Abstract
Pectinases are a group of enzymes with broad application, including in plant fiber processing, pectic wastewater treatment, paper pulping, fruit juice extraction, and clarification. With an increasing industrial demand for these enzymes, it is useful to isolate organisms that produce large amounts of pectinase and possess wide ranges of stability factors like temperature and pH. In this study, 17 out of 29 bacteria (58.62%) from forest soil samples were pectinolytic. However, only four bacteria (S-5, S-10, S-14, and S-17) showed high pectin hydrolysis zones (ranging from 0.2 cm to 1.7 cm). These four bacteria were identified based on colony morphology, microscopic characterization, biochemical characteristics, and 16S rDNA sequencing. They were designated as Streptomyces sp. (S-5, S-14), Cellulomonas sp. (S-10), and Bacillus sp. (S-17). Interestingly, bacteria showed cellulase and xylanase activity in addition to pectinase. The quantitative assay for pectinase activity of the four isolates provided proof that they are pectinase producers and can be considered potential candidates for industrial uses. The crude enzyme extracts of these bacteria are applicable in oil and juice extraction from sesame seeds and apples, respectively.
1. Introduction
Pectinases catalyze the degradation of pectic polysaccharides into simpler molecules like galacturonic acids [1]. Pectinases are found in bacteria, fungi, yeasts, plants, and insects [2]. The biotechnological potentials of pectinase from microorganisms are of great interest due to their broad substrate specificity and versatility [1]. Pectinases are widely used in industrial applications such as processing fruits and vegetables, production and clarification of juice, fermentation of tea and coffee. In addition, pectinases are exploited for bleaching pulp and recycling wastepaper, animal feed, vegetable oil extraction, and pretreatment of wastewater produced from different fruit juice industries. Moreover, pectinases are applied for bioethanol production, liquefaction and saccharification of biomass, bio-scouring of cotton fiber, retting and degumming plant fiber, and oil extraction [3,4,5]. A recent study also showed that pectinase-treated fruit juice has inhibitory effects on colorectal cancer proliferation [6]. Due to the extensive applications of pectinase in different avenues, the demand for pectinase enzymes is increasing continuously [7]. With growing opportunities and the need for pectinase use worldwide, it is beneficial to isolate and characterize new pectinase-producing bacteria. Different microbes, including bacteria, fungi, yeast, insects, are habituated in soil, and it is the most readily available source for microbes [8]. Thus, the present study focuses on isolating and identifying the pectinase-producing bacteria from the forest soil, understanding their phylogenetic relationship with each other, and studying their application in oil extraction from sesame seeds and juice extraction from apples.
2. Materials and Methods
2.1. Collection of Samples
The soil sample was collected during the end of spring and beginning of summer 2019 from the three nearby boreal forests in Thunder Bay, Ontario, Canada. The topsoil was collected with sterile spatulas, kept in clean zip lock bags, and transferred to the laboratory as soon as possible after collection.
2.2. Isolation of Pure Bacterial Strains and Preservation
Initially, 5 g of soil sample was taken in a sterilized Erlenmeyer flask (250 mL), and 45 mL of 0.9% (w/v) autoclaved NaCl solution in distilled water was added. The homogenized samples were agitated for an hour at 120 rpm in a shaking incubator and then serially diluted until 10−4 and 10−5. For the isolation of a single pure bacterium, a sample of 0.1 mL aliquots from each dilution was spread onto sterilized and solidified nutrient agar (NA) plates with the help of sterile disposable spreaders. Then these plates were cultured at 35 °C for 24–48 h. Different colonies from countable plates were selected and sub-cultured on NA plate by streak plate technique until a pure isolated colony was observed. Pure cultures of microorganisms were stored at 4 °C after streaking in NA slant for further studies.
