C. pasteurianum (blue solid column), R. palustris (pink column), S. wolfei (crosse black column); and by the microbial consortiums: (B) H2VPR of “NPC-C4” (
); (C) PHA C. pasteurianum (blue solid column), R. palustris (pink column), S. wolfei (crosse black column); and by the microbial consortiums: (B) H2VPR of “NPC-C4” ( ), H2VPR of “BNPC-C4” (
), H2VPR of “BNPC-C4” ( ) and H2VPR of the “design consortium” (
) and H2VPR of the “design consortium” ( ); (D) PHA “NPC-C4” (solid golden column), “BNPC-C4” (brown column), “design consortium” (cross green column).
); (D) PHA “NPC-C4” (solid golden column), “BNPC-C4” (brown column), “design consortium” (cross green column). Author Contributions
A.G., writing—review and editing, formal analysis; E.S., conceptualization, writing—review and editing, funding acquisition; Z.V., C.N.-N. and O.C., investigation process, data collection, writing—original draft; I.C., supervision, writing—original draft; E.I.G.-P., conceptualization, writing—review and editing—including pre- or postpublication stages, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by CONACYT Grant Frontera 682137 and by IPN grant 20220846.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations and Acronyms
Dark fermentation (DF). Photo-fermentation (PF). Biohydrogen (bioH2). Poly-hydroxy-alkanoates (PHA). Volumetric production rates (VPR). Lactate (HLac). Acetate (HAc). Butyrate (HBu). Propionate (HPr). Purple non-sulphur bacteria (PNSB). Poly-hydroxy-butyrate- Poly-hydroxy-valerate (PHB-PHV). Cost (USD)/Giga Joule (USD GJ−1). Real-time quantitative PCR (RT-QPCR). nouF gene (NADH ubiquinone dehydrogenase). Dark fermentation effluents (DFE). Natural photoheterotrophic consortium (“NPC-C4”). Bioaugmented natural photoheterotrophic consortium (“BNPC-C4”, NPC-C4 + S. wolfei). Designed consortium (R.palustris, C.pasteurianum and S.wolfei). Hydrogen nuclear magnetic resonance (HNMR). Thermal conductivity detector gas chromatography (TCD-GC). Total organic acids (TOAs) including lactate, acetate, butyrate, and propionate.
References
- Schwartz, E.; Fritsch, J.; Friedrich, B. H2-Metabolizing Prokaryotes. Prokaryotes: Prokaryotic Physiol. Biochem. 2013, 2, 119–199. [Google Scholar] [CrossRef]
- Niño-Navarro, C.; Chairez, I.; Christen, P.; Canul-Chan, M.; Garcia-Peña, E.I. Enhanced Hydrogen Production by a Sequential Dark and Photo Fermentation Process: Effects of Initial Feedstock Composition, Dilution and Microbial Population. Renew. Energy 2020, 147, 924–936. [Google Scholar] [CrossRef]
- Cheng, D.; Ngo, H.H.; Guo, W.; Chang, S.W.; Nguyen, D.D.; Bui, X.T.; Wei, W.; Ni, B.; Varjani, S.; Hoang, N.B. Enhanced Photo-Fermentative Biohydrogen Production from Biowastes: An Overview. Bioresour. Technol. 2022, 357, 127341. [Google Scholar] [CrossRef]
- Zong, W.; Yu, R.; Zhang, P.; Fan, M.; Zhou, Z. Efficient Hydrogen Gas Production from Cassava and Food Waste by a Two-Step Process of Dark Fermentation and Photo-Fermentation. Biomass Bioenergy 2009, 33, 1458–1463. [Google Scholar] [CrossRef]
