Antimicrobial Activity of Se-Nanoparticles from Bacterial Biotransformation
Abstract
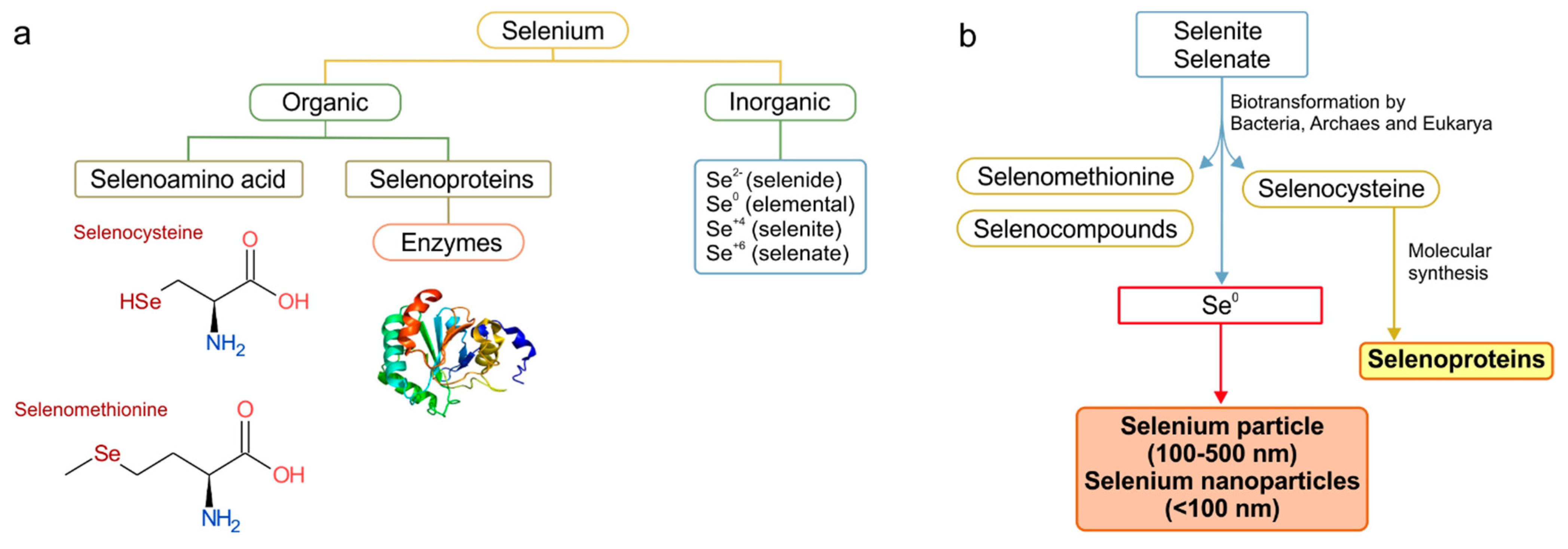
:1. Introduction
2. Elemental Selenium as the Best Form for the Antimicrobial Activity
3. Bacterial SeNPs Synthesis
3.1. Microbial Growth and Its Relation with SeNPs Synthesis
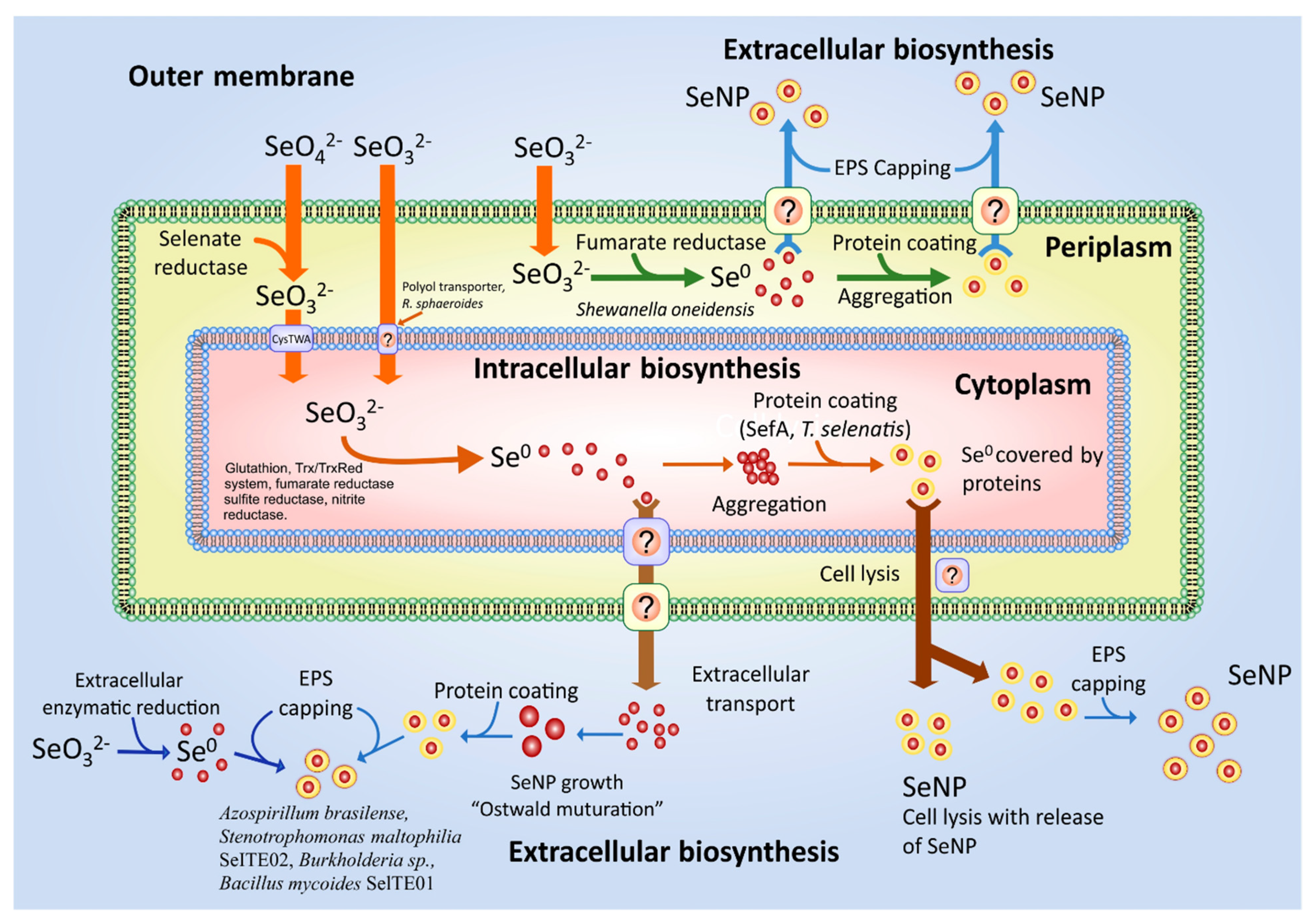
3.2. General Mechanism of SeNPs Synthesis
3.2.1. Se0 Formation: Selenate Reduction to Selenite
3.2.2. Se0 Formation: Selenite Reduction to Se0
3.2.3. SeNPs Formation
3.2.4. SeNPs Formation: Assembly
4. SeNPs Morphology and the Antimicrobial Activity
4.1. SeNPs Concentration
4.2. Coating Surface and Charge
4.3. Size and Shape
4.3.1. Physicochemical Parameters That Affect Shape and Size of SeNPs
4.3.2. Microbiological Parameters That Affect SeNPs Size and Shape
5. Antimicrobial Action Mechanisms
5.1. Cell Wall and Membrane Damage
5.2. Intracellular Penetration and Damage
5.3. Oxidative Stress
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Greeshma, B.C.; Mahesh, M. Biosynthesis of selenium nanoparticles from Bacillus species and its applications. J. Appl. Nat. Sci. 2019, 11, 810–815. [Google Scholar] [CrossRef]
- Strambeanu, N.; Demetrovici, L.; Dragos, D.; Lungu, M. Nanoparticles: Definition, Classification and General Physical Properties. In Nanoparticles’ Promises and Risks; Lungu, M., Neculae, A., Bunoiu, M.B.C., Eds.; Springer: Cham, Switzerland, 2015; pp. 3–8. ISBN 9783319117287. [Google Scholar]
- El-Batal, A.I.; Mosallam, F.M.; Ghorab, M.M.; Hanora, A.; Gobara, M.; Baraka, A.; Elsayed, M.A.; Pal, K.; Fathy, R.M.; Abd Elkodous, M.; et al. Factorial design-optimized and gamma irradiation-assisted fabrication of selenium nanoparticles by chitosan and Pleurotus ostreatus fermented fenugreek for a vigorous in vitro effect against carcinoma cells. Int. J. Biol. Macromol. 2020, 156, 1584–1599. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.D.; Tian, Y.Q.; Wu, J.L.; Wang, S.Y. Synthesis, characterization, and biological activity of selenium nanoparticles conjugated with polysaccharides. Crit. Rev. Food Sci. Nutr. 2020, 61, 2225–2236. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, J.J.; Merroun, M.L.; Tugarova, A.V.; Lampis, S.; Kamnev, A.A.; Gardiner, P.H.E. Developments in the study and applications of bacterial transformations of selenium species. Crit. Rev. Biotechnol. 2020, 40, 1250–1264. [Google Scholar] [CrossRef]
- Zambonino, M.C.; Quizhpe, E.M.; Jaramillo, F.E.; Rahman, A.; Santiago-Vispo, N.; Jeffryes, C.; Dahoumane, S.A. Green synthesis of selenium and tellurium nanoparticles: Current trends, biological properties and biomedical applications. Int. J. Mol. Sci. 2021, 22, 989. [Google Scholar] [CrossRef]
- Ranjitha, V.R.; Rai, V.R. Selenium nanostructure: Progress towards green synthesis and functionalization for biomedicine. J. Pharm. Investig. 2021, 51, 117–135. [Google Scholar] [CrossRef]
- Kessi, J.; Ramuz, M.; Wehrli, E.; Spycher, M.; Bachofen, R. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl. Environ. Microbiol. 1999, 65, 4734–4740. [Google Scholar] [CrossRef] [Green Version]
- Palomo-Siguero, M.; Gutiérrez, A.M.; Pérez-Conde, C.; Madrid, Y. Effect of selenite and selenium nanoparticles on lactic bacteria: A multi-analytical study. Microchem. J. 2016, 126, 488–495. [Google Scholar] [CrossRef]
- Wang, Y.; Shu, X.; Zhou, Q.; Fan, T.; Wang, T.; Chen, X.; Li, M.; Ma, Y.; Ni, J.; Hou, J.; et al. Selenite reduction and the biogenesis of selenium nanoparticles by Alcaligenes faecalis se03 isolated from the gut of Monochamus alternatus (Coleoptera: Cerambycidae). Int. J. Mol. Sci. 2018, 19, 2799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Wang, X.; Xu, T. Elemental selenium at nano size (Nano-Se) as a potential chemopreventive agent with reduced risk of selenium toxicity: Comparison with se-methylselenocysteine in mice. Toxicol. Sci. 2008, 101, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maseko, T.; Callahan, D.L.; Dunshea, F.R.; Doronila, A.; Kolev, S.D.; Ng, K. Chemical characterisation and speciation of organic selenium in cultivated selenium-enriched Agaricus bisporus. Food Chem. 2013, 141, 3681–3687. [Google Scholar] [CrossRef]
- Mangiapane, E.; Pessione, A.; Pessione, E. Selenium and selenoproteins: An overview on different biological systems. Curr. Protein Pept. Sci. 2014, 15, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Gladyshev, V.N. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005, 3, 2080–2089. [Google Scholar] [CrossRef] [Green Version]
- Scortecci, J.F.; Serrão, V.H.B.; Fernandes, A.F.; Basso, L.G.M.; Gutierrez, R.F.; Araujo, A.P.U.; Neto, M.O.; Thiemann, O.H. Initial steps in selenocysteine biosynthesis: The interaction between selenocysteine lyase and selenophosphate synthetase. Int. J. Biol. Macromol. 2020, 156, 18–26. [Google Scholar] [CrossRef]
- Stock, T.; Rother, M. Selenoproteins in Archaea and Gram-positive bacteria. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Sumner, S.E.; Markley, R.L.; Kirimanjeswara, G.S. Role of selenoproteins in bacterial pathogenesis. Biol. Trace Elem. Res. 2019, 192, 69–82. [Google Scholar] [CrossRef]
- Lobanov, A.V.; Hatfield, D.L.; Gladyshev, V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 1424–1428. [Google Scholar] [CrossRef] [Green Version]
- Avery, J.C.; Hoffmann, P.R. Selenium, selenoproteins, and immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, L.; Neves, C.; Melo, M.; Soares, P. Selenium and selenoproteins in immune mediated thyroid disorders. Diagnostics 2018, 8, 70. [Google Scholar] [CrossRef] [Green Version]
- Winther, K.H.; Rayman, M.P.; Bonnema, S.J.; Hegedüs, L. Selenium in thyroid disorders—Essential knowledge for clinicians. Nat. Rev. Endocrinol. 2020, 16, 165–176. [Google Scholar] [CrossRef]
- Roman, M.; Jitaru, P.; Barbante, C. Selenium biochemistry and its role for human health. Metallomics 2014, 6, 25–54. [Google Scholar] [CrossRef]