2.3. Screening of Isolates for Pectinase Activity
The isolates were primarily screened for pectinase activity by point inoculation on 1% (w/v) pectin agar, i.e., pectinase screening agar medium (PSAM), pH 5.5 ± 0.5, and incubated at 35 °C for 24–48 h. After 48 h of incubation, when colonies of 2–4 mm were observed, the plates were flooded with 50 mM iodine potassium iodide solution. A transparent halo zone around the colonies indicated the isolates could produce pectinase [9]. Once a halo zone around the colonies was noted, further different tests were performed on those bacteria as described below. Similarly, screening for cellulase, xylanase, and amylase activity was performed by flooding congo red and iodine solution over the colonies isolated in the agar plate containing 1% (w/v) CMC, 1% (w/v) xylan, and 1% (w/v) starch respectively. The halo or clear zone was the indication of pectin degradation. Pectin degradation index (PDI)% was calculated as PDI% = (colony diameter + clear zone diameter)/clear zone diameter [10]. The composition of different media is described in Supplementary Materials.
2.4. Identification
After selecting bacteria with pectinolytic properties, they were streaked on NA plates and incubated at 35 °C to produce isolated colonies. From the pure individual growth, colony morphology, biochemical studies, and molecular analysis were performed.
2.5. Colony Morphology and Different Tests
Once the pure isolated colonies were observed in NA agar plates, size, shape, elevation, color, consistency, and transparency-like characteristics of each colony were noted. From each different colony, various tests were performed, such as Gram’s staining, capsule staining, biofilm production, catalase, oxidase, indole, methyl red (MR), Voges-Proskauer (VP), citrate use, DNase, urease, hemolysis, antibiotic susceptibility, etc. Additional information on the tests is presented in the Supplementary Materials.
2.6. Genomic DNA Extraction, 16S rDNA Amplification, and Extraction of DNA from Gel
Pure isolated colonies were picked up from NA plates with a sterile toothpick and resuspended in 50 mL of Luria-Bertani (LB) broth and incubated at 35 °C for 18–24 h. From this suspension, the genomic DNA of selected isolates was extracted by the freeze-thaw cycles method as described by Chen et al. [11]. The 16S rDNA gene of the isolates was amplified by Taq DNA polymerase with a universal eubacterial primer set, (Forward Primer) 27F-5′-AGAGTTTGATCCTGGCTCAG-3′ and (Reverse Primer) 1492R-5′-GGTTACCTTGTTACGACTT-3′.
The amplification system contained 2 × Taq PCR Master Mix of 10 μL (10 × Taq DNA polymerase buffer, 10 mM dNTPs, 25 mM of MgCl2, 1 U of Taq DNA polymerase), 1 μL of 10 μM forward and reverse primers respectively, 1 μL of the genomic DNA template, and 7 μL of distilled water making a total volume of 20 μL. The PCR reaction conditions used were as follows: denaturation at 94 °C for 5 min and a cycle starting at 94 °C for 30 s, followed by annealing at 55 °C for 30 s, extending at 72 °C for 1.5 min for 33 cycles, and finally extending at 72 °C for 10 min. The PCR products were determined by 1% (w/v) agarose gel electrophoresis. The target fragments from the gel were cut, and DNA was extracted from the gel with a gel extraction minipreps kit (Biobasic) and then sent for DNA sequencing.
2.7. Gene Sequencing and Phylogenetic Analysis
The 16S rDNA sequences of the isolates were compared with known sequences found in the GenBank operating the basic local alignment search tool (BLASTn) of the National Center for Biotechnology Information (NCBI). Isolates were identified based on the percentage similarity with the known species sequences in the database. New sequences of those isolates were deposited in GenBank (accession numbers W547427- MW547430). For molecular analyses, the accessible sequence data for all related species of Cellulomonas sp., Streptomyces sp., and Bacillus sp. were downloaded from the NCBI database. All the sequences were congregated and parallelized using the Clustalw module in BioEdit v. 7.0.9.0 [12] with default settings. Phylogenetic analysis was constructed using a Neighbor-Joining (NJ) tree with 1000 bootstrap using MEGA 7 [13].
2.8. Growth at Different Temperature and pH
The selected isolates were cultured in NA and incubated at different temperatures ranging from 25 °C to 50 °C. They were also cultivated in NA with different pH levels (5 to 9) and at 35 °C. The colonies of the isolates were observed and noted after 24 h of incubation.
