Anaerobic Membrane Bioreactor Effluent Reuse: A Review of Microbial Safety Concerns
Abstract
:1. Shifting Paradigms in Wastewater Treatment
1.1. The Anaerobic MBR as an Alternative to Conventional Wastewater Treatment
1.2. The Need to Consider Emerging Contaminants in Wastewater
1.2.1. Non-Microbial Contaminants
1.2.2. Microbial-Associated Contaminants
2. Antibiotic-Associated Microbial Risks and Impact
2.1. The Effect of Anaerobic Digestion on ARB
2.2. The Effect of Anaerobic Digestion on ARGs
2.3. Impact of Antibiotics on The Anaerobic Digestion Process
2.4. The Role of Membrane-Based Treatment on Antibiotic Resistance Reduction
3. Pathogen-Associated Microbial Risks
3.1. Pathogens in Wastewater Treatment
3.2. The Role of Anaerobic Digestion in Pathogen and Indicator Microorganism Removal
3.3. Pathogen-Associated Risk Reduction by Membrane Bioreactors
4. Wastewater Treatment Sustainability: The Role of AnMBRs in Reducing Effluent Reuse Risk
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Haandel, A.; Kato, M.T.; Cavalcanti, P.F.F.; Florencio, L. Anaerobic reactor design concepts for the treatment of domestic wastewater. Rev. Environ. Sci. Bio/Technol. 2006, 5, 21–38. [Google Scholar] [CrossRef]
- Smith, A.L.; Stadler, L.B.; Love, N.G.; Skerlos, S.J.; Raskin, L. Perspectives on anaerobic membrane bioreactor treatment of domestic wastewater: A critical review. Bioresour. Technol. 2012, 122, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Arthurson, V. Proper sanitization of sewage sludge: A critical issue for a sustainable society. Appl. Environ. Microbiol. 2008, 74, 5267–5275. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jia, X.; Gao, B.; Bo, L.; Wang, L. Membrane fouling behavior in anaerobic baffled membrane bioreactor under static operating condition. Bioresour. Technol. 2016, 214, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Aiyuk, S.; Forrez, I.; Lieven de, K.; van Haandel, A.; Verstraete, W. Anaerobic and complementary treatment of domestic sewage in regions with hot climates—A review. Bioresour. Technol. 2006, 97, 2225–2241. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Castro, C.; Lopes, A.R.; Vaz-Moreira, I.; Silva, E.F.; Manaia, C.M.; Nunes, O.C. Wastewater reuse in irrigation: A microbiological perspective on implications in soil fertility and human and environmental health. Environ. Int. 2015, 75, 117–135. [Google Scholar] [CrossRef] [PubMed]
- González, O.; Bayarri, B.; Aceña, J.; Pérez, S.; Barceló, D. Treatment technologies for wastewater reuse: Fate of contaminants of emerging concern. In Advanced Treatment Technologies for Urban Wastewater Reuse; Fatta-Kassinos, D., Dionysiou, D., Kummerer, K., Eds.; Springer: Berlin, Germany, 2015; Volume 45, pp. 5–37. [Google Scholar]
- Grassi, M.; Rizzo, L.; Farina, A. Endocrine disruptors compounds, pharmaceuticals and personal care products in urban wastewater: Implications for agricultural reuse and their removal by adsorption process. Environ. Sci. Pollut. Res. Int. 2013, 20, 3616–3628. [Google Scholar] [CrossRef] [PubMed]
- Gehr, R.; Wagner, M.; Veerasubramanian, P.; Payment, P. Disinfection efficiency of peracetic acid, uv and ozone after enhanced primary treatment of municipal wastewater. Water Res. 2003, 37, 4573–4586. [Google Scholar] [CrossRef]
- Pollice, A.; Lopez, A.; Laera, G.; Rubino, P.; Lonigro, A. Tertiary filtered municipal wastewater as alternative water source in agriculture: A field investigation in southern italy. Sci. Total Environ. 2004, 324, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Watson, K.; Shaw, G.; Leusch, F.; Knight, N. Chlorine disinfection by-products in wastewater effluent: Bioassay-based assessment of toxicological impact. Water Res. 2012, 46, 6069–6083. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.D.; Mitch, W.A. Halonitroalkanes, halonitriles, haloamides, and n-nitrosamines: A critical review of nitrogenous disinfection byproduct formation pathways. Environ. Sci. Technol. 2011, 46, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Bolong, N.; Ismail, A.; Salim, M.R.; Matsuura, T. A review of the effects of emerging contaminants in wastewater and options for their removal. Desalination 2009, 239, 229–246. [Google Scholar] [CrossRef]
- Xu, J.; Xu, Y.; Wang, H.; Guo, C.; Qiu, H.; He, Y.; Zhang, Y.; Li, X.; Meng, W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 2015, 119, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Homem, V.; Santos, L. Degradation and removal methods of antibiotics from aqueous matrices–A review. J. Environ. Manag. 2011, 92, 2304–2347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, B. Occurrence, transformation, and fate of antibiotics in municipal wastewater treatment plants. Crit. Rev. Environ. Sci. Technol. 2011, 41, 951–998. [Google Scholar] [CrossRef]
- Le-Minh, N.; Khan, S.; Drewes, J.; Stuetz, R. Fate of antibiotics during municipal water recycling treatment processes. Water Res. 2010, 44, 4295–4323. [Google Scholar] [CrossRef] [PubMed]