- Qazi, I.H.; Angel, C.; Yang, H.; Zoidis, E.; Pan, B.; Wu, Z.; Ming, Z.; Zeng, C.J.; Meng, Q.; Han, H.; et al. Role of selenium and selenoproteins in male reproductive function: A review of past and present evidences. Antioxidants 2019, 8, 268. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhou, H.; Bai, J.; Li, Y.; Yang, J.; Ma, Q.; Qu, Y. Biosynthesis of selenium nanoparticles mediated by fungus Mariannaea sp. HJ and their characterization. Colloids Surf. A Physicochem. Eng. Asp. 2019, 571, 9–16. [Google Scholar] [CrossRef]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Biosynthesis of selenium nanoparticles by Azoarcus sp. CIB. Microb. Cell Fact. 2016, 15, 109. [Google Scholar] [CrossRef]
- Staicu, L.C.; van Hullebusch, E.D.; Lens, P.N.L. Production, recovery and reuse of biogenic elemental selenium. Environ. Chem. Lett. 2015, 13, 89–96. [Google Scholar] [CrossRef]
- Tugarova, A.V.; Mamchenkova, P.V.; Khanadeev, V.A.; Kamnev, A.A. Selenite reduction by the rhizobacterium Azospirillum brasilense, synthesis of extracellular selenium nanoparticles and their characterisation. New Biotechnol. 2020, 58, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.J.; Weiner, J.H.; Taylor, D.E. Selenium metabolism in Escherichia coli. BioMetals 1998, 11, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Debieux, C.M.; Dridge, E.J.; Mueller, C.M.; Splatt, P.; Paszkiewicz, K.; Knight, I.; Florance, H.; Love, J.; Titball, R.W.; Lewis, R.J.; et al. A bacterial process for selenium nanosphere assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 13480–13485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehan, M.; Alsohim, A.S.; El-Fadly, G.; Tisa, L.S. Detoxification and reduction of selenite to elemental red selenium by Frankia. Antonie Van Leeuwenhoek 2019, 112, 127–139. [Google Scholar] [CrossRef]
- Huang, X.; Chen, X.; Chen, Q.; Yu, Q.; Sun, D.; Liu, J. Investigation of functional selenium nanoparticles as potent antimicrobial agents against superbugs. Acta Biomater. 2016, 30, 397–407. [Google Scholar] [CrossRef]
- Sánchez-López, E.; Gomes, D.; Esteruelas, G.; Bonilla, L.; Lopez-Machado, A.L.; Galindo, R.; Cano, A.; Espina, M.; Ettcheto, M.; Camins, A.; et al. Metal-based nanoparticles as antimicrobial agents: An overview. Nanomaterials 2020, 10, 292. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, Z.; Dai, C.; Wang, P.; Fan, S.; Yu, B.; Qu, Y. Antibacterial properties and mechanism of selenium nanoparticles synthesized by Providencia sp. DCX. Environ. Res. 2021, 194, 110630. [Google Scholar] [CrossRef]
- Khiralla, G.M.; El-Deeb, B.A. Antimicrobial and antibiofilm effects of selenium nanoparticles on some foodborne pathogens. LWT Food Sci. Technol. 2015, 63, 1001–1007. [Google Scholar] [CrossRef]
- Nguyen, T.H.D.; Vardhanabhuti, B.; Lin, M.; Mustapha, A. Antibacterial properties of selenium nanoparticles and their toxicity to Caco-2 cells. Food Control 2017, 77, 17–24. [Google Scholar] [CrossRef]
- San Keskin, N.O.S.; Vural, O.A.; Abaci, S. Biosynthesis of noble selenium nanoparticles from Lysinibacillus sp. NOSK for antimicrobial, antibiofilm activity, and biocompatibility. Geomicrobiol. J. 2020, 37, 919–928. [Google Scholar] [CrossRef]
- Dayem, A.A.; Hossain, M.K.; Lee, S.B.; Kim, K.; Saha, S.K.; Yang, G.M.; Choi, H.Y.; Cho, S.G. The role of reactive oxygen species (ROS) in the biological activities of metallic nanoparticles. Int. J. Mol. Sci. 2017, 18, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zonaro, E.; Lampis, S.; Turner, R.J.; Qazi, S.J.S.; Vallini, G. Biogenic selenium and tellurium nanoparticles synthesized by environmental microbial isolates efficaciously inhibit bacterial planktonic cultures and biofilms. Front. Microbiol. 2015, 6, 584. [Google Scholar] [CrossRef] [Green Version]
- Alam, H.; Khatoon, N.; Khan, M.A.; Husain, S.A.; Saravanan, M.; Sardar, M. Synthesis of selenium nanoparticles using probiotic bacteria Lactobacillus acidophilus and their enhanced antimicrobial activity against resistant bacteria. J. Clust. Sci. 2020, 31, 1003–1011. [Google Scholar] [CrossRef]
- Medina-Cruz, D.; Mi, G.; Webster, T.J. Synthesis and characterization of biogenic selenium nanoparticles with antimicrobial properties made by Staphylococcus aureus, methicillin-resistant Staphylococcus aureus (MRSA), Escherichia coli, and Pseudomonas aeruginosa. J. Biomed. Mater. Res. Part A 2018, 106, 1400–1412. [Google Scholar] [CrossRef] [PubMed]
- Akçay, F.A.; Avcı, A. Effects of process conditions and yeast extract on the synthesis of selenium nanoparticles by a novel indigenous isolate Bacillus sp. EKT1 and characterization of nanoparticles. Arch. Microbiol. 2020, 202, 2233–2243. [Google Scholar] [CrossRef] [PubMed]
- Piacenza, E.; Presentato, A.; Zonaro, E.; Lemire, J.A.; Demeter, M.; Vallini, G.; Turner, R.J.; Lampis, S. Antimicrobial activity of biogenically produced spherical Se-nanomaterials embedded in organic material against Pseudomonas aeruginosa and Staphylococcus aureus strains on hydroxyapatite-coated surfaces. Microb. Biotechnol. 2017, 10, 804–818. [Google Scholar] [CrossRef] [Green Version]
- Khoei, N.S.; Lampis, S.; Zonaro, E.; Yrjälä, K.; Bernardi, P.; Vallini, G. Insights into selenite reduction and biogenesis of elemental selenium nanoparticles by two environmental isolates of Burkholderia fungorum. New Biotechnol. 2017, 34, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Qiao, L.; Ma, L.; Yan, S.; Guo, Y.; Dou, X.; Zhang, B.; Roman, A. Biosynthesis of polysaccharides-capped selenium nanoparticles using Lactococcus lactis NZ9000 and their antioxidant and anti-inflammatory activities. Front. Microbiol. 2019, 10, 1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerrard, T.L.; Telford, J.N.; Williams, H.H. Detection of selenium deposits in Escherichia coli by electron microscopy. J. Bacteriol. 1974, 119, 1057–1060. [Google Scholar] [CrossRef] [Green Version]
- Bharathi, S.; Kumaran, S.; Suresh, G.; Ramesh, M.; Thangamani, V.; Pugazhvendan, S.R.; Sathiyamurthy, K. Extracellular synthesis of nanoselenium from fresh water bacteria Bacillus sp., and its validation of antibacterial and cytotoxic potential. Biocatal. Agric. Biotechnol. 2020, 27, 101655. [Google Scholar] [CrossRef]
- Fischer, S.; Krause, T.; Lederer, F.; Merroun, M.L.; Shevchenko, A.; Hübner, R.; Firkala, T.; Stumpf, T.; Jordan, N.; Jain, R. Bacillus safensis JG-B5T affects the fate of selenium by extracellular production of colloidally less stable selenium nanoparticles. J. Hazard. Mater. 2020, 384, 121146. [Google Scholar] [CrossRef]