2.9. Quantitative Determination of Pectinase Enzyme Activity
The concentration of reducing sugar was estimated by the dinitrosalicylic acid (DNS) method [14]. The reaction products after the degradation of pectin by a pectinase were reducing sugars. The quantity of reducing sugars obtained by the sample was calculated with the standard graph curve obtained from different galacturonic acid concentrations versus absorbance following the same procedure [14]. The enzyme activity (U/mL) was calculated according to Equation (1) [15]:
where V is the total volume of solution, v is the volume of the crude enzyme used in the assay, 194.1 is the molecular weight of galacturonic acid, and t is the reaction time in minutes.
One unit of enzymatic activity (U) was defined as the amount of enzyme required to release 1 μmol of reducing sugars per minute [14].
For enzyme activity assay, 1 mL samples of freshly grown culture were centrifuged at 10,000 rpm for 5 min. The supernatant obtained represented the crude enzyme extract, and 1% (w/v) pectin in phosphate buffer (0.1 M, pH 7) served as the enzyme assay substrate. From the prepared substrate, 20 µL was taken into a well, and 10 µL of enzyme extract was added, then kept in a water bath (50 °C) for 10 min. The mixture temperature was then lowered to room temperature, 60 µL of DNS reagent was added, and the solutions were kept in a boiling water bath for 5 min. Finally, the absorbance of the solutions was measured at 540 nm using a spectrophotometer. Enzyme blank and reagent blank were also measured for quantitative analysis of pectinase.
2.10. Application in Oil and Juice Extraction
Oil extraction was performed as described by Demir et al. with slight modification [16]. Sesame seeds were dried, cleaned, and stored at room temperature. Two grams of sesame seeds were weighed and ground with mortar and pestle. When the seeds were ground, 2 mL of the crude enzyme was added (for control samples, distilled water was added instead of enzyme extract). The paste formed was transferred into 50 mL centrifuge tube by adding 10 mL of water. The mixture was incubated at 50 °C for 1 h and centrifuged at 4000 rpm for 20 min. The floating layer of emulsified oil was collected in another test tube, and the volume was measured.
For juice extraction, an apple was washed, cut into small pieces, and weighed (25 g). Thereafter, apple pieces were ground with mortar and pestle by adding 4 mL of crude enzyme. The apple paste was transferred into a 50 mL centrifuge tube by rinsing the mortar and pestle with 10 mL of distilled water. The mixture was incubated at 50 °C for 1 h and centrifuged at 5000 rpm for 20 min. The amount of clear supernatant was measured and collected in another tube. The percentage juice recovery, clarity, and relative viscosity of the recovered apple juices were analyzed. The clarity of the apple juice was analyzed by taking absorbance at 660 nm using a UV visible spectrophotometer. Furthermore, the relative viscosity of the juice was measured at room temperature using an Ostwald viscometer. The percentage of juice recovery was calculated as ‘(apple weight − total solid waste) × 100/apple weight’.
2.11. Statistical Analysis
The tests in this study were performed in triplicate, and the values are expressed as mean with standard deviation. The statistical significances were assessed using one-way analysis of variance (ANOVA) followed by the Tukey–Kramer comparison. A p-value less than 0.05 is regarded as statistically significant for the experimental data.
3. Results
3.1. Isolation of Pure Bacterial Strain and Preservation
The forest soil was serially diluted, spread on NA plates, and incubated at 35 °C for 24 to 48 h. After this incubation, we observed different isolates with different colony morphologies. From these various colonies, 29 different isolates were selected based on their color, elevation, consistency, transparency, edges, etc. They were subcultured on the NA plate until the pure colonies were isolated. Screening of pectin hydrolyzing activity was based on observing halo zones around the microbial colonies. Among 29 isolates, only 17 (58.62%) isolates produced a halo zone around the colonies and were considered as having pectin hydrolyzing properties. These 17 bacterial strains were preserved with streaking on the nutrient slant and storing them at 4 °C for further study.