- Stasinakis, A.S. Review on the fate of emerging contaminants during sludge anaerobic digestion. Bioresour. Technol. 2012, 121, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Larsson, D.J.; Amézquita, A.; Collignon, P.; Brandt, K.K.; Graham, D.W.; Lazorchak, J.M.; Suzuki, S.; Silley, P.; Snape, J.R. Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 2013, 121, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Hollender, J.; Zimmermann, S.G.; Koepke, S.; Krauss, M.; McArdell, C.S.; Ort, C.; Singer, H.; von Gunten, U.; Siegrist, H. Elimination of organic micropollutants in a municipal wastewater treatment plant upgraded with a full-scale post-ozonation followed by sand filtration. Environ. Sci. Technol. 2009, 43, 7862–7869. [Google Scholar] [CrossRef] [PubMed]
- Cirja, M.; Ivashechkin, P.; Schäffer, A.; Corvini, P.F. Factors affecting the removal of organic micropollutants from wastewater in conventional treatment plants (CTP) and membrane bioreactors (MBR). Rev. Environ. Sci. Bio/Technol. 2008, 7, 61–78. [Google Scholar] [CrossRef]
- De Wever, H.; Weiss, S.; Reemtsma, T.; Vereecken, J.; Müller, J.; Knepper, T.; Rörden, O.; Gonzalez, S.; Barcelo, D.; Hernando, M.D. Comparison of sulfonated and other micropollutants removal in membrane bioreactor and conventional wastewater treatment. Water Res. 2007, 41, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Radjenović, J.; Petrović, M.; Barceló, D. Fate and distribution of pharmaceuticals in wastewater and sewage sludge of the conventional activated sludge (CAS) and advanced membrane bioreactor (MBR) treatment. Water Res. 2009, 43, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Zuehlke, S.; Duennbier, U.; Lesjean, B.; Gnirss, R.; Buisson, H. Long-term comparison of trace organics removal performances between conventional and membrane activated sludge processes. Water Environ. Res. 2006, 2480–2486. [Google Scholar] [CrossRef]
- Monsalvo, V.M.; McDonald, J.A.; Khan, S.J.; Le-Clech, P. Removal of trace organics by anaerobic membrane bioreactors. Water Res. 2014, 49, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Wijekoon, K.C.; McDonald, J.A.; Khan, S.J.; Hai, F.I.; Price, W.E.; Nghiem, L.D. Development of a predictive framework to assess the removal of trace organic chemicals by anaerobic membrane bioreactor. Bioresour. Technol. 2015, 189, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; Santos, J.L.; Aparicio, I.; Alonso, E. Pharmaceutically active compounds in sludge stabilization treatments: Anaerobic and aerobic digestion, wastewater stabilization ponds and composting. Sci. Total Environ. 2015, 503, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Su, C.; Li, B.; Qian, Y. Treatment of high-strength pharmaceutical wastewater and removal of antibiotics in anaerobic and aerobic biological treatment processes. J. Environ. Eng-Asce 2006, 132, 129–136. [Google Scholar] [CrossRef]
- Michael, I.; Rizzo, L.; McArdell, C.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–995. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Ju, F.; Zhang, T. Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ. Sci. Technol. 2013, 47, 10197–10205. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Burch, T.R.; Sadowsky, M.J.; LaPara, T.M. Fate of antibiotic resistance genes and class 1 integrons in soil microcosms following the application of treated residual municipal wastewater solids. Environ. Sci. Technol. 2014, 48, 5620–5627. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, J.; Chen, P.; Ding, R.; Zhang, P.; Li, X. Occurrence of antibiotics and antibiotic resistances in soils from wastewater irrigation areas in beijing and tianjin, china. Environ. Pollut. 2014, 193, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Jjemba, P.K.; Weinrich, L.A.; Cheng, W.; Giraldo, E.; LeChevallier, M.W. Regrowth of potential opportunistic pathogens and algae in reclaimed-water distribution systems. Appl. Environ. Microbiol. 2010, 76, 4169–4178. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zeng, S.; Gu, A.Z.; He, M.; Shi, H. Inactivation, reactivation and regrowth of indigenous bacteria in reclaimed water after chlorine disinfection of a municipal wastewater treatment plant. J. Environ. Sci. 2013, 25, 1319–1325. [Google Scholar] [CrossRef]
- Giannakis, S.; Darakas, E.; Escalas-Canellas, A.; Pulgarin, C. Elucidating bacterial regrowth: Effect of disinfection conditions in dark storage of solar treated secondary effluent. J. Photochem. Photobiol. A Chem. 2014, 290, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Al-Jassim, N.; Mantilla-Calderon, D.; Wang, T.; Hong, P.Y. Inactivation and gene expression of a virulent wastewater Escherichia coli strain and the nonvirulent commensal Escherichia coli DSM1103 strain upon solar irradiation. Environ. Sci. Technol. 2017, 51, 3649–3659. [Google Scholar] [CrossRef] [PubMed]