- Ruiz-Fresneda, M.A.; Eswayah, A.S.; Romero-González, M.; Gardiner, P.H.E.; Solari, P.L.; Merroun, M.L. Chemical and structural characterization of Se (IV) biotransformations by Stenotrophomonas bentonitica into Se0 nanostructures and volatiles Se species. Environ. Sci. Nano 2020, 7, 2140–2155. [Google Scholar] [CrossRef]
- Wadhwani, S.A.; Gorain, M.; Banerjee, P.; Shedbalkar, U.U.; Singh, R.; Kundu, G.C.; Chopade, B.A. Green synthesis of selenium nanoparticles using Acinetobacter sp. SW30: Optimization, characterization and its anticancer activity in breast cancer cells. Int. J. Nanomed. 2017, 12, 6841–6855. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, M.; Schmidt, S.; Winter, J. Formation of Se (0) Nanoparticles by Duganella sp. and Agrobacterium sp. isolated from Se-laden soil of North-East Punjab, India. Microb. Cell Fact. 2012, 11, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tugarova, A.V.; Vetchinkina, E.P.; Loshchinina, E.A.; Burov, A.M.; Nikitina, V.E.; Kamnev, A.A. Reduction of selenite by Azospirillum brasilense with the formation of selenium nanoparticles. Microb. Ecol. 2014, 68, 495–503. [Google Scholar] [CrossRef]
- Zheng, S.; Su, J.; Wang, L.; Yao, R.; Wang, D.; Deng, Y.; Wang, R.; Wang, G.; Rensing, C. Selenite reduction by the obligate aerobic bacterium Comamonas testosteroni S44 isolated from a metal-contaminated soil. BMC Microbiol. 2014, 14, 204. [Google Scholar] [CrossRef] [PubMed]
- Dobias, J.; Suvorova, E.I.; Bernier-Latmani, R. Role of proteins in controlling selenium nanoparticle size. Nanotechnology 2011, 22, 195605. [Google Scholar] [CrossRef]
- Kazempour, Z.B.; Yazdi, M.H.; Rafii, F.; Shahverdi, A.R. Sub-inhibitory concentration of biogenic selenium nanoparticles lacks post antifungal effect for Aspergillus niger and Candida albicans and stimulates the growth of Aspergillus niger. Iran. J. Microbiol. 2013, 5, 81–85. [Google Scholar] [PubMed]
- Torres, S.K.; Campos, V.L.; León, C.G.; Rodríguez-Llamazares, S.M.; Rojas, S.M.; González, M.; Smith, C.; Mondaca, M.A. Biosynthesis of selenium nanoparticles by Pantoea agglomerans and their antioxidant activity. J. Nanopart. Res. 2012, 14, 1236. [Google Scholar] [CrossRef]
- Wang, Y.; Shu, X.; Hou, J.; Lu, W.; Zhao, W.; Huang, S.; Wu, L. Selenium nanoparticle synthesized by Proteus mirabilis YC801: An efficacious pathway for selenite biotransformation and detoxification. Int. J. Mol. Sci. 2018, 19, 3809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Deeb, B.; Al-Talhi, A.; Mostafa, N.; Abou-assy, R. Biological synthesis and structural characterization of selenium nanoparticles and assessment of their antimicrobial properties. Am. Sci. Res. J. Eng. Technol. Sci. 2018, 45, 135–170. [Google Scholar]
- Dwivedi, S.; AlKhedhairy, A.A.; Ahamed, M.; Musarrat, J. Biomimetic synthesis of selenium nanospheres by bacterial strain JS-11 and its role as a biosensor for nanotoxicity assessment: A Novel Se-Bioassay. PLoS ONE 2013, 8, e57404. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, Z.; Liu, H.; Zhang, L.; Gao, P.; Li, D. Biosynthesis and structural characteristics of selenium nanoparticles by Pseudomonas alcaliphila. Colloids Surf. B Biointerfaces 2011, 88, 196–201. [Google Scholar] [CrossRef]
- Avendaño, R.; Chaves, N.; Fuentes, P.; Sánchez, E.; Jiménez, J.I.; Chavarría, M. Production of selenium nanoparticles in Pseudomonas putida KT2440. Sci. Rep. 2016, 6, 37155. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Liu, N.; Li, Y.; Jing, W.; Fan, J.; Li, D.; Zhang, L.; Zhang, X.; Zhang, Z.; Wang, L. Reduction of selenite to red elemental selenium by Rhodopseudomonas palustris strain N. PLoS ONE 2014, 9, e95955. [Google Scholar] [CrossRef]
- Oremland, R.S.; Herbel, M.J.; Blum, J.S.; Langley, S.; Beveridge, T.J.; Ajayan, P.M.; Sutto, T.; Ellis, A.V.; Curran, S. Structural and spectral features of selenium nanospheres produced by Se-respiring bacteria. Appl. Environ. Microbiol. 2004, 70, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Tam, K.; Ho, C.T.; Lee, J.H.; Lai, M.; Chang, C.H.; Rheem, Y.; Chen, W.; Hur, H.G.; Myung, N.V. Growth mechanism of amorphous selenium nanoparticles synthesized by Shewanella sp. HN-41. Biosci. Biotechnol. Biochem. 2010, 74, 696–700. [Google Scholar] [CrossRef] [Green Version]
- Dungan, R.S.; Yates, S.R.; Frankenberger, W.T. Transformations of selenate and selenite by Stenotrophomonas maltophilia isolated from a seleniferous agricultural drainage pond sediment. Environ. Microbiol. 2003, 5, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Cremonini, E.; Zonaro, E.; Donini, M.; Lampis, S.; Boaretti, M.; Dusi, S.; Melotti, P.; Lleo, M.M.; Vallini, G. Biogenic selenium nanoparticles: Characterization, antimicrobial activity and effects on human dendritic cells and fibroblasts. Microb. Biotechnol. 2016, 9, 758–771. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Cecconi, D.; Monti, F.; Micaroni, M.; Turner, R.J.; Butler, C.S.; Vallini, G. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles. J. Hazard. Mater. 2017, 324, 3–14. [Google Scholar] [CrossRef]
- Ramya, S.; Shanmugasundaram, T.; Balagurunathan, R. Biomedical potential of actinobacterially synthesized selenium nanoparticles with special reference to anti-biofilm, anti-oxidant, wound healing, cytotoxic and anti-viral activities. J. Trace Elem. Med. Biol. 2015, 32, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Hnain, A.; Brooks, J.; Lefebvre, D.D. The synthesis of elemental selenium particles by Synechococcus leopoliensis. Appl. Microbiol. Biotechnol. 2013, 97, 10511–10519. [Google Scholar] [CrossRef]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci. Rep. 2017, 7, 16046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, N.; Mukhopadhyay, M. Biosynthesis and structural characterization of selenium nanoparticles mediated by Zooglea ramigera. Powder Technol. 2013, 244, 26–29. [Google Scholar] [CrossRef]
- Dhanjal, S.; Cameotra, S.S. Aerobic biogenesis of selenium nanospheres by Bacillus cereus isolated from coalmine soil. Microb. Cell Fact. 2010, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Pouri, S.; Motamedi, H.; Honary, S.; Kazeminezhad, I. Biological synthesis of selenium nanoparticles and evaluation of their bioavailability. Braz. Arch. Biol. Technol. 2017, 60, e170452. [Google Scholar] [CrossRef]