- Liu, B.F.; Ren, N.Q.; Tang, J.; Ding, J.; Liu, W.Z.; Xu, J.F.; Cao, G.L.; Guo, W.Q.; Xie, G.J. Bio-Hydrogen Production by Mixed Culture of Photo- and Dark-Fermentation Bacteria. Int. J. Hydrogen Energy 2010, 35, 2858–2862. [Google Scholar] [CrossRef]
- Lo, Y.C.; Chen, C.Y.; Lee, C.M.; Chang, J.S. Photo Fermentative Hydrogen Production Using Dominant Components (Acetate, Lactate, and Butyrate) in Dark Fermentation Effluents. Int. J. Hydrogen Energy 2011, 36, 14059–14068. [Google Scholar] [CrossRef]
- Hitit, Z.Y.; Lazaro, C.Z.; Hallenbeck, P.C. Hydrogen Production by Co-Cultures of Clostridium butyricum and Rhodospeudomonas palustris: Optimization of Yield Using Response Surface Methodology. Int. J. Hydrogen Energy 2017, 42, 6578–6589. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Oda, Y.; Ruhl, M.; Posto, A.L.; Sauer, U.; Harwood, C.S. Non-Growing Rhodopseudomonas palustris Increases the Hydrogen Gas Yield from Acetate by Shifting from the Glyoxylate Shunt to the Tricarboxylic Acid Cycle. J. Biol. Chem. 2014, 289, 1960–1970. [Google Scholar] [CrossRef] [PubMed]
- Riedel, S.; Brigham, C. Inexpensive and Waste Raw Materials for PHAProduction. In The Handbook of Polyhydroxyalkanoates; Koller, M., Ed.; CRC Press: Boca Raton, FL, USA, 2020; pp. 203–220. [Google Scholar]
- McKinlay, J.B.; Harwood, C.S. Photobiological Production of Hydrogen Gas as a Biofuel. Curr. Opin. Biotechnol. 2010, 21, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Vincenzini, M.; Marchini, A.; Ena, A.; de Philippis, R. H2 and Poly-b-Hydroxybutyrate, Two Alternative Chemicals from Purple Non Sulfur Bacteria. Biotechnol. Lett. 1997, 19, 759–762. [Google Scholar] [CrossRef]
- Thulasidharan, D.; Arumugam, A.; Uppuluri, K.B. Research and Economic Perspectives on an Integrated Biorefinery Approach for the Simultaneous Production of Polyhydroxyalkanoates and Biohydrogen. Int. J. Biol. Macromol. 2021, 193, 1937–1951. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-Y.; Shi, Z.-Y.; Chen, J.-C.; Wu, Q.; Chen, G.-Q. Enhanced Co-Production of Hydrogen and Poly-(R)-3-Hydroxybutyrate by Recombinant PHB Producing E. Coli over-Expressing Hydrogenase 3 and Acetyl-CoA Synthetase. Metab. Eng. 2012, 14, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.F.; Rafa, N.; Mofijur, M.; Badruddin, I.A.; Inayat, A.; Ali, M.S.; Farrok, O.; Yunus Khan, T.M. Biohydrogen Production From Biomass Sources: Metabolic Pathways and Economic Analysis. Front. Energy Res. 2021, 9, 753878. [Google Scholar] [CrossRef]
- Fernández-Dacosta, C.; Posada, J.A.; Kleerebezem, R.; Cuellar, M.C.; Ramirez, A. Microbial Community-Based Polyhydroxyalkanoates (PHAs) Production from Wastewater: Techno-Economic Analysis and Ex-Ante Environmental Assessment. Bioresour. Technol. 2015, 185, 368–377. [Google Scholar] [CrossRef]
- Manikandan, N.A.; Pakshirajan, K.; Pugazhenthi, G. Techno-Economic Assessment of a Sustainable and Cost-Effective Bioprocess for Large Scale Production of Polyhydroxybutyrate. Chemosphere 2021, 284, 131371. [Google Scholar] [CrossRef] [PubMed]