3.2. Screening, Identification, and Growth Conditions of Pectinolytic Isolates
Of 17 pectin hydrolyzing isolates, four isolates (S-5, S-10, S-14, and S-17) showed the high hydrolysis zone and were selected for the identification (see Supplementary Materials; Figure S1). The isolates were identified based on colony morphologies, biochemical tests, and 16S rDNA sequencing. After reading the colony characteristics, they were subjected to various tests, including capsule staining, starch hydrolysis, gelatin hydrolysis, antibiotic susceptibility, biofilm production, etc., and the results, along with the colony characteristics and their PDI%, are shown in Table 1.

Table 1.
Different tests result of the four selected isolates with pectinolytic properties.
The screening test for cellulase and xylanase of the four isolates was performed by flooding congo red over the colonies. Similarly, the screening test for amylase was conducted by flooding with iodine solution. The results of the screening tests are shown in Table 1. When the isolates were cultured at different temperatures (25 to 50 °C) and pH levels (5 to 9), all the bacteria were able to grow at 35 °C and pH 7, as depicted in Table 1.
The sequence similarity of isolates was analyzed, blasted, and compared with the known probable sequences in the NCBI database. The phylogenetic tree of 16S rDNA sequences was constructed using the Neighbor-joining algorithm, as shown in Figure 1. The phylogenetic results identified the isolates S-5, S-10, S-14, and S-17 as Streptomyces sp., Cellulomonas sp., Streptomyces sp., and Bacillus sp., respectively.
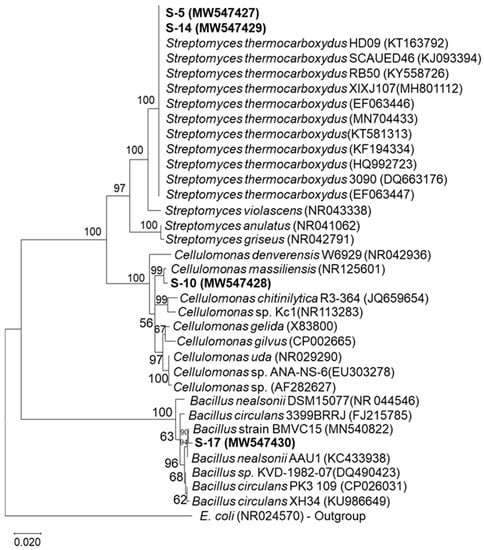
Figure 1.
Neighbor-joining (NJ) tree based on DNA sequences of the 16S rRNA gene showing the phylogenetic relationships. Numbers present on branches of the NJ tree are bootstrap support values.
3.3. Quantitative Analysis of Pectinase Enzyme Activity of Different Isolates
For quantitative analysis of pectinase activity, all isolates were grown at 35 °C, pH 7, and for 120 h in a pectinase production media. Every 24 h, 1 mL of the cultured broth was taken out aseptically and centrifuged to get cell-free crude enzyme extract for quantitative analysis of enzyme activity. The bacterial growth (OD600) and enzyme activity of the isolates were studied, as shown in Figure 2.
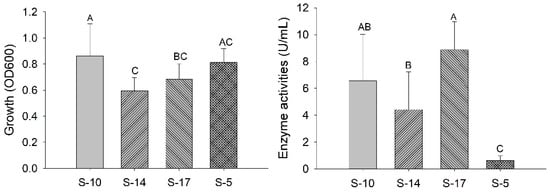
Figure 2.
Bacterial growth and enzyme activity produced by isolates. The bar represents the mean of bacterial growth and enzyme activity. The statistical analysis was performed with one-way ANOVA followed by a post hoc Tukey comparison test. Letters above each bar represent statistical significance at p < 0.05.