- Ali, E.N.; El-Sonbaty, S.M.; Salem, F.M. Evaluation of selenium nanoparticles as a potential chemopreventive agent against lung carcinoma. Int. J. Pharm. Biol. Sci. 2013, 2, 38–46. [Google Scholar]
- Sonkusre, P.; Nanduri, R.; Gupta, P.; Cameotra, S.S. Improved extraction of intracellular biogenic selenium nanoparticles and their specificity for cancer chemoprevention. J. Nanomed. Nanotechnol. 2014, 5, 1000194. [Google Scholar] [CrossRef] [Green Version]
- Mishra, R.R.; Prajapati, S.; Das, J.; Dangar, T.K.; Das, N.; Thatoi, H. Reduction of selenite to red elemental selenium by moderately halotolerant Bacillus megaterium strains isolated from Bhitarkanika mangrove soil and characterization of reduced product. Chemosphere 2011, 84, 1231–1237. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Bernardi, P.; Butler, C.S.; Vallini, G. Delayed formation of zero-valent selenium nanoparticles by Bacillus mycoides SeiTE01 as a consequence of selenite reduction under aerobic conditions. Microb. Cell Fact. 2014, 13, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Shakibaie, M.; Khorramizadeh, M.R.; Faramarzi, M.A.; Sabzevari, O.; Shahverdi, A.R. Biosynthesis and recovery of selenium nanoparticles and the effects on matrix metalloproteinase-2 expression. Biotechnol. Appl. Biochem. 2010, 56, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yang, L.; Zhang, B.; Liu, J. Extracellular biosynthesis and transformation of selenium nanoparticles and application in H2O2 biosensor. Colloids Surf. B Biointerfaces 2010, 80, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Eszenyi, P.; Sztrik, A.; Babka, B.; Prokisch, J. Production of Lactomicrosel® and nanosize (100–500 nm) selenium spheres by probiotic lactic acid bacteria. Int. Conf. Food Eng. Biotechnol. 2011, 9, 97–101. [Google Scholar]
- Shoeibi, S.; Mashreghi, M. Biosynthesis of selenium nanoparticles using Enterococcus faecalis and evaluation of their antibacterial activities. J. Trace Elem. Med. Biol. 2017, 39, 135–139. [Google Scholar] [CrossRef]
- Rajasree, R.S.R.; Gayathri, S. Extracellular biosynthesis of Selenium nanoparticles using some species of Lactobacillus. Indian J. Geo Mar. Sci. 2015, 43, 766–775. [Google Scholar]
- Moreno-Martin, G.; Pescuma, M.; Pérez-Corona, T.; Mozzi, F.; Madrid, Y. Determination of size and mass-and number-based concentration of biogenic SeNPs synthesized by lactic acid bacteria by using a multimethod approach. Anal. Chim. Acta 2017, 992, 34–41. [Google Scholar] [CrossRef]
- Yazdi, M.H.; Mahdavi, M.; Setayesh, N.; Esfandyar, M.; Shahverdi, A.R. Selenium nanoparticle-enriched Lactobacillus brevis causes more efficient immune responses in vivo and reduces the liver metastasis in metastatic form of mouse breast cancer. DARU J. Pharm. Sci. 2013, 21, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Qiao, L.; Guo, Y.; Ma, L.; Cheng, Y. Preparation, characteristics and antioxidant activity of polysaccharides and proteins-capped selenium nanoparticles synthesized by Lactobacillus casei ATCC 393. Carbohydr. Polym. 2018, 195, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Li, D.B.; Cheng, Y.Y.; Wu, C.; Li, W.W.; Li, N.; Yang, Z.C.; Tong, Z.H.; Yu, H.Q. Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci. Rep. 2014, 4, 3735. [Google Scholar] [CrossRef] [PubMed]
- Van Fleet-Stalder, V.; Chasteen, T.G.; Pickering, I.J.; George, G.N.; Prince, R.C. Fate of selenate and selenite metabolized by Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2000, 66, 4849–4853. [Google Scholar] [CrossRef] [Green Version]
- Wells, M.; McGarry, J.; Gaye, M.M.; Basu, P.; Oremland, R.S.; Stolz, J.F. Respiratory selenite reductase from Bacillus selenitireducens strain MLS10. J. Bacteriol. 2019, 201, e00614-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugarova, A.V.; Kamnev, A.A. Proteins in microbial synthesis of selenium nanoparticles. Talanta 2017, 174, 539–547. [Google Scholar] [CrossRef]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef]
- Tan, Y.; Wang, Y.; Wang, Y.; Xu, D.; Huang, Y.; Wang, D.; Wang, G.; Rensing, C.; Zheng, S. Novel mechanisms of selenate and selenite reduction in the obligate aerobic bacterium Comamonas testosteroni S44. J. Hazard. Mater. 2018, 359, 129–138. [Google Scholar] [CrossRef]
- Nancharaiah, Y.V.; Lens, P.N.L. Selenium biomineralization for biotechnological applications. Trends Biotechnol. 2015, 33, 323–330. [Google Scholar] [CrossRef]
- Bébien, M.; Kirsch, J.; Méjean, V.; Verméglio, A. Involvement of a putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology 2002, 148, 3865–3872. [Google Scholar] [CrossRef] [Green Version]
- Ridley, H.; Watts, C.A.; Richardson, D.J.; Butler, C.S. Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl. Environ. Microbiol. 2006, 72, 5173–5180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schröder, I.; Rech, S.; Krafft, T.; Macy, J.M. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 1997, 272, 23765–23768. [Google Scholar] [CrossRef] [Green Version]
- Dridge, E.J.; Watts, C.A.; Jepson, B.J.N.; Line, K.; Santini, J.M.; Richardson, D.J.; Butler, C.S. Investigation of the redox centres of periplasmic selenate reductase from Thauera selenatis by EPR spectroscopy. Biochem. J. 2007, 408, 19–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krafft, T.; Bowen, A.; Theis, F.; Macy, J.M. Cloning and sequencing of the genes encoding the periplasmic-cytochrome b-containing selenate reductase of Thauera selenatis. DNA Seq. 2000, 10, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Lowe, E.C.; Bydder, S.; Hartshorne, R.S.; Tape, H.L.U.; Dridge, E.J.; Debieux, C.M.; Paszkiewicz, K.; Singleton, I.; Lewis, R.J.; Santini, J.M.; et al. Quinol-cytochrome c oxidoreductase and cytochrome c4 mediate electron transfer during selenate respiration in Thauera selenatis. J. Biol. Chem. 2010, 285, 18433–18442. [Google Scholar] [CrossRef] [Green Version]