- Hai, F.I.; Riley, T.; Shawkat, S.; Magram, S.F.; Yamamoto, K. Removal of pathogens by membrane bioreactors: A review of the mechanisms, influencing factors and reduction in chemical disinfectant dosing. Water 2014, 6, 3603–3630. [Google Scholar] [CrossRef]
- Avery, L.M.; Anchang, K.Y.; Tumwesige, V.; Strachan, N.; Goude, P.J. Potential for pathogen reduction in anaerobic digestion and biogas generation in sub-saharan africa. Biomass Bioenerg. 2014, 70, 112–124. [Google Scholar] [CrossRef]
- Pretel, R.; Shoener, B.; Ferrer, J.; Guest, J. Navigating environmental, economic, and technological trade-offs in the design and operation of submerged anaerobic membrane bioreactors (AnMBRs). Water Res. 2015, 87, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Wegener, H.C. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 2003, 6, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Abdul, P.; Lloyd, D. The survival of antibiotic resistant and sensitive Escherichia coli strains during anaerobic digestion. Appl. Microbiol. Biotechnol. 1985, 22, 373–377. [Google Scholar] [CrossRef]
- Resende, J.A.; Silva, V.L.; de Oliveira, T.L.R.; de Oliveira Fortunato, S.; da Costa Carneiro, J.; Otenio, M.H.; Diniz, C.G. Prevalence and persistence of potentially pathogenic and antibiotic resistant bacteria during anaerobic digestion treatment of cattle manure. Bioresour. Technol. 2014, 153, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Beneragama, N.; Iwasaki, M.; Lateef, S.A.; Yamashiro, T.; Ihara, I.; Umetsu, K. The survival of multidrug-resistant bacteria in thermophilic and mesophilic anaerobic co-digestion of dairy manure and waste milk. Anim. Sci. J. 2013, 84, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Han, I.; Congeevaram, S.; Park, J. Improved control of multiple-antibiotic-resistance-related microbial risk in swine manure wastes by autothermal thermophilic aerobic digestion. Water Sci. Technol. 2009, 59, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H.; Novak, J.T.; Knocke, W.R.; Pruden, A. Survival of antibiotic resistant bacteria and horizontal gene transfer control antibiotic resistance gene content in anaerobic digesters. Front. Microbiol. 2016, 7, 263. [Google Scholar] [CrossRef] [PubMed]
- Mantilla-Calderon, D.; Hong, P.Y. Fate and persistence of a pathogenic NDM-1-positive Escherichia coli strain in anaerobic and aerobic sludge microcosms. Appl. Environ. Microbiol. 2017, 83, e00640-17. [Google Scholar] [CrossRef] [PubMed]
- Dodd, M.C. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment. J. Environ. Monit. 2012, 14, 1754–1771. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 1992, 5, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Ramsden, S.J.; LaPara, T.M. The role of anaerobic digestion in controlling the release of tetracycline resistance genes and class 1 integrons from municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2009, 84, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Diehl, D.L.; LaPara, T.M. Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids. Environ. Sci. Technol. 2010, 44, 9128–9133. [Google Scholar] [CrossRef] [PubMed]
- Sköld, O. Sulfonamide resistance: Mechanisms and trends. Drug Resist. Updat. 2000, 3, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H.; Novak, J.T.; Knocke, W.R.; Young, K.; Hong, Y.; Vikesland, P.J.; Hull, M.S.; Pruden, A. Effect of silver nanoparticles and antibiotics on antibiotic resistance genes in anaerobic digestion. Water Environ. Res. 2013, 85, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wilson, C.A.; Novak, J.T.; Riffat, R.; Aynur, S.; Murthy, S.; Pruden, A. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class i integrons. Environ. Sci. Technol. 2011, 45, 7855–7861. [Google Scholar] [CrossRef] [PubMed]
- Resende, J.A.; Diniz, C.; Silva, V.; Otenio, M.; Bonnafous, A.; Arcuri, P.; Godon, J.J. Dynamics of antibiotic resistance genes and presence of putative pathogens during ambient temperature anaerobic digestion. J. Appl. Microbiol. 2014, 117, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Ince, B.; Ince, O. Assessment of anaerobic bacterial diversity and its effects on anaerobic system stability and the occurrence of antibiotic resistance genes. Bioresour. Technol. 2016, 207, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Harb, M.; Hong, P.Y. Performance and microbial community variations of anaerobic digesters under increasing tetracycline concentrations. Appl. Microbiol. Biotechnol. 2017, 101, 5505–5517. [Google Scholar] [CrossRef] [PubMed]
- McNamara, P.J.; LaPara, T.M.; Novak, P.J. The impacts of triclosan on anaerobic community structures, function, and antimicrobial resistance. Environ. Sci. Technol. 2014, 48, 7393–7440. [Google Scholar] [CrossRef] [PubMed]