3.4. Application of Pectinase in Oil Extraction and Juice Extraction
A crude enzyme extract of 2 mL was added to ground sesame seeds and was transferred into the 50 mL centrifuge tube, and 10 mL of water added. After incubation at 50 °C for 1 h and centrifugation, an emulsified oil appeared as a floating layer at the top of the tube and was collected and measured. The emulsified oil extracted from enzyme extract of isolate S-10 was significantly higher in volume than the other samples (Table 2). Similarly, the crude enzyme extract was added to ground apples for juice extraction, incubated, and centrifuged. The amount of juice extracted was measured and compared with control (no enzyme extract), as shown in Table 2.

Table 2.
Application of pectinase produced from those isolates in oil and juice extraction.
4. Discussion
Pectinase has been used in the fruit and juice industries for decades, and in the present biotechnological era, the industrial applications of pectinase and its market demand are in increasing even more. Thus, it is beneficial to isolate novel microorganisms with higher pectinolytic properties and with stability at a wide range of temperature and pH levels. In our study, 58.6% of isolates demonstrating pectin hydrolyzing properties were isolated from forest soil samples, the majority of which were Gram-positive cocci. Aislabie and Deslippe [8] report that soil contains various microorganisms that contribute to different soil services such as recycling wastes, nitrogen cycle, detoxification of pollutants, etc. The types and mass of the microorganisms in the soil depend upon soil properties and the carbon source(s) available for energy and cell synthesis. Aaisha and Bharate [17] isolated 51.4% of pectinolytic bacteria from different soil samples and observed Bacillus sp. to be the most prominent pectinase-producing isolates. In another study, ten bacteria were isolated from agricultural waste dump soil, and 30% were pectinolytic bacteria [18]. Similarly, a study performed by Oumer and Abate [19] isolated 31.6% of isolates with pectinase activity from the coffee pulp, and Ajobiewe et al. [20] isolated five pectinolytic bacteria from soil containing decaying fruits and vegetables.
A pure culture of isolates was obtained only after several subculturing on NA. A pure culture of isolates was considered usable if there was no contamination and the colonies were all similar in color, size, shape, elevation, consistency, etc. After obtaining a pure culture of isolates, further studies were possible, such as screening tests, identification of isolates, DNA extraction, and more.
Screening of pectin hydrolysis was performed within the screening agar plate containing pectin and when a hydrolysis zone was observed after the addition of potassium iodide solution (Supplementary Figure S1). The hydrolysis area looked clear because the pectinase produced by bacteria hydrolyzed the pectin and made the pectin incapable of binding with iodine that formed a complex. The highest hydrolysis area was an indication of the most pectinase activity [9]. In this study, the hydrolysis zone ranged from 2 mm to 17 mm. However, the four isolates with a hydrolysis zone of more than 10 mm were chosen for further study, including identification.
Xylanase and cellulase enzymes were present in addition to pectinase in isolates S-5 and S-14. The isolate S-10 had pectinase and cellulase, while isolate S-17 had pectinase and amylase. Beg et al. [21] isolated Streptomyces sp. with thermostable pectinase and xylanase. Similarly, Kaur et al. [22] isolated Bacillus pumilus, which produced xylanase and pectinase from soil contaminated with paper and pulp industry effluents.
Production of more than one commercial enzyme from a single microorganism is rare, but is economical and feasible for industrial applications. For example, the combination of xylanase and pectinase effectively removes bark from wood, processing plant fibers [21], and bio-scouring fibers in textile industries [23]. Thus, detailed studies on the production of various industrially essential enzymes from a single microbe for commercial benefit and their industrial applications are needed.
The detailed morphological characteristics and different biochemical test results for the isolates were potentially helpful in identifying them based on Bergy’s Manual of Systemic Bacteriology. Additionally, 16S rDNA sequencing was performed to determine the isolates. The genomic DNA of the four isolates produced one distinct band on an agarose gel. The 16S rDNA amplification of the genomic DNA was subjected to PCR by using universal primers. The amplified 16S rDNA was extracted from the gel using the Gel extraction minipreps kit (Biobasic) and sent for sequencing. The sequence results were analyzed and blasted with the probable sequences in NCBI.