- Watts, C.A.; Ridley, H.; Condie, K.L.; Leaver, J.T.; Richardson, D.J.; Butler, C.S. Selenate reduction by Enterobacter cloacae SLD1a-1 is catalysed by a molybdenum-dependent membrane-bound enzyme that is distinct from the membrane-bound nitrate reductase. FEMS Microbiol. Lett. 2003, 228, 273–279. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Kobayashi, D.Y.; Yee, N. Chemical kinetic and molecular genetic study of selenium oxyanion reduction by Enterobacter cloacae SLD1a-1. Environ. Sci. Technol. 2007, 41, 7795–7801. [Google Scholar] [CrossRef]
- Kuroda, M.; Yamashita, M.; Miwa, E.; Imao, K.; Fujimoto, N.; Ono, H.; Nagano, K.; Sei, K.; Ike, M. Molecular cloning and characterization of the srdBCA operon, encoding the respiratory selenate reductase complex, from the selenate-reducing bacterium Bacillus selenatarsenatis SF-1. J. Bacteriol. 2011, 193, 2141–2148. [Google Scholar] [CrossRef] [Green Version]
- Butler, C.S.; Debieux, C.M.; Dridge, E.J.; Splatt, P.; Wright, M. Biomineralization of selenium by the selenate-respiring bacterium Thauera selenatis. Biochem. Soc. Trans. 2012, 40, 1239–1243. [Google Scholar] [CrossRef] [Green Version]
- Yee, N.; Ma, J.; Dalia, A.; Boonfueng, T.; Kobayashi, D.Y. Se(VI) reduction and the precipitation of Se(0) by the facultative bacterium Enterobacter cloacae SLD1a-1 are regulated by FNR. Appl. Environ. Microbiol. 2007, 73, 1914–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Kobayashi, D.Y.; Yee, N. Role of menaquinone biosynthesis genes in selenate reduction by Enterobacter cloacae SLD1a-1 and Escherichia coli K12. Environ. Microbiol. 2009, 11, 149–158. [Google Scholar] [CrossRef]
- Guymer, D.; Maillard, J.; Sargent, F. A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 2009, 191, 519–528. [Google Scholar] [CrossRef] [PubMed]
- DeMoll-Decker, H.; Macy, J.M. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch. Microbiol. 1993, 160, 241–247. [Google Scholar] [CrossRef]
- Basaglia, M.; Toffanin, A.; Baldan, E.; Bottegal, M.; Shapleigh, J.P.; Casella, S. Selenite-reducing capacity of the copper-containing nitrite reductase of Rhizobium sullae. FEMS Microbiol. Lett. 2007, 269, 124–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, D.; Li, X.; Cheng, Y.; Xiao, X.; Lu, Z.; Wang, Y.; Wang, F. Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci. Rep. 2017, 7, 3239. [Google Scholar] [CrossRef] [PubMed]
- Painter, E.P. The chemistry and toxicity of selenium compounds, with special reference to the selenium problem. Chem. Rev. 1941, 28, 179–213. [Google Scholar] [CrossRef]
- Kessi, J.; Hanselmann, K.W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 2004, 279, 50662–50669. [Google Scholar] [CrossRef]
- Ni, T.W.; Staicu, L.C.; Nemeth, R.S.; Schwartz, C.L.; Crawford, D.; Seligman, J.D.; Hunter, W.J.; Pilon-Smits, E.A.H.; Ackerson, C.J. Progress toward clonable inorganic nanoparticles. Nanoscale 2015, 7, 17320–17327. [Google Scholar] [CrossRef] [Green Version]
- Dungan, R.S.; Frankenberger, W.T. Reduction of selenite to elemental selenium by Enterobacter cloacae SLD1a-1. J. Environ. Qual. 1998, 27, 1301–1306. [Google Scholar] [CrossRef]
- Xia, X.; Wu, S.; Li, N.; Wang, D.; Zheng, S.; Wang, G. Novel bacterial selenite reductase CsrF responsible for Se(IV) and Cr(VI) reduction that produces nanoparticles in Alishewanella sp. WH16-1. J. Hazard. Mater. 2018, 342, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Garbisu, C.; Carlson, D.; Adamkiewicz, M.; Yee, B.C.; Wong, J.H.; Resto, E.; Leighton, T.; Buchanan, B.B. Morphological and biochemical responses of Bacillus subtilis to selenite stress. BioFactors 1999, 10, 311–319. [Google Scholar] [CrossRef]
- Zonaro, E.; Piacenza, E.; Presentato, A.; Monti, F.; Dell’Anna, R.; Lampis, S.; Vallini, G. Ochrobactrum sp. MPV1 from a dump of roasted pyrites can be exploited as bacterial catalyst for the biogenesis of selenium and tellurium nanoparticles. Microb. Cell Fact. 2017, 16, 215. [Google Scholar] [CrossRef] [Green Version]
- Hunter, W.J. Pseudomonas seleniipraecipitans proteins potentially involved in selenite reduction. Curr. Microbiol. 2014, 69, 69–74. [Google Scholar] [CrossRef]
- Bebien, M.; Chauvin, J.P.; Adriano, J.M.; Grosse, S.; Verméglio, A. Effect of selenite on growth and protein synthesis in the phototrophic bacterium Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2001, 67, 4440–4447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledgham, F.; Quest, B.; Vallaeys, T.; Mergeay, M.; Covès, J. A probable link between the DedA protein and resistance to selenite. Res. Microbiol. 2005, 156, 367–374. [Google Scholar] [CrossRef]
- Gaballa, A.; Newton, G.L.; Antelmann, H.; Parsonage, D.; Upton, H.; Rawat, M.; Claiborne, A.; Fahey, R.C.; Helmann, J.D. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. USA 2010, 107, 6482–6486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, P.; Ding, X.; Yang, Y.Y.; Xu, Q.H. Metal nanoparticles for diagnosis and therapy of bacterial infection. Adv. Healthc. Mater. 2018, 7, 1701392. [Google Scholar] [CrossRef] [PubMed]
- Zannoni, D.; Borsetti, F.; Harrison, J.J.; Turner, R.J. The bacterial response to the chalcogen metalloids Se and Te. Adv. Microb. Physiol. 2007, 53, 1–312. [Google Scholar] [CrossRef]
- Helmann, J.D. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox Signal. 2011, 15, 123–133. [Google Scholar] [CrossRef]
- Nancharaiah, Y.V.; Lens, P.N.L. Ecology and biotechnology of selenium-respiring bacteria. Microbiol. Mol. Biol. Rev. 2015, 79, 61–80. [Google Scholar] [CrossRef] [Green Version]
- Jain, R.; Jordan, N.; Weiss, S.; Foerstendorf, H.; Heim, K.; Kacker, R.; Hübner, R.; Kramer, H.; van Hullebusch, E.D.; Farges, F.; et al. Extracellular polymeric substances govern the surface charge of biogenic elemental selenium nanoparticles. Environ. Sci. Technol. 2015, 49, 1713–1720. [Google Scholar] [CrossRef]
- Lenz, M.; Kolvenbach, B.; Gygax, B.; Moes, S.; Corvini, P.F.X. Shedding light on selenium biomineralization: Proteins associated with bionanominerals. Appl. Environ. Microbiol. 2011, 77, 4676–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamnev, A.A.; Dyatlova, Y.A.; Kenzhegulov, O.A.; Vladimirova, A.A.; Mamchenkova, P.V.; Tugarova, A.V. Fourier transform infrared (FTIR) spectroscopic analyses of microbiological samples and biogenic selenium nanoparticles of microbial origin: Sample preparation effects. Molecules 2021, 26, 1146. [Google Scholar] [CrossRef]