- Carey, D.E.; Zitomer, D.; Hristova, K.R.; Kappell, A.D.; McNamara, P.J. Triclocarban influences antibiotic resistance and alters anaerobic digester microbial community structure. Environ. Sci. Technol. 2016, 50, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.F.; Chang, X.; Ye, Y.; Wang, Z.X.; Shao, Y.B.; Shi, W.; Li, X.; Li, J.B. Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfra genes in a plasmid-mediated class 1 integron. Int. J. Antimicrob. Agents. 2011, 37, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Li, B.; Ma, L.; Wang, Y.; Huang, D.; Zhang, T. Antibiotic resistance genes and human bacterial pathogens: Co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res. 2016, 91, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yang, Y.; Pruden, A. Effect of temperature on removal of antibiotic resistance genes by anaerobic digestion of activated sludge revealed by metagenomic approach. Appl. Microbiol. Biotechnol. 2015, 99, 7771–7779. [Google Scholar] [CrossRef] [PubMed]
- Christgen, B.; Yang, Y.; Ahammad, S.; Li, B.; Rodriquez, D.C.; Zhang, T.; Graham, D.W. Metagenomics shows that low-energy anaerobic—Aerobic treatment reactors reduce antibiotic resistance gene levels from domestic wastewater. Environ. Sci. Technol. 2015, 49, 2577–2584. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Cetecioglu, Z.; Arikan, O.; Ince, B.; Ozbayram, E.G.; Ince, O. Inhibitory effects of antibiotic combinations on syntrophic bacteria, homoacetogens and methanogens. Chemosphere 2015, 120, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Ince, B.; Cetecioglu, Z.; Arikan, O.; Ozbayram, E.G.; Shahi, A.; Ince, O. Combined effect of erythromycin, tetracycline and sulfamethoxazole on performance of anaerobic sequencing batch reactors. Bioresour. Technol. 2015, 186, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Ince, B.; Ince, O. Application of real-time PCR to determination of combined effect of antibiotics on bacteria, methanogenic archaea, archaea in anaerobic sequencing batch reactors. Water Res. 2015, 76, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Shahi, A.; Ozbayram, E.G.; Ince, B.; Ince, O. Use of PCR-DGGE based molecular methods to assessment of microbial diversity during anaerobic treatment of antibiotic combinations. Bioresour. Technol. 2015, 192, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Gottschall, N.; Topp, E.; Metcalfe, C.; Edwards, M.; Payne, M.; Kleywegt, S.; Russell, P.; Lapen, D. Pharmaceutical and personal care products in groundwater, subsurface drainage, soil, and wheat grain, following a high single application of municipal biosolids to a field. Chemosphere 2012, 87, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Mao, D.; Mu, Q.; Luo, Y. Fate and proliferation of typical antibiotic resistance genes in five full-scale pharmaceutical wastewater treatment plants. Sci. Total Environ. 2015, 526, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Harb, M.; Wei, C.H.; Wang, N.; Amy, G.; Hong, P.Y. Organic micropollutants in aerobic and anaerobic membrane bioreactors: Changes in microbial communities and gene expression. Bioresour. Technol. 2016, 218, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Jia, R.; Feng, F.; Xie, K.; Li, H.; Jing, D.; Xu, X. Effect of solids retention time on antibiotics removal performance and microbial communities in an A/O-MBR process. Bioresour. Technol. 2012, 106, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Sahar, E.; Messalem, R.; Cikurel, H.; Aharoni, A.; Brenner, A.; Godehardt, M.; Jekel, M.; Ernst, M. Fate of antibiotics in activated sludge followed by ultrafiltration (CAS-UF) and in a membrane bioreactor (MBR). Water Res. 2011, 45, 4827–4836. [Google Scholar] [CrossRef] [PubMed]
- Sipma, J.; Osuna, B.; Collado, N.; Monclús, H.; Ferrero, G.; Comas, J.; Rodriguez-Roda, I. Comparison of removal of pharmaceuticals in MBR and activated sludge systems. Desalination 2010, 250, 653–659. [Google Scholar] [CrossRef]
- Wei, C.H.; Hoppe-Jones, C.; Amy, G.; Leiknes, T. Organic micro-pollutants’ removal via anaerobic membrane bioreactor with ultrafiltration and nanofiltration. J. Water Reuse. Desalination 2015, jwrd2015138. [Google Scholar] [CrossRef]
- Wang, J.; Li, K.; Wei, Y.; Cheng, Y.; Wei, D.; Li, M. Performance and fate of organics in a pilot MBR-NF for treating antibiotic production wastewater with recycling nf concentrate. Chemosphere 2015, 121, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Breazeal, M.V.R.; Novak, J.T.; Vikesland, P.J.; Pruden, A. Effect of wastewater colloids on membrane removal of antibiotic resistance genes. Water Res. 2013, 47, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Skerlos, S.J.; Raskin, L. Membrane biofilm development improves COD removal in anaerobic membrane bioreactor wastewater treatment. Microb. Biotechnol. 2015, 8, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Rysz, M.; Mansfield, W.R.; Fortner, J.D.; Alvarez, P.J. Tetracycline resistance gene maintenance under varying bacterial growth rate, substrate and oxygen availability, and tetracycline concentration. Environ. Sci. Technol. 2013, 47, 6995–7001. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Tallon, P.; Magajna, B.; Lofranco, C.; Leung, K.T. Microbial indicators of faecal contamination in water: A current perspective. Water Air Soil Pollut. 2005, 166, 139–166. [Google Scholar] [CrossRef]