The phylogenetic tree was constructed with the closest sequences found within the NCBI GenBank and E. coli as an out-group. The phylogenetic analysis indicated that the Cellulomonas strain (isolate S-10) and Bacillus sp. (isolate S-17) were clustered with other strains of Cellulomonas massiliensis and Bacillus strain, respectively, with high bootstrap support value. Similarly, Streptomyces strains (isolate S-5 and isolate S-14) were grouped with Streptomyces thermocarboxydus (Figure 1).
In a study conducted by Bharadwaj and Udupa [24], 50 °C and pH 4 were the optimal growth temperature and pH for Streptomyces thermocarboxydus isolated from soil. However, the maximum enzyme activity of partially purified pectinase was found at 60 °C and neutral to alkaline pH [24]. In another study, Streptomyces fumigatiscleroticus VIT-SP4 showed the optimum values at an incubation period of 48 h, pH level 6, and temperature of 35 °C [25]. Thus, in our study, the four isolates were cultured in broth media containing 1% (w/v) pectin at 35 °C and pH 7 for the quantitative analysis of pectinase activity. Additionally, the temperature and pH were selected after observing the results of the isolates cultured in different temperatures and pH (Table 1). Our study showed the isolate S-17 had the highest pectinase activity, followed by S-10, within the conditions as shown Figure 2. However, the enzyme activities of S-10 and S-17 were not statistically different from each other but were statistically different from other isolates (p < 0.05). Similarly, bacterial growth was higher for S-10, and its growth was statistically significant (p < 0.05). Enzyme activity and growth depend on various factors, including microorganisms, carbon and nitrogen sources, incubation temperature, pH, hours, fermentation process, etc. Thus, further detailed studies are needed for the optimization of the cultural conditions of the isolates to maximize pectinase production.
An antibiotic susceptibility test was conducted against the isolates, and it was observed that most of them were susceptible to the seven most commonly used antibiotics (see Supplementary Materials: Table S1). Furthermore, those isolates were tested for hemolysis, capsule, and biofilm production (Supplementary Materials), and it was found that they were non-capsular, non-hemolytic, and non-producers of biofilm (Table 1).
Furthermore, the cell-free supernatants (crude enzyme extracts) were used to study their application in oil and apple juice extraction. The amount of emulsified oil obtained from aqueous extraction of the crude enzyme-treated sesame seeds and apple juice extracted was measured and considered to reflect the enzyme’s ability to extract oil and juice, respectively. Table 2 shows that enzymes stimulated oil extraction from sesame seeds and accelerated the juice extraction from apples. The oil and juice were extracted in higher volume compared to the control without enzyme extract. Of the four bacteria, the amount of emulsified oil volume and juice amount was greater from isolate S-10 followed by S-17, and the extraction was significant statistically (p < 0.05). Moreover, apple juice extracted by treatment with the crude enzyme extract from isolate S-10 had a much lower relative viscosity compared to the control. The clarity of apple juice was affected by the enzymatic treatment, which was determined in terms of absorbance and transmittance at 660 nm.
Pectinase facilitates oil extraction from sesame seeds and juice extraction from apples by breaking pectin present in seeds and apples [16]. The synergistic action of different hydrolytic enzymes such as pectinase, cellulase, xylanase, and amylase plays an important role in extracting and clarifying juice [26]. Enzyme treatment degraded any pectin present in the fruit, leading to a decrease in the water holding capacity of pectin, and water was released to the system, increasing juice yield [4]. Moreover, a combination of enzymes increased clarity, juice recovery, and decreased viscosity and turbidity of juice and oil. The enzymatic extraction of fruit and oil is a novel, green and beneficial technology [26,27]. This study suggests that isolates from forest soil can effectively extract oil from sesame seeds and juice from apples. The pectinase produced by these isolates may also have potential in various industries besides fruit juice and oil extraction and merits further study.