- Tugarova, A.V.; Mamchenkova, P.V.; Dyatlova, Y.A.; Kamnev, A.A. FTIR and Raman spectroscopic studies of selenium nanoparticles synthesised by the bacterium Azospirillum thiophilum. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 192, 458–463. [Google Scholar] [CrossRef]
- Sinharoy, A.; Saikia, S.; Pakshirajan, K. Biological removal of selenite from wastewater and recovery as selenium nanoparticles using inverse fluidized bed bioreactor. J. Water Process Eng. 2019, 32, 100988. [Google Scholar] [CrossRef]
- Chandramohan, S.; Sundar, K.; Muthukumaran, A. Reducing agents influence the shapes of selenium nanoparticles (SeNPs) and subsequently their antibacterial and antioxidant activity. Mater. Res. Express 2019, 6, 0850i2. [Google Scholar] [CrossRef]
- Rangrazi, A.; Bagheri, H.; Ghazvini, K.; Boruziniat, A.; Darroudi, M. Synthesis and antibacterial activity of colloidal selenium nanoparticles in chitosan solution: A new antibacterial agent. Mater. Res. Express 2019, 6, 1250h3. [Google Scholar] [CrossRef]
- Chung, Y.C.; Su, Y.P.; Chen, C.C.; Jia, G.; Wang, H.L.; Wu, J.C.G.; Lin, J.G. Relationship between antibacterial activity of chitosan and surface characteristics of cell wall. Acta Pharmacol. Sin. 2004, 25, 932–936. [Google Scholar]
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15. [Google Scholar] [CrossRef]
- Gottenbos, B.; Grijpma, D.W.; Van Der Mei, H.C.; Feijen, J.; Busscher, H.J. Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2001, 48, 7–13. [Google Scholar] [CrossRef]
- Dickson, J.S.; Koohmaraie, M. Cell surface charge characteristics and their relationship to bacterial attachment to meat surfaces. Appl. Environ. Microbiol. 1989, 55, 832–836. [Google Scholar] [CrossRef] [Green Version]
- Tran, P.A.; O’Brien-Simpson, N.; Reynolds, E.C.; Pantarat, N.; Biswas, D.P.; O’Connor, A.J. Low cytotoxic trace element selenium nanoparticles and their differential antimicrobial properties against S. aureus and E. coli. Nanotechnology 2015, 27, 045101. [Google Scholar] [CrossRef]
- Boroumand, S.; Safari, M.; Shaabani, E.; Shirzad, M.; Faridi-Majidi, R. Selenium nanoparticles: Synthesis, characterization and study of their cytotoxicity, antioxidant and antibacterial activity. Mater. Res. Express 2019, 6, 0850d8. [Google Scholar] [CrossRef]
- Galić, E.; Ilić, K.; Hartl, S.; Tetyczka, C.; Kasemets, K.; Kurvet, I.; Milić, M.; Barbir, R.; Pem, B.; Erceg, I.; et al. Impact of surface functionalization on the toxicity and antimicrobial effects of selenium nanoparticles considering different routes of entry. Food Chem. Toxicol. 2020, 144, 111621. [Google Scholar] [CrossRef]
- Filipović, N.; Ušjak, D.; Milenković, M.T.; Zheng, K.; Liverani, L.; Boccaccini, A.R.; Stevanović, M.M. Comparative study of the antimicrobial activity of selenium nanoparticles with different surface chemistry and structure. Front. Bioeng. Biotechnol. 2021, 8, 624621. [Google Scholar] [CrossRef]
- Tran, P.A.; Webster, T.J. Selenium nanoparticles inhibit Staphylococcus aureus growth. Int. J. Nanomed. 2011, 6, 1553–1558. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.; Holden, J.A.; Heath, D.E.; O’Brien-Simpson, N.M.; O’Connor, A.J. Engineering highly effective antimicrobial selenium nanoparticles through control of particle size. Nanoscale 2019, 11, 14937–14951. [Google Scholar] [CrossRef]
- Srivastava, N.; Mukhopadhyay, M. Green synthesis and structural characterization of selenium nanoparticles and assessment of their antimicrobial property. Bioprocess Biosyst. Eng. 2015, 38, 1723–1730. [Google Scholar] [CrossRef]
- Decuzzi, P.; Pasqualini, R.; Arap, W.; Ferrari, M. Intravascular delivery of particulate systems: Does geometry really matter? Pharm. Res. 2009, 26, 235–243. [Google Scholar] [CrossRef] [Green Version]
- Presentato, A.; Piacenza, E.; Anikovskiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Biosynthesis of selenium-nanoparticles and nanorods as a product of selenite bioconversion by the aerobic bacterium Rhodococcus aetherivorans BCP1. New Biotechnol. 2018, 41, 1–8. [Google Scholar] [CrossRef]
- Guisbiers, G.; Wang, Q.; Khachatryan, E.; Mimun, L.C.; Mendoza-Cruz, R.; Larese-Casanova, P.; Webster, T.J.; Nash, K.L. Inhibition of E. coli and S. aureus with selenium nanoparticles synthesized by pulsed laser ablation in deionized water. Int. J. Nanomed. 2016, 11, 3731–3736. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [Green Version]
- Calvo, J.; Martínez-Martínez, L. Mecanismos de acción de los antimicrobianos. Enferm. Infecc. Microbiol. Clín. 2009, 27, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Bruslind, L. General Microbiology; Oregon State University: Corvallis, OR, USA, 2017. [Google Scholar]
- Gopinath, V.; Priyadarshini, S.; Loke, M.F.; Arunkumar, J.; Marsili, E.; MubarakAli, D.; Velusamy, P.; Vadivelu, J. Biogenic synthesis, characterization of antibacterial silver nanoparticles and its cell cytotoxicity. Arab. J. Chem. 2017, 10, 1107–1117. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Yuan, J.; Gao, H. Microbial oxidative stress response: Novel insights from environmental facultative anaerobic bacteria. Arch. Biochem. Biophys. 2015, 584, 28–35. [Google Scholar] [CrossRef]
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium nanoparticles as a nutritional supplement. Nutrition 2017, 33, 83–90. [Google Scholar] [CrossRef]
- Johnson, L.A.; Hug, L.A. Distribution of reactive oxygen species defense mechanisms across domain bacteria. Free Radic. Biol. Med. 2019, 140, 93–102. [Google Scholar] [CrossRef]

| Gram-Negative Bacteria | Shape | Size (nm) | [Se] | Culture Conditions | Location | Ref. |
|---|---|---|---|---|---|---|
| Acinetobacter sp. SW30 | Spherical/rod | 78–126 | 0.3–2 mM Na2SeO3 | Aerobic, 30 °C, 24 h | NS | [49] |
| Agrobacterium | Spherical | 185–190 | 40 mg/L Na2SeO4 | Aerobic, 28 °C, 8 h | Ex | [50] |
| Alcaligenes faecalis | Spherical | 273.8 | 5 mM Na2SeO3 | Aerobic, 30 °C, 36 h | Ex | [10] |
| Azoarcus sp. | Spherical | 123 | 1 mM Na2SeO3 | Aerobic, 30 °C, 7 days | Ex | [25] |
| Azospirillum brasilense Sp245 | Spherical | 78–84/40–50/25–28 | 10/25/50 mM Na2SeO3 | Aerobic, 31–32 °C, 24 h | Ex | [27] |