- Gilbride, K.A.; Lee, D.-Y.; Beaudette, L. Molecular techniques in wastewater: Understanding microbial communities, detecting pathogens, and real-time process control. J. Microbiol. Methods 2006, 66, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Girones, R.; Ferrus, M.A.; Alonso, J.L.; Rodriguez-Manzano, J.; Calgua, B.; de Abreu Correˆa, A.; Hundesa, A.; Carratala, A.; Bofill-Mas, S. Molecular detection of pathogens in water—The pros and cons of molecular techniques. Water Res. 2010, 44, 4325–4339. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Fujimura, R.; Agata, K.; Ohta, H. Molecular characterization of viable Legionella spp. In cooling tower water samples by combined use of ethidium monoazide and PCR. Microbes. Environ. 2015, 30, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Camper, A.K. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol. Lett. 2009, 291, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Ju, F.; Zhang, T. Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl. Microbiol. Biotechnol. 2014, 98, 3317–3326. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Zhang, T. Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environ. Sci. Technol. 2013, 47, 5433–5441. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-Y.; Shannon, K.; Beaudette, L.A. Detection of bacterial pathogens in municipal wastewater using an oligonucleotide microarray and real-time quantitative PCR. J. Microbiol. Methods 2006, 65, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Shannon, K.; Lee, D.Y.; Trevors, J.; Beaudette, L. Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci. Total Environ. 2007, 382, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, T. Pathogenic bacteria in sewage treatment plants as revealed by 454 pyrosequencing. Environ. Sci. Technol. 2011, 45, 7173–7179. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.L.; Gattie, D.K. Pathogen risks from applying sewage sludge to land. Int. Sci. Technol. 2002, 36, 286–293. [Google Scholar]
- Brooks, J.P.; McLaughlin, M.R.; Gerba, C.P.; Pepper, I.L. Land application of manure and class b biosolids: An occupational and public quantitative microbial risk assessment. J. Environ. Qual. 2012, 41, 2009–2023. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Stafford, D.; Hughes, D.; Clarkson, J. The reduction of three plant pathogens (fusarium, corynebacterium and globodera) in anaerobic digesters. Agric. Wastes 1983, 6, 1–11. [Google Scholar] [CrossRef]
- Abdul, P.; Lloyd, D. Pathogen survival during anaerobic digestion: Fatty acids inhibit anaerobic growth of escherichia coli. Biotechnol. Lett. 1985, 7, 125–128. [Google Scholar] [CrossRef]
- Horan, N.; Fletcher, L.; Betmal, S.; Wilks, S.; Keevil, C. Die-off of enteric bacterial pathogens during mesophilic anaerobic digestion. Water Res. 2004, 38, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Pepper, I.L.; Brooks, J.P.; Sinclair, R.G.; Gurian, P.L.; Gerba, C.P. Pathogens and indicators in united states class b biosolids: National and historic distributions. J. Environ. Qual. 2010, 39, 2185–2190. [Google Scholar] [CrossRef] [PubMed]
- Sahlström, L. A review of survival of pathogenic bacteria in organic waste used in biogas plants. Bioresour. Technol. 2003, 87, 161–166. [Google Scholar] [CrossRef]
- Smith, S.; Lang, N.; Cheung, K.; Spanoudaki, K. Factors controlling pathogen destruction during anaerobic digestion of biowastes. Waste Manag. 2005, 25, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Oropeza, M.R.; Cabirol, N.; Ortega, S.; Ortiz, L.C.; Noyola, A. Removal of fecal indicator organisms and parasites (fecal coliforms and helminth eggs) from municipal biologic sludge by anaerobic mesophilic and thermophilic digestion. Water Sci. Technol. 2001, 44, 97–101. [Google Scholar]
- Pandey, P.K.; Soupir, M.L. Escherichia coli inactivation kinetics in anaerobic digestion of dairy manure under moderate, mesophilic and thermophilic temperatures. AMB Express 2011, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Watcharasukarn, M.; Kaparaju, P.; Steyer, J.P.; Krogfelt, K.A.; Angelidaki, I. Screening Escherichia coli, Enterococcus faecalis, and Clostridium perfringens as indicator organisms in evaluating pathogen-reducing capacity in biogas plants. Microb. Ecol. 2009, 58, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Ziemba, C.; Peccia, J. Net energy production associated with pathogen inactivation during mesophilic and thermophilic anaerobic digestion of sewage sludge. Water Res. 2011, 45, 4758–4768. [Google Scholar] [CrossRef] [PubMed]