5. Conclusions
Novel pectinase-producing bacteria were isolated from boreal forest soil in Thunder Bay, Ontario. Four bacteria showing high pectinolytic activity in the screening test were identified as Streptomyces sp. (S-5), Cellulomonas sp. (S-10), Streptomyces sp. (S-14), and Bacillus sp. (S-17). Identification was based on the morphology of the bacteria, various biochemical tests, and molecular analyses. The bacteria all had Gram-positive reactions, rod-shapes, and formed spores, except S-10. The enzymes produced from the isolates have the potential for oil and juice extraction, and could therefore be alternatives for commercial pectinase production. However, further studies are needed to evaluate the optimization of cultural conditions for enzyme production and optimization of oil and juice extraction conditions to maximize oil production and juice yield. The characterization of enzymes, oil, and juice and their physicochemical properties are essential to study for industrial and economic development. The isolates studied here demonstrated multi-enzyme activities, indicating that they are potentially viable candidates for various industrial applications and worthy of further exploration.
Supplementary Materials
The following are available online at https://www.mdpi.com/2311-5637/7/1/40/s1.
Author Contributions
Conceptualization, S.S.; methodology, S.S. and F.C.; software, C.C., J.R.K. and X.Z.; validation, S.S. and J.R.K.; formal analysis, S.S.; investigation, S.S.; resources, W.Q.; data curation, S.S. and A.L.M.K.; writing—original draft preparation, S.S.; writing—review and editing, S.S., A.L.M.K., F.C., X.Z., X.C., C.C., S.H. and W.Q.; visualization, S.S.; supervision, W.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2017-05366) to W.Q.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data can be available upon request.
Acknowledgments
The authors would like to acknowledge all the individuals who directly or indirectly helped to complete this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pedrolli, D.B.; Monteiro, A.C.; Gomes, E.; Carmona, E.C. Pectin and pectinases: Production, characterization and industrial application of microbial pectinolytic enzymes. Open Biotechnol. J. 2009, 3, 9–18. [Google Scholar] [CrossRef]
- Frati, F.; Galletti, R.; De Lorenzo, G.; Salerno, G.; Conti, E. Activity of endo-polygalacturonases in mirid bugs (Heteroptera: Miridae) and their inhibition by plant cell wall proteins (PGIPs). Eur. J. Entomol. 2006, 103, 515–522. [Google Scholar] [CrossRef]
- Kubra, K.T.; Ali, S.; Walait, M.; Sundus, H. Potential applications of pectinases in food, agricultural and environmental sectors. J. Pharm. Chem. Biol. Sci. 2017, 6, 23–34. [Google Scholar]
- Kashyap, D.R.; Vohra, P.K.; Chopra, S.; Tewari, R. Applications of pectinases in the commercial sector: A review. Bioresour. Technol. 2001, 77, 215–227. [Google Scholar] [CrossRef]
- Garg, G.; Singh, A.; Kaur, A.; Singh, R.; Kaur, J.; Mahajan, R. Microbial pectinases: An ecofriendly tool of nature for industries. 3 Biotech. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.D.; Kim, J.H.; Won, Y.S.; Moon, K.D.; Seo, K. Il Inhibitory Effects of Pectinase-Treated Prunus Mume Fruit Concentrate on Colorectal Cancer Proliferation and Angiogenesis of Endothelial Cells. J. Food Sci. 2019, 84, 3284–3295. [Google Scholar] [CrossRef]
- Oumer, O.J. Pectinase: Substrate, Production and their Biotechnological Applications. Int. J. Environ. Agric. Biotechnol. 2017, 2, 1007–1014. [Google Scholar] [CrossRef]
- Aislabie, J.; Deslippe, J.R. Soil Microbes and Their Contribution to Soil Services; Manaaki Whenua Press: Lincoln, New Zealand, 2013. [Google Scholar]