| Azospirillum brasilense Sp7 and Sp245 | Spherical | 50–400 | 0.3 mM Na2SeO3 | Aerobic, 31–32 °C, 7 days | In | [51] |
| Burkholderia fungorum 95 | Spherical | 170 | 2 mM Na2SeO3 | Anaerobic, 27 °C, 24/48/72 h | In and Ex | [44] |
| Burkholderia fungorum DBT1 | Spherical | 200 | 2 mM Na2SeO3 | Anaerobic, 27 °C, 24/48/72 h | In and Ext | [44] |
| Comamonas testosteroni | Spherical | 100–200 | 1–20 mM Na2SeO3 | Aerobic, 28 °C, 24 h | In | [52] |
| Duganella sp. | Spherical | 140–200 | 40 mg/L Na2SeO4 | Aerobic, 28 °C, 8 h | Ex and cell bound | [50] |
| E. coli K-12 | Spherical | 24–122 | 4 mM H2SeO3 | Condition not specified, 48 h | Ex | [53] |
| E. coli | Spherical | 120 | 2 mM Na2SeO3 | Aerobic, 37 °C, 72 h | NS | [40] |
| Klebsiella pneumonia | Spherical | 90–320 | 200 mg/L SeCl4 | Aerobic, 37 °C, 24 h | In | [54] |
| Lysinibacillus sp. NOSK | Spherical | 150/130/145 | 1 mM Na2SeO3 | Aerobic, 37 °C, 24/48/72 h | NS | [36] |
| Methicillin-resistance Staphylococcus aureus | Spherical/rod | 121 | 2 mM Na2SeO3 | Aerobic, 37 °C, 72 h | NS | [40] |
| Pantoea agglomerans | Spherical | 30–300 | 1 mM Na2SeO3 | Aerobic, 25 °C, 15/20/24 h | In | [55] |
| Proteus mirabilis YC0801 | Spherical | 178.3 | 5 mM Na2SeO3 | Aerobic, 30 °C, 24–36 h | Ex | [56] |
| Providencia vermicola BGRW | Hexagonal | 28 | 1 mM SeO2 | Aerobic, 37 °C, 24 h | In and Ex | [57] |
| Pseudomonas aeruginosa | Spherical | 140 | 2 mM Na2SeO3 | Aerobic, 37 °C, 72 h | Ex | [58] |
| Pseudomonas aeruginosa | Spherical | 171 | 2 mM Na2SeO3 | Aerobic, 37 °C, 72 h | NS | [40] |
| Pseudomonas alcaliphila | Spherical | 50–500 | 0.1 mM Na2SeO3 | Anaerobic, 28 °C, 48 h | In | [59] |
| Pseudomonas putida | Spherical | 100–500 | 1 mM Na2SeO3 | Aerobic, 30 °C, 24 h | In and Ex | [60] |
| Rhodopseudomonas palustris strain N | Spherical | 80–200 | 1 mM Na2SeO3 | Anaerobic, 30 °C, 8 days | Ex | [61] |
| Selenihalanaerobacter shriftii | Spherical | ~300 | 3 mM Na2SeO3 | Anaerobic, 25 °C, 72 h | In and Ex | [62] |
| Shewanella sp. | Spherical | <103 | 0.01–1 mM Na2SeO3 | 30 °C, 72 h | In | [63] |
| Staphylococcus aureus | Spherical | 180 | 2 mM Na2SeO3 | Aerobic, 37 °C, 72 h | NS | [40] |
| Stenotrophomonas bentonitica | Spherical/Trigonal Se crystals | 100–400 | 2 mM Na2SeO3 | Aerobic, 48–145 h | In and Ex | [48] |
| Stenotrophomonas maltophilia | Spherical | ≤ 270 | 0.5 mM Na2SeO4 0.5 mM Na2SeO3 | Aerobic, room temp, 48 h | Near periphery of cell wall | [64] |
| Stenotrophomonas maltophilia | Spherical | 170.6 | 2 mM Na2SeO3 | Anaerobic, 27 °C, 24 h | Ex | [65] |
| Stenotrophomonas maltophilia SeITE02 | Spherical | 221/345/357 | 0.5 mM Na2SeO3 | Aerobic, 27 °C, 6/24/48 h | NS | [38] |
| Stenotrophomonas maltophilia SeITE02 | Spherical | 100–300 | 0.5 mM Na2SeO3 | Aerobic, 27 °C, 48 h | Ex | [66] |
| Streptomyces minutiscleroticus M10A62 | Spherical | 100–250 | 2 mM Na2SeO3 | Aerobic, room temperature, 72 h | Ex | [67] |
| Sulfurospirillum barnesii | Spherical | ~300 | 3 mM Na2SeO3 | Anaerobic, 25 °C, 72 h | In and Ex | [62] |
| Synechococcus leopoliensis | Spherical | 174–348 | 5 mM Na2SeO3 | Aerobic, 22 °C, 9 days | On cell surface | [68] |
| Thauera selenatis | Spherical | 150 | 10 mM Selenate | Anaerobic, condition not specified | In | [29] |
| Vibrio natriegen | Spherical | 100–400 | 1 mM Na2SeO3 | Aerobic, 30 °C, 24 h | In | [69] |
| Zooglea ramigera | Spherical, Trigonal selenium nanorods | 30–150 | 3 mM Na2SeO3 | Aerobic, 30 °C, 48 h | Ex | [70] |
| Gram positive bacteria | ||||||
| Bacillus cereus | Spherical | 150–200 | 2 mM Na2SeO3 | Aerobic, 37 °C, 24 h | In and cell bound | [71] |
| Bacillus cereus | Spherical | 400–600 | 100 mg/mL Na2SeO3 | Condition not specified, 30 °C, 24 h | NS | [72] |
| Bacillus licheniformis ATCC 10716 | Spherical | 50–80 | 1 mM SeO2 | Aerobic, 37 °C, 24 h | In, on the bacterial cell | [73] |
| Bacillus licheniformis JS2 | Spherical | 120 | 1.8 mM Na2SeO3 | Aerobic, 37 °C, 15 h | In | [74] |
| Bacillus megaterium | Spherical | 200 | 0.05–2 mM Na2SeO3 | Aerobic, 37 °C, 80 h | Ex | [75] |
| Bacillus mycoides | Spherical | 50–400 | 2 mM Na2SeO3 | Aerobic, 28 °C, 48 h | Ex | [76] |
| Bacillus mycoides | Spherical | 160 | 2 mM Na2SeO3 | Aerobic, 27 °C, 6 h | NS | [65] |
| Bacillus mycoides SelTE01 | Spherical | <100 | 100 mM Na2SeO3 | Aerobic, 27 °C, 24 h | NS | [42] |
| Bacillus safensis JG-B5T | Spherical | 85–450 | 2.5 mM Na2SeO3 | Anaerobic, 30 °C, 24 h | Ex | [47] |
| Bacillus selenitireducens | Spherical | 300 | 3 mM Na2SeO3 | Aerobic, 25 °C, 3 days | In and Ex | [62] |
| Bacillus sp. | Spherical | 31–335 | 6.4 mM Na2SeO3 | Aerobic, 33 °C, 72 h | Ex | [41] |
| Bacillus sp. | Oval | 209–748 | 30 mM Na2SeO3 | Condition not specified, 37 °C | NS | [1] |
| Bacillus sp. B2 | Spherical | 20–50 | 5 mM Na2SeO3 | Condition not specified. room temperature, 24 h | Ex | [46] |
| Bacillus sp. MSh-1 | Spherical | 80–220 | 1.26 mM SeO2 | Aerobic, 30 °C, 14 h | In | [77] |
| Bacillus subtilis | Spherical | 50–400 | 4 mM Na2SeO3 | Aerobic, 35 °C, 48 h | Ex | [78] |
| Bifidobacterium sp. | Spherical | 400–500 | 200 mg/L NaHSeO3 | Aerobic, 37 °C, 36–48 h | NS | [79] |
| Enterococcus faecalis | Spherical | 29–195 | 0.19–2.97 mM Na2SeO3 | Aerobic, 37–42 °C, 24 and 48 h | Ex | [80] |
| Lactobacillus acidophilus | Spherical | 10–20 | 4 mM Na2SeO3 | Aerobic, 35 °C, 48 h | NS | [81] |
| Lactobacillus acidophilus | Spherical | 2–15 | 15 mM Na2SeO3 | Aerobic, 37 °C, 48 h | Ex | [39] |
| Lactobacillus acidophilus CRL 636 | Spherical | 176 | 25 mM Na2SeO3 | Aerobic, 37 °C 24 h | In | [82] |
| Lactobacillus brevis | − | >250 | 2.54 mM SeO2 | Condition not specified, 37 °C, 72 h | In | [83] |
| Lactobacillus bulgaricus CRL 656 | Spherical | 160 | 25 mM Na2SeO3 | Aerobic, 37 °C 24 h | In | [82] |
| Lactobacillus casei 393 | Spherical | 50–80 | 1.2 mM Na2SeO3 | Aerobic, 37 °C, 24 h | In | [84] |
| Lactobacillus lactis NZ9000 | Spherical | 143 | 0.6 mM Na2SeO3 | Anaerobic, 30 °C, 48 h | NS | [44] |
| Lactobacillus plantarum | Spherical | 60–80 | 4 mM Na2SeO3 | Aerobic, 37 °C, 36–48 h | NS | [81] |
| Lactobacillus reuteri CRL 1101 | Spherical | 130 | 25 mM Na2SeO3 | Aerobic, 37 °C, 24 h | In | [82] |
| Lactobacillus rhamnosus | Spherical | 60–80 | 4 mM Na2SeO3 | Aerobic, 37 °C, 36–48 h | NS | [81] |