- Burtscher, C.; Wuertz, S. Evaluation of the use of PCR and reverse transcriptase PCR for detection of pathogenic bacteria in biosolids from anaerobic digestors and aerobic composters. Appl. Environ. Microbiol. 2003, 69, 4618–4627. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fu, B.; Wang, Y.; Jiang, Q.; Liu, H. Reactor performance and bacterial pathogen removal in response to sludge retention time in a mesophilic anaerobic digester treating sewage sludge. Bioresour. Technol. 2012, 106, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Viau, E.; Peccia, J. Evaluation of the enterococci indicator in biosolids using culture-based and quantitative PCR assays. Water Res. 2009, 43, 4878–4887. [Google Scholar] [CrossRef] [PubMed]
- Viau, E.; Peccia, J. Survey of wastewater indicators and human pathogen genomes in biosolids produced by class a and class b stabilization treatments. Appl. Environ. Microbiol. 2009, 75, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Bibby, K.; Viau, E.; Peccia, J. Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Res. 2010, 44, 4252–4260. [Google Scholar] [CrossRef] [PubMed]
- Saddoud, A.; Ellouze, M.; Dhouib, A.; Sayadi, S. A comparative study on the anaerobic membrane bioreactor performance during the treatment of domestic wastewaters of various origins. Environ. Technol. 2006, 27, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.; Xagoraraki, I.; Wallace, J.; Bickert, W.; Srinivasan, S.; Rose, J.B. Removal of viruses and indicators by anaerobic membrane bioreactor treating animal waste. J. Environ. Qual. 2009, 38, 1694–1699. [Google Scholar] [CrossRef] [PubMed]
- Jong, J.; Lee, J.; Kim, J.; Hyun, K.; Hwang, T.; Park, J.; Choung, Y. The study of pathogenic microbial communities in graywater using membrane bioreactor. Desalination 2010, 250, 568–572. [Google Scholar] [CrossRef]
- Francy, D.S.; Stelzer, E.A.; Bushon, R.N.; Brady, A.M.; Williston, A.G.; Riddell, K.R.; Borchardt, M.A.; Spencer, S.K.; Gellner, T.M. Comparative effectiveness of membrane bioreactors, conventional secondary treatment, and chlorine and uv disinfection to remove microorganisms from municipal wastewaters. Water Res. 2012, 46, 4164–4178. [Google Scholar] [CrossRef] [PubMed]
- Ottoson, J.; Hansen, A.; Björlenius, B.; Norder, H.; Stenström, T. Removal of viruses, parasitic protozoa and microbial indicators in conventional and membrane processes in a wastewater pilot plant. Water Res. 2006, 40, 1449–1457. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, B.; Fru Ngwa, F.; Deconinck, S.; Verstraete, W. Effluent quality of a conventional activated sludge and a membrane bioreactor system treating hospital wastewater. Environ. Technol. 2006, 27, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Winward, G.P.; Avery, L.M.; Frazer-Williams, R.; Pidou, M.; Jeffrey, P.; Stephenson, T.; Jefferson, B. A study of the microbial quality of grey water and an evaluation of treatment technologies for reuse. Ecol. Eng. 2008, 32, 187–197. [Google Scholar] [CrossRef]
- United States EPA. Guidelines for Water Reuse. Washington, DC 2012, 450. Available online: https://watereuse.org/wp-content/uploads/2015/04/epa-2012-guidelines-for-water-reuse.pdf (accessed on 3 August 2017).
- Nolde, E. Greywater reuse systems for toilet flushing in multi-storey buildings—Over ten years experience in berlin. Urban. Water 2000, 1, 275–284. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, X.C.; Ji, Z.; Xu, L.; Yu, Z. Source identification of bacterial and viral pathogens and their survival/fading in the process of wastewater treatment, reclamation, and environmental reuse. World J. Microb. Biot. 2015, 31, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Monclús, H.; Jofre, J.; Rodriguez-Roda, I.; Comas, J.; Balcázar, J.L. Removal of microbial indicators from municipal wastewater by a membrane bioreactor (MBR). Bioresour. Technol. 2011, 102, 5004–5009. [Google Scholar] [CrossRef] [PubMed]
- Harb, M.; Hong, P.Y. Molecular-based detection of potentially pathogenic bacteria in membrane bioreactor (MBR) systems treating municipal wastewater: A case study. Environ. Sci. Pollut. Res. Int. 2016, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Harb, M.; Hong, P.Y. Characterization of biofoulants illustrates different membrane fouling mechanisms for aerobic and anaerobic membrane bioreactors. Sep. Purif. Technol. 2016, 157, 192–202. [Google Scholar] [CrossRef]
- Van den Akker, B.; Trinh, T.; Coleman, H.M.; Stuetz, R.M.; Le-Clech, P.; Khan, S.J. Validation of a full-scale membrane bioreactor and the impact of membrane cleaning on the removal of microbial indicators. Bioresour. Technol. 2014, 155, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, Z.; Zang, L.; Huang, J.; Wu, Z. Occurrence and fate of potential pathogenic bacteria as revealed by pyrosequencing in a full-scale membrane bioreactor treating restaurant wastewater. RSC Adv. 2015, 5, 24469–24478. [Google Scholar] [CrossRef]