- Montiel-Corona, V.; Revah, S.; Morales, M. Hydrogen Production by an Enriched Photoheterotrophic Culture Using Dark Fermentation Effluent as Substrate: Effect of Flushing Method, Bicarbonate Addition, and Outdoor–Indoor Conditions. Int. J. Hydrogen Energy 2015, 40, 9096–9105. [Google Scholar] [CrossRef]
- Xu, C.; Yu, H. Insights into Constructing a Stable and Efficient Microbial Consortium. Chin. J. Chem. Eng. 2021, 30, 112–120. [Google Scholar] [CrossRef]
- Guerra-Blanco, P.; Cortes, O.; Poznyak, T.; Chairez, I.; García-Peña, E.I. Polyhydroxyalkanoates (PHA) Production by Photoheterotrophic Microbial Consortia: Effect of Culture Conditions over Microbial Population and Biopolymer Yield and Composition. Eur. Polym. J. 2018, 98, 94–104. [Google Scholar] [CrossRef]
- Lopez-Romero, J.; Salgado-Manjarrez, E.; Torres, L.; Garcia-Peña, E.I. Enhanced Carotenoid Production by Rhodopseudomonas palustris ATCC 17001 under Low Light Conditions. J. Biotechnol. 2020, 323, 159–165. [Google Scholar] [CrossRef]
- Cabrol, L.; Marone, A.; Tapia-Venegas, E.; Steyer, J.-P.; Ruiz-Filippi, G.; Trably, E. Microbial Ecology of Fermentative Hydrogen Producing Bioprocesses: Useful Insights for Driving the Ecosystem Function. FEMS Microbiol. Rev. 2017, 41, 158–181. [Google Scholar] [CrossRef]
- Ergal, İ.; Bochmann, G.; Fuchs, W.; Rittmann, S.K.-M. Design and Engineering of Artificial Microbial Consortia for Biohydrogen Production. Curr. Opin. Biotechnol. 2022, 73, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.E.; Oda, Y.; Harwood, C.S. Regulation of Uptake Hydrogenase and Effects of Hydrogen Utilization on Gene Expression in Rhodopseudomonas palustris. J. Bacteriol. 2006, 188, 6143–6151. [Google Scholar] [CrossRef]
- Hitit, Z.Y.; Zampol Lazaro, C.; Hallenbeck, P.C. Increased Hydrogen Yield and COD Removal from Starch/Glucose Based Medium by Sequential Dark and Photo-Fermentation Using Clostridium butyricum and Rhodopseudomonas palustris. Int. J. Hydrogen Energy 2017, 42, 18832–18843. [Google Scholar] [CrossRef]
- Kao, P.-M.; Hsu, B.-M.; Huang, K.-H.; Tao, C.-W.; Chang, C.-M.; Ji, W.-T. Biohydrogen Production by Immobilized Co-Culture of Clostridium butyricum and Rhodopseudomonas palustris. Energy Procedia 2014, 61, 834–837. [Google Scholar] [CrossRef]
- Jurado-Marban, V.H.; Tapia-Bustos, M.A.; Gonzalez-Garcia, R.A.; Salgado, E.; Garcia-Peña, E.I. Hydrogen Production by a Mixed Photoheterotrophic Culture: Correlation between Gene Expression Analysis and Physiological Behavior. Int. J. Hydrogen Energy 2019, 44, 641–651. [Google Scholar] [CrossRef]
- Gomez-Romero, J.; Gonzalez-Garcia, A.; Chairez, I.; Torres, L.; Garcia-Peña, E.I. Selective Adaptation of an Anaerobic Microbial Community: Biohydrogen Production by Co-Digestion of Cheese Whey and Vegetables Fruit Waste. Int. J. Hydrogen Energy 2014, 39, 12541–12550. [Google Scholar] [CrossRef]