- Takcı, H.A.M.; Turkmen, F.U. Extracellular pectinase production and purification from a newly isolated Bacillus subtilis strain. Int. J. Food Prop. 2016, 19, 2443–2450. [Google Scholar] [CrossRef]
- Haile, M.; Kang, W.H. Isolation, identification, and characterization of pectinolytic yeasts for starter culture in coffee fermentation. Microorganisms 2019, 7, 16. [Google Scholar] [CrossRef]
- Chen, F.; Ye, J.; Chio, C.; Liu, W.; Shi, J.; Qin, W. A simplified quick microbial genomic DNA extraction via freeze-thawing cycles. Mol. Biol. Rep. 2020, 47, 703–709. [Google Scholar] [CrossRef]
- Hall, T.A. BIoEdit: A user-friendly bilogical sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Kavuthodi, B.; Thomas, S.; Sebastian, D. Co-production of pectinase and biosurfactant by the newly isolated strain Bacillus subtilis BKDS1. Br. Microbiol. Res. J. 2015, 10, 1–12. [Google Scholar] [CrossRef]
- Demir, N.; Nadaroglu, H.; Demir, Y.; Isik, C.; Taskin, E.; Adiguzel, A.; Gulluce, M. Purification and characterization of an alkaline pectin lyase produced by a newly isolated Brevibacillus borstelensis (p35) and its applications in fruit juice and oil extraction. Eur. Food Res. Technol. 2014, 239, 127–135. [Google Scholar] [CrossRef]
- Aaisha, G.A.; Barate, D.L. Isolation and Identification of Pectinolytic Bacteria from Soil Samples of Akola Region, India. Int. J. Curr. Microbiol. App. Sci. 2016, 5, 514–521. [Google Scholar] [CrossRef]
- Karthik, J.L.; Kumar, G.; Rao, K.V.B. Screening of pectinase producing microorganisms from agricultural waste dump soil. Asian J. Biochem. Pharm. Res. 2011, 1, 329–337. [Google Scholar]
- Oumer, O.J.; Abate, D. Screening and molecular identification of pectinase producing microbes from coffee pulp. Biomed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Ajobiewe, H.F.; Ajobiewe, J.O.; Mbagwu, T.T.; Ale, T.; Yakubu, G.T. Demonstration of Pectinase Enzyme Activity from Soil Isolated Bacillus spp. in Karu Nasarawa State of Nigeria Using Orange Peels Substrate. J. Microbiol. Res. 2019, 9, 6–11. [Google Scholar] [CrossRef]
- Beg, Q.; Bhushan, B.; Kapoor, M.; Hoondal, G. Production and characterization of thermostable xylanase and pectinase from Streptomyces sp. QG-11-3. J. Ind. Microbiol. Biotechnol. 2000, 24, 396–402. [Google Scholar] [CrossRef]
- Kaur, A.; Singh, A.; Dua, A.; Mahajan, R. Cost-effective and concurrent production of industrially valuable xylano-pectinolytic enzymes by a bacterial isolate Bacillus pumilus AJK. Prep. Biochem. Biotechnol. 2017, 47, 8–18. [Google Scholar] [CrossRef]
- Singh, A.; Varghese, L.M.; Battan, B.; Patra, A.K.; Mandhan, R.P.; Mahajan, R. Eco-friendly scouring of ramie fibers using crude xylano-pectinolytic enzymes for textile purpose. Environ. Sci. Pollut. Res. 2020, 27, 6701–6710. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, P.S.; Udupa, P.M. Isolation, purification and characterization of pectinase enzyme from Streptomyces thermocarboxydus. J. Clin. Microbiol. Biochem. Technol. 2019, 5, 1–6. [Google Scholar] [CrossRef]
- Govindaraji, P.K.; Vuppu, S. Characterisation of pectin and optimization of pectinase enzyme from novel Streptomyces fumigatiscleroticus VIT-SP4 for drug delivery and concrete crack-healing applications: An eco-friendly approach. Saudi J. Biol. Sci. 2020, 27, 3529–3540. [Google Scholar] [CrossRef]
- Sharma, H.P.; Patel, H.; Sharma, S. Enzymatic extraction and clarification of juice from various Fruits. Trends Post Harvest Technol. J. 2016, 2, 1–14. [Google Scholar]
- Mwaurah, P.W.; Kumar, S.; Kumar, N.; Attkan, A.K.; Panghal, A.; Singh, V.K.; Garg, M.K. Novel oil extraction technologies: Process conditions, quality parameters, and optimization. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3–20. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).