- Zanetti, F.; De Luca, G.; Sacchetti, R. Performance of a full-scale membrane bioreactor system in treating municipal wastewater for reuse purposes. Bioresour. Technol. 2010, 101, 3768–3771. [Google Scholar] [CrossRef] [PubMed]
- De Luca, G.; Sacchetti, R.; Leoni, E.; Zanetti, F. Removal of indicator bacteriophages from municipal wastewater by a full-scale membrane bioreactor and a conventional activated sludge process: Implications to water reuse. Bioresour. Technol. 2013, 129, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Cramer, C.; Mauer, C.; Köster, S.; Schröder, H.F.; Pinnekamp, J. MBR technology: A promising approach for the (pre-) treatment of hospital wastewater. Water Sci. Technol. 2012, 65, 1648–1653. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.; van den Akker, B.; Coleman, H.; Stuetz, R.; Le-Clech, P.; Khan, S. Removal of endocrine disrupting chemicals and microbial indicators by a decentralised membrane bioreactor for water reuse. J. Water Reuse. Desalination 2012, 2, 67–73. [Google Scholar] [CrossRef]
- Zhang, K.; Farahbakhsh, K. Removal of native coliphages and coliform bacteria from municipal wastewater by various wastewater treatment processes: Implications to water reuse. Water Res. 2007, 41, 2816–2824. [Google Scholar] [CrossRef] [PubMed]
- Alaboud, T.M. Membrane bioreactor for wastewater reclamation-pilot plant study in jeddah, saudi arabia. Res. J. Environ. Sci. 2009, 3, 267–277. [Google Scholar] [CrossRef]
- Bolzonella, D.; Fatone, F.; di Fabio, S.; Cecchi, F. Application of membrane bioreactor technology for wastewater treatment and reuse in the mediterranean region: Focusing on removal efficiency of non-conventional pollutants. J. Environ. Manag. 2010, 91, 2424–2431. [Google Scholar] [CrossRef] [PubMed]
- Branch, A.; Trinh, T.; Carvajal, G.; Leslie, G.; Coleman, H.M.; Stuetz, R.M.; Drewes, J.E.; Khan, S.J.; Le-Clech, P. Hazardous events in membrane bioreactors—Part 3: Impacts on microorganism log removal efficiencies. J. Membr. Sci. 2016, 497, 514–523. [Google Scholar] [CrossRef]
- Purnell, S.; Ebdon, J.; Buck, A.; Tupper, M.; Taylor, H. Removal of phages and viral pathogens in a full-scale MBR: Implications for wastewater reuse and potable water. Water Res. 2016, 100, 20–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirani, Z.M.; DeCarolis, J.F.; Adham, S.S.; Jacangelo, J.G. Peak flux performance and microbial removal by selected membrane bioreactor systems. Water Res. 2010, 44, 2431–2440. [Google Scholar] [CrossRef] [PubMed]
- Hirani, Z.M.; DeCarolis, J.F.; Lehman, G.; Adham, S.S.; Jacangelo, J.G. Occurrence and removal of microbial indicators from municipal wastewaters by nine different MBR systems. Water Sci. Technol. 2012, 66, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A. Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ. Sci. Technol. 2014, 48, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Harder, R.; Heimersson, S.; Svanström, M.; Peters, G.M. Including pathogen risk in life cycle assessment of wastewater management. 1. Estimating the burden of disease associated with pathogens. Environ. Sci. Technol. 2014, 48, 9438–9445. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Kurola, J.; Lähde, K.; Kymäläinen, M.; Sinkkonen, A.; Romantschuk, M. Biogas production and methanogenic archaeal community in mesophilic and thermophilic anaerobic co-digestion processes. J. Environ. Manag. 2014, 143, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, J.; Plaza, F.; Garralon, G.; Fdz-Polanco, F.; Peña, M. Long-term operation of a pilot scale anaerobic membrane bioreactor (AnMBR) for the treatment of municipal wastewater under psychrophilic conditions. Bioresour. Technol. 2015, 185, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Skerlos, S.J.; Raskin, L. Psychrophilic anaerobic membrane bioreactor treatment of domestic wastewater. Water Res. 2013, 47, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
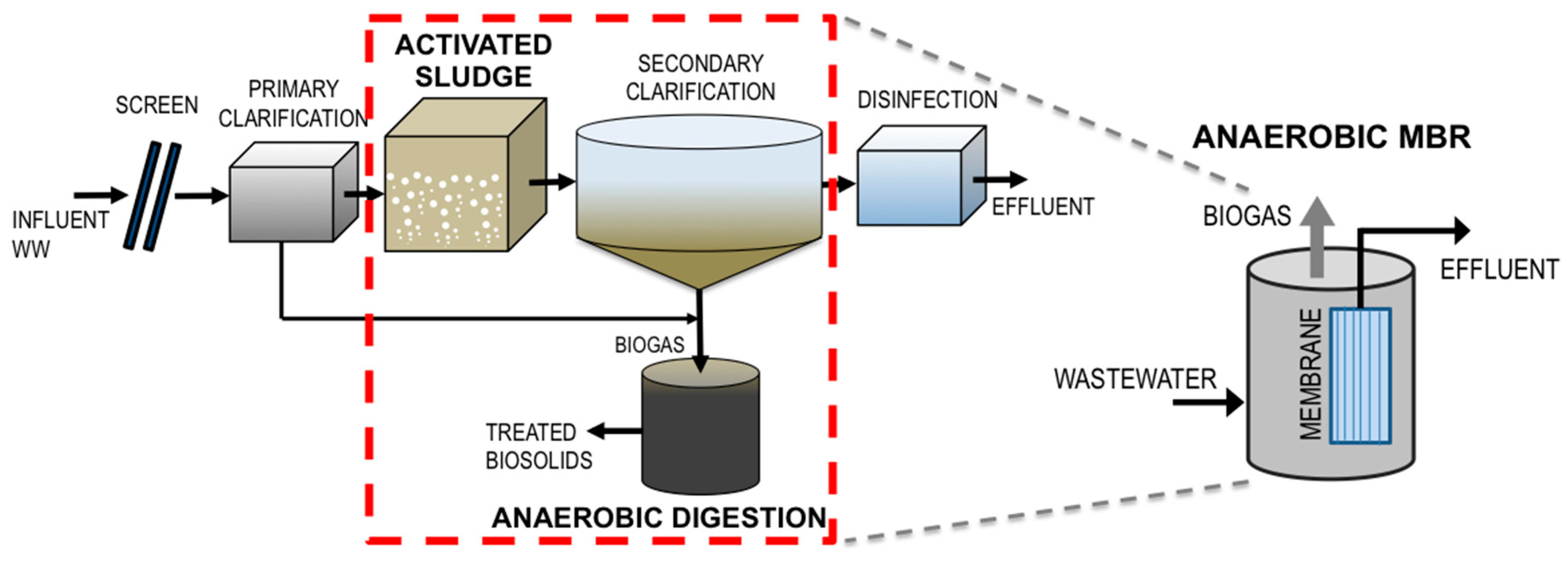
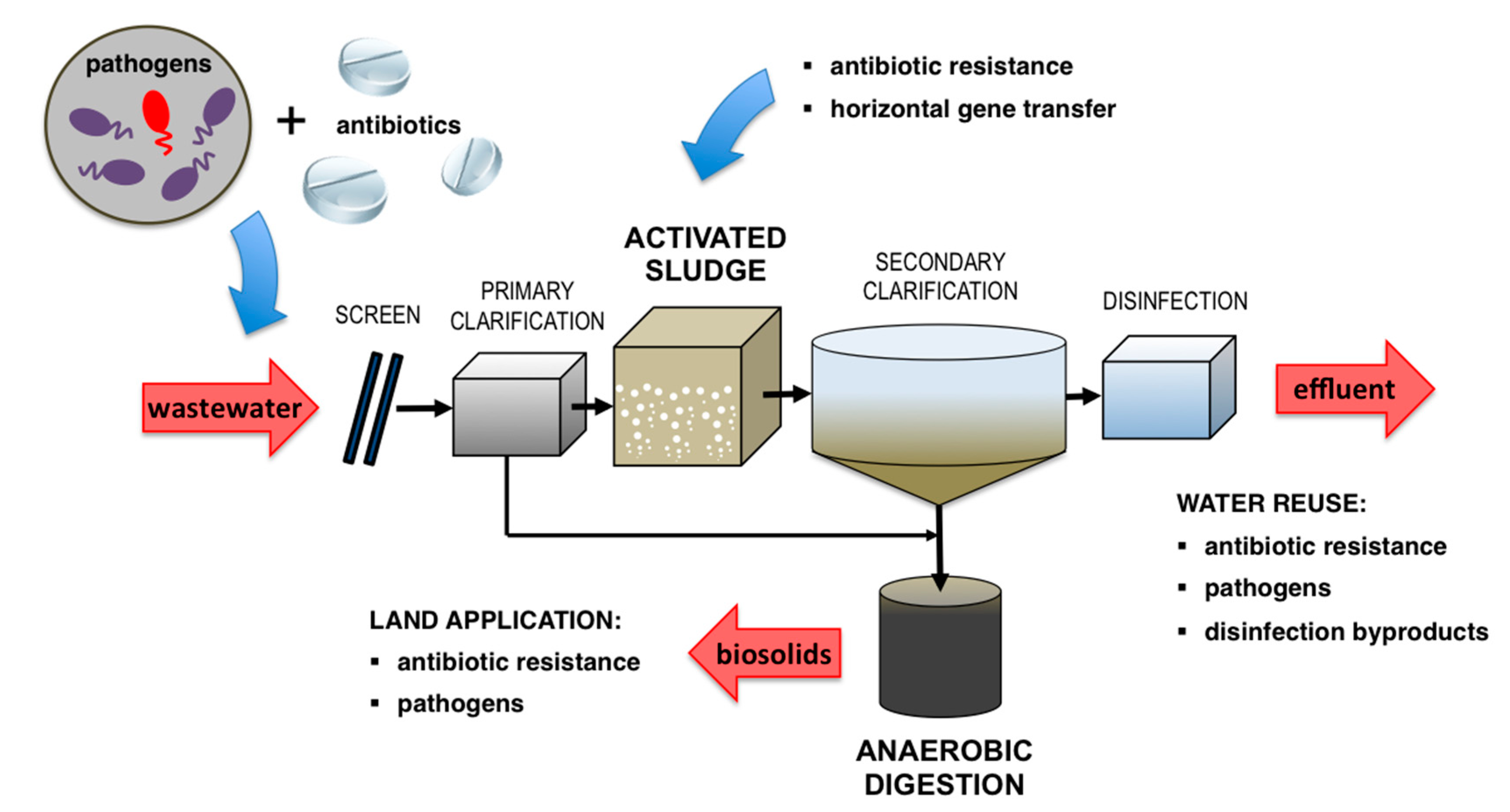
| Treatment Method | Source | Associated Antibiotic | Gene Class | Average Gene Abundance from Source | Average Observed Gene Abundance Post-Treatment | Comments | Ref. | |
|---|---|---|---|---|---|---|---|---|
| Thermophilic-mesophilic AD | WWTP biosolids | tetracycline | tetA | 8 × 10−5–4 × 10−4 (/16S) | 4 × 10−5–6 × 10−4 (/16S) | Thermophilic-mesophilic more effective than mesophilic alone. | [53] | |
| tetO | 4 × 10−6–3 × 10−5 | 1 × 10−6–2 × 10−5 | ||||||
| tetX | 2 × 10−3–1 × 10−2 | 5 × 10−5–3 × 10−4 | ||||||
| class 1 integron | intl1 | 4 × 10−4–2 × 10−3 | 1 × 10−4–2 × 10−4 | |||||
| Thermophilic AD | WWTP biosolids | Tetracycline | tetA | 6.0 × 104 (μL) | 5.0 × 103 (μL) | Gene reduction by anaerobic digestion effective at 37, 47, and 55 °C while aerobic digestion was ineffective. | [54] | |
| tetL | 4.2 × 105 | 1.0 × 104 | ||||||
| tetO | 1.8 × 105 | 2.0 × 104 | ||||||
| tetW | 8.4 × 101 | 1.0 × 101 | ||||||