- Cheng, Y.; Wei, H.; Sun, R.; Tian, Z.; Zheng, X. Rapid Method for Protein Quantitation by Bradford Assay after Elimination of the Interference of Polysorbate 80. Anal. Biochem. 2016, 494, 37–39. [Google Scholar] [CrossRef]
- Govindaraju, A.; McKinlay, J.B.; LaSarre, B. Phototrophic Lactate Utilization by Rhodopseudomonas palustris Is Stimulated by Coutilization with Additional Substrates. Appl. Environ. Microbiol. 2019, 85, e00048-19. [Google Scholar] [CrossRef]
- Diez-Gonzalez, F.; Russell, J.B.; Hunter, J.B. The Role of an NAD-Independent Lactate Dehydrogenase and Acetate in the Utilization of Lactate by Clostridium acetobutylicum Strain P262. Arch. Microbiol. 1995, 164, 36–42. [Google Scholar] [CrossRef]
- Buckel, W.; Thauer, R.K. Flavin-Based Electron Bifurcation, A New Mechanism of Biological Energy Coupling. Chem. Rev. 2018, 118, 3862–3886. [Google Scholar] [CrossRef]
- Demmer, J.K.; Pal Chowdhury, N.; Selmer, T.; Ermler, U.; Buckel, W. The Semiquinone Swing in the Bifurcating Electron Transferring Flavoprotein/Butyryl-CoA Dehydrogenase Complex from Clostridium difficile. Nat. Commun. 2017, 8, 1577. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Lu, W.-B.; Liu, C.-H.; Chang, J.-S. Improved Phototrophic H2 Production with Rhodopseudomonas palustris WP3-5 Using Acetate and Butyrate as Dual Carbon Substrates. Bioresour. Technol. 2008, 99, 3609–3616. [Google Scholar] [CrossRef] [PubMed]
- Cardeña, R.; Valdez-Vazquez, I.; Buitrón, G. Effect of Volatile Fatty Acids Mixtures on the Simultaneous Photofermentative Production of Hydrogen and Polyhydroxybutyrate. Bioprocess Biosyst. Eng. 2017, 40, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Sarma, S.; Anand, A.; Dubey, V.K.; Moholkar, V.S. Metabolic Flux Network Analysis of Hydrogen Production from Crude Glycerol by Clostridium pasteurianum. Bioresour. Technol. 2017, 242, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.J.; Rocha, J.M.S.; Tramper, J.; Wijffels, R.H. Acetate as a Carbon Source for Hydrogen Production by Photosynthetic Bacteria. J. Biotechnol. 2001, 85, 25–33. [Google Scholar] [CrossRef]
- Coma, M.; Vilchez-Vargas, R.; Roume, H.; Jauregui, R.; Pieper, D.H.; Rabaey, K. Product Diversity Linked to Substrate Usage in Chain Elongation by Mixed-Culture Fermentation. Environ. Sci. Technol. 2016, 50, 6467–6476. [Google Scholar] [CrossRef]
- Liu, B.; Popp, D.; Sträuber, H.; Harms, H.; Kleinsteuber, S. Draft Genome Sequences of Three Clostridia Isolates Involved in Lactate-Based Chain Elongation. Microbiol. Resour. Announc. 2020, 9, e00679-20. [Google Scholar] [CrossRef] [PubMed]
- Doi, H.; Hoshino, Y.; Nakase, K.; Usuda, Y. Reduction of Hydrogen Peroxide Stress Derived from Fatty Acid Beta-Oxidation Improves Fatty Acid Utilization in Escherichia coli. Appl. Microbiol. Biotechnol. 2014, 98, 629–639. [Google Scholar] [CrossRef]
- Pinske, C.; Jaroschinsky, M.; Linek, S.; Kelly, C.L.; Sargent, F.; Sawers, R.G. Physiology and Bioenergetics of [NiFe]-Hydrogenase 2-Catalyzed H2 -Consuming and H2 -Producing Reactions in Escherichia coli. J. Bacteriol. 2015, 197, 296–306. [Google Scholar] [CrossRef]