| Lactobacillus sp. | Spherical | 100–200 | 200 mg/L NaHSeO3 | Aerobic, 37 °C, 36–48 h | NS | [79] |
| Streptococcus thermophilus | Spherical | 50–100 | 200 mg/L NaHSeO3 | Aerobic, 37 °C, 36–48 h | NS | [79] |
| Bacteria | Selenite mM | Reduction of Na2SeO3 | Selenite Reduction Detection (Time) | Reference |
|---|---|---|---|---|
| Alcaligenes faecalis Se03 | 5 | > 73% | End-exponential growth phase and stretched into the stationary phase (18 and 42 h). | [10] |
| Alcaligenes faecalis Se03 | 1 | > 90% | Mid-exponential growth phase (12 and 24 h) | [10] |
| Azoarcus sp. CIB | 8 | − | Stationary growth phase (48 h) | [25] |
| Bacillus mycoides Sel TE01 | 0.5 | 50% | Early-exponential growth phase (5 h) | [76] |
| Bacillus mycoides Sel TE01 | 2 | 25% | Early-exponential growth phase (5 h) | [76] |
| Burkholderia fungorum DBT1 | 2 | 79% of 5 mM | Mid-exponential growth phase (24 h) | [43] |
| Lactococcus lactis NZ9000 | 0.6 | 100% | Stationary growth phase (48 h) | [44] |
| Proteus mirabilis YC801 | 1 | 81% | Mid-exponential growth phase (12 and 24 h) | [56] |
| Proteus mirabilis YC802 | 5 | 59% | End-exponential growth phase (24 h) | [56] |
| Pseudomonas moraviensis | 10 | − | Stationary growth est (12 h) | [26] |
| Pseudomonas putida KT2440 | 1 | − | Mid-exponential growth phase (12 h) | [60] |
| Rhodopseudomonas palustris N | 8 | − | Stationary growth est (50 h) | [61] |
| Rhodospirillum rubrum | 1.5 | − | Late-exponential growth phase (70 h) | [8] |
| Shewanella oneidensis MR-1 | 0.5 | 82% | Mid-exponential growth phase (12 h) | [85] |
| Shewanella sp. HN-41 | 1 | − | Mid-exponential growth phase (12 h) | [63] |
| Stenotrophomonas maltophilia SelTE02 | 0.5 | 100% | Early-exponential growth phase (80 h) | [66] |
| Stenotrophomonas maltophilia SelTE02 | 2 | 86% | Stationary growth phase (92 h) | [66] |
| Synechococcus leopoliensis | 5 | − | Mid-exponential growth phase (24 h) | [68] |
| Vibrio natriegens | 100 | − | Early-exponential growth phase (3 h) | [69] |
| Bacteria | Compartment | Selenate Reductase | Additional Requirements for Selenate Reduction Pathway | Ref. |
|---|---|---|---|---|
| Thauera selenatis | Periplasm | Trimeric molybdoenzyme: SerA, catalytic subunit; SerB, iron-sulfur protein and SerC heme b protein. serABCD operon. | [29,96,101] | |
| Enterobacter cloacae SLD1a-1 | Membrane-bound | Trimeric molybdoenzyme: molybdenum, heme, and non-heme iron | Global transcriptional regulatory gene fnr; tatABC translocation pathway: menaquinone biosynthetic pathway menFDHBCE | [93,98,99,102,103] |
| E. coli | Periplasm | Molybdoenzyme: YgfK, YgfN and YgfM protein. ygfKLMN operon. | [92] | |
| E. coli | Associated with the periplasmic face of the cytoplasmic membrane | Molybdo-enzyme: YnfE and YnfF as putative Tat-targeted selenate reductases. ynfEFGH operon | TAT translocase apparatus and TorD-like chaperone | [104] |
| Bacteria | Growing Conditions | Cell Compartment | Enzymes and Biomolecules Involved in Selenite Reduction | Ref. |
|---|---|---|---|---|
| Thauera selenatis | Anaerobic | Cytoplasm | Periplasmic nitrite reductases or glutathione | [29,101,105] |
| Enterobacter cloacae SLD1a-1 | Anaerobic | Cytoplasm | Nitrite reductases or glutathione | [101,111] |
| E. coli | Anaerobic | Cytoplasm | Glutathione, thioredoxin reductase | [43,50] |
| E. cloacae Z0206 | Aerobic | Cytoplasm | Fumarate reductase, glutathione | [63] |
| Alcaligenes faecalis Se03 | Anaerobic | Cytoplasm | Sulfite reductase, thioredoxin reductase, NADPH or NADH as electron donors | [10] |
| Alishewanella sp. WH16–1 | Aerobic | - | Selenite reductase called CsrF | [112] |
| Burkholderia fungorum DBT1 | Aerobic | Cytoplasm | Glutathione, cytoplasmic reductases | [43] |
| Bacillus mycoides SeITE01 | Aerobic | Cytoplasm | Sulfhydryl groups on peptide thiols, membrane reductases, bacilithiols | [76] |
| Bacillu safensis JG B5T | Aerobic | Outside the cells | Succinate dehydrogenase (membrane-associated proteins) | [47] |
| Bacillus subtilis | Aerobic | - | Thioredoxin reductase | [113] |
| Ochrobactrum sp. MPV1 | Aerobic | Cytoplasm | Glutathione | [114] |
| Proteus mirabilis YC801 | Aerobic | Cytoplasm | Thioredoxin reductase and fumarate reductase | [56] |
| Pseudomonas seleniipraecipitans | Aerobic | - | Thioredoxin reductase | [115] |
| Rhizobium sullae | Aerobic | - | Nitrite reductases | [106] |
| Rhodobacter sphaeroides | Aerobic and anaerobic | Cytoplasm | Glutathione a | [116] |
| Rhodospirillum rubrum | Anaerobic | Cytoplasm | Glutathione | [109] |
| Shewanella oneidensis MR-1 | Anaerobic | Periplasm | Fumarate reductase FccA | [85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar-Ramírez, M.C.; Castañeda-Ovando, A.; Pérez-Escalante, E.; Rodríguez-Serrano, G.M.; Ramírez-Moreno, E.; Quintero-Lira, A.; Contreras-López, E.; Añorve-Morga, J.; Jaimez-Ordaz, J.; González-Olivares, L.G. Antimicrobial Activity of Se-Nanoparticles from Bacterial Biotransformation. Fermentation 2021, 7, 130. https://doi.org/10.3390/fermentation7030130
Escobar-Ramírez MC, Castañeda-Ovando A, Pérez-Escalante E, Rodríguez-Serrano GM, Ramírez-Moreno E, Quintero-Lira A, Contreras-López E, Añorve-Morga J, Jaimez-Ordaz J, González-Olivares LG. Antimicrobial Activity of Se-Nanoparticles from Bacterial Biotransformation. Fermentation. 2021; 7(3):130. https://doi.org/10.3390/fermentation7030130
Chicago/Turabian StyleEscobar-Ramírez, Meyli Claudia, Araceli Castañeda-Ovando, Emmanuel Pérez-Escalante, Gabriela Mariana Rodríguez-Serrano, Esther Ramírez-Moreno, Aurora Quintero-Lira, Elizabeth Contreras-López, Javier Añorve-Morga, Judith Jaimez-Ordaz, and Luis Guillermo González-Olivares. 2021. "Antimicrobial Activity of Se-Nanoparticles from Bacterial Biotransformation" Fermentation 7, no. 3: 130. https://doi.org/10.3390/fermentation7030130
APA StyleEscobar-Ramírez, M. C., Castañeda-Ovando, A., Pérez-Escalante, E., Rodríguez-Serrano, G. M., Ramírez-Moreno, E., Quintero-Lira, A., Contreras-López, E., Añorve-Morga, J., Jaimez-Ordaz, J., & González-Olivares, L. G. (2021). Antimicrobial Activity of Se-Nanoparticles from Bacterial Biotransformation. Fermentation, 7(3), 130. https://doi.org/10.3390/fermentation7030130