- Schut, G.J.; Adams, M.W.W. The Iron-Hydrogenase of Thermotoga maritima Utilizes Ferredoxin and NADH Synergistically: A New Perspective on Anaerobic Hydrogen Production. J. Bacteriol. 2009, 191, 4451–4457. [Google Scholar] [CrossRef]
- Weghoff, M.C.; Bertsch, J.; Müller, V. A Novel Mode of Lactate Metabolism in Strictly Anaerobic Bacteria. Environ. Microbiol. 2015, 17, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Beaty, P.S.; McInerney, M.J. Effects of Organic Acid Anions on the Growth and Metabolism of Syntrophomonas wolfei in Pure Culture and in Defined Consortia. Appl. Environ. Microbiol. 1989, 55, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Detman, A.; Mielecki, D.; Pleśniak, Ł.; Bucha, M.; Janiga, M.; Matyasik, I.; Chojnacka, A.; Jędrysek, M.-O.; Błaszczyk, M.K.; Sikora, A. Methane-Yielding Microbial Communities Processing Lactate-Rich Substrates: A Piece of the Anaerobic Digestion Puzzle. Biotechnol. Biofuels 2018, 11, 116. [Google Scholar] [CrossRef] [PubMed]
- Detman, A.; Mielecki, D.; Chojnacka, A.; Salamon, A.; Błaszczyk, M.K.; Sikora, A. Cell Factories Converting Lactate and Acetate to Butyrate: Clostridium Butyricum and Microbial Communities from Dark Fermentation Bioreactors. Microb. Cell Fact 2019, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Sieber, J.R.; Sims, D.R.; Han, C.; Kim, E.; Lykidis, A.; Lapidus, A.L.; McDonnald, E.; Rohlin, L.; Culley, D.E.; Gunsalus, R.; et al. The Genome of Syntrophomonas wolfei: New Insights into Syntrophic Metabolism and Biohydrogen Production. Environ. Microbiol. 2010, 12, 2289–2301. [Google Scholar] [CrossRef]
- Beaty, P.S.; McInerney, M.J. Growth of Syntrophomonas wolfei in Pure Culture on Crotonate. Arch. Microbiol. 1987, 147, 389–393. [Google Scholar] [CrossRef]
- McInerney, M.J.; Bryant, M.P.; Hespell, R.B.; Costerton, J.W. Syntrophomonas wolfei Gen. Nov. Sp. Nov., an Anaerobic, Syntrophic, Fatty Acid-Oxidizing Bacterium. Appl. Environ. Microbiol. 1981, 41, 1029–1039. [Google Scholar] [CrossRef]
- Lo, Y.-C.; Chen, S.-D.; Chen, C.-Y.; Huang, T.-I.; Lin, C.-Y.; Chang, J.-S. Combining Enzymatic Hydrolysis and Dark–Photo Fermentation Processes for Hydrogen Production from Starch Feedstock: A Feasibility Study. Int. J. Hydrogen Energy 2008, 33, 5124–5233. [Google Scholar] [CrossRef]
- Su, H.; Cheng, J.; Zhou, J.; Song, W.; Cen, K. Improving Hydrogen Production from Cassava Starch by Combination of Dark and Photo Fermentation. Int. J. Hydrogen Energy 2009, 34, 1780–1786. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Yang, M.-H.; Yeh, K.-L.; Liu, C.-H.; Chang, J.-S. Biohydrogen Production Using Sequential Two-Stage Dark and Photo Fermentation Processes. Int. J. Hydrogen Energy 2008, 33, 4755–4762. [Google Scholar] [CrossRef]
- Kremp, F.; Roth, J.; Müller, V. The Sporomusa Type Nfn Is a Novel Type of Electron-Bifurcating Transhydrogenase That Links the Redox Pools in Acetogenic Bacteria. Sci. Rep. 2020, 10, 14872. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).