| tetX | 3.3 × 104 | 1.0 × 103 | ||||||
| class 1 integron | intl1 | 1.0 × 106 | 5.0 × 103 | |||||
| Mesophilic or thermophilic AD | WWTP biosolids | Sulfonamide | sulI | 2 × 10−2–2 × 10−1 (/16S) | 5 × 10−3–4 × 10−2 (/16S) | Thermophilic more effective than mesophilic. sulI correlated with intl1. | [48] | |
| tetracycline | tetO | 5 × 10−4–3 × 10−3 | 1 × 10−4–4 × 10−4 | |||||
| tetW | 5 × 10−4–4 × 10−3 | 1 × 10−4–1 × 10−3 | ||||||
| class 1 integron | intl1 | 1 × 10−3–5 × 10−2 | 1 × 10−3–5 × 10−3 | |||||
| Mesophilic or thermophilic AD | WWTP biosolids | Sulfonamide | sulI | 1 × 1010 (g) | 8 × 108 (thermophilic) | Sul and erm genes more effectively reduced by thermophilic treatment while mesophilic treatment more effectively reduced three tet resistance genes and intl1. | [57] | |
| sulII | 6 × 109 | 7 × 108 (thermophilic) | ||||||
| macrolide | ermB | 1 × 109 | 2 × 108 (thermophilic) | |||||
| ermF | 1 × 109 | 7 × 107 (thermophilic) | ||||||
| tetracycline | tetO | 1 × 109 | 4 × 108 (thermophilic) | |||||
| tetW | 2 × 109 | 3 × 108 (thermophilic) | ||||||
| tetC | 4 × 109 | 5 × 108 (mesophilic) | ||||||
| tetG | 1 × 1010 | 1 × 109 (mesophilic) | ||||||
| tetX | 2 × 108 | 1 × 107 (mesophilic) | ||||||
| class 1 integron | intl1 | 6 × 109 | 5 × 108 (mesophilic) | |||||
| Thermophilic AD | WWTP biosolids | Sulfonamide | sulI | 1 × 107 (μL) | 1 × 106 (μL) | Silver nanoparticles/sulfamethoxazole had no effect on ARG abundance. Tet abundances were higher in mesophilic. | [56] | |
| sulII | 1 × 106 | 1 × 105 | ||||||
| tetracycline | tetO | 1 × 105 | 1 × 104 | |||||
| tetW | 1 × 105 | 3 × 104 | ||||||
| class 1 integron | intl1 | 4 × 106 | 2 × 105 | |||||
| Mesophilic AD | cattle manure | Macrolide | ermB | 106 (g) | 105 (g) | ARG decreases were greater during summer month ambient AD. | [58] | |
| aminoglycoside | aphA2 | 104 | 102–103 | |||||
| beta-lactam | blaTEM-1 | 107 | 104–106 | |||||
| Mesophilic AD | synthetic mixture | Multidrug | mexB | NR | 105–106 (g) | mexB increased with high levels of triclosan and intl1 decreased. | [61] | |
| class 1 integron | intl1 | NR | 10−6–10−2/16S | |||||
| Mesophilic AD | synthetic mixture | Erythromycin | ermA, B, F | NR | 101–103 (mL) | Increasing conc. of erythromycin and tetracycline lead to increases in their associated ARGs. | [59] | |
| ereA | ||||||||
| tetracycline | tetA, B, C, D, E, M, S, Q | NR | 102–104 | |||||
| Mesophilic AD | synthetic sludge | Erythromycin | ermF | NR | 10−5–10−2 (per 16S) | Triclocarban conc. Increases mexB and tetL genes while reducing ermF genes. | [62] | |
| tetracycline | tetL | NR | 10−6–10−2 | |||||
| multidrug | mexB | NR | 10−7–10−4 | |||||
| class 1 integron | intl1 | NR | 10−5–10−4 | |||||
| Mesophilic AD | synthetic mixture | Tetracycline | tetW | NR | 4 × 105–2 × 106 (mg) | Increasing propionate conc. with high tetracycline increased gene abundance. | [60] | |
| tetQ | NR | 2 × 102–8 × 104 | ||||||
| Anaerobic EGSB-aerobic MBR | pharmac-eutical WW | Sulfonamide | sulI | 105 (mL) | 106 (EGSB) | 107 (MBR) | EGSB and anaerobic sludge had the lowest ARG concentrations while aerobic activated sludge had higher abundances. | [73] |
| sulII | 104 | 106 (mL) | 107 (mL) | |||||
| tetracycline | tetM | 106 | 107 | 108 | ||||
| tetO | <103 | 104 | 105 | |||||
| tetQ | 104 | 107 | 107 | |||||
| tetW | 104 | 106 | 107 | |||||
| erythromycin | ermB | 105 | 106 | 107 | ||||
| class 1 integron | intl1 | 107 | 107 | 108 | ||||
| AnMBR AeMBR | synthetic mixture | Sulfonamide | sulI | NR | 10−4 (Ana.) | 10−2 (Aer.) | Lower ARG conc. than aerobic MBR. sulI correlated with intl1 abundance. | [74] |
| sulII | NR | 10−4 (/16S) | 10−2 (/16S) | |||||
| trimethoprim | dfrA | NR | 10−3 | 10−2 | ||||
| class 1 integron | intl1 | NR | 10−4 | 10−3 | ||||
| Anoxic-oxic-MBR | Synthetic mixture | Sulfonamide | sulI | NR | 3 x 107–2 × 108 (mL) | tet genes incr. at lower SRTs, sulI incr. then decr. and sulII decr. with lower SRT. | [75] | |
| sulII | NR | 1 x 107–1 × 109 | ||||||
| tetracycline | tetC | NR | 3 x 106–1 × 107 | |||||
| tetE | NR | 6 x 107–1 × 108 | ||||||
| Aerobic MBR | domestic WW | Sulfonamide | sulI | 108 (100 mL) | 105.5 (100 mL) | MBR had higher LRV of tet genes than other treatment processes. | [76] | |
| tetracycline | tetO | 109 | 102 | |||||
| tetW | 109 | 103 | ||||||
| System Type | Membrane Nominal Pore Size | Wastewater Type | Detection Method | Indicator/Pathogen | Abundance in Influent | LRV | Ref. |
|---|---|---|---|---|---|---|---|
| Aerobic MBR | 0.4 μm | Municipal wastewater | Culture-based plate count | E. coli | N.A. | 5.0 | [116] |
| Enterococci | N.A. | 4.5 | |||||
| C. perfringens | N.A. | 3.0 | |||||
| Aerobic MBR | 0.4 μm | Municipal wastewater | Culture-based plate count | Total coliform | ~108 CFU/100 mL | 6.0 | [127] |
| Fecal coliform | ~107 CFU/100 mL | 6.9 | |||||
| E. coli | ~106.9 CFU/100 mL | 6.7 | |||||
| Enterococci | ~105.9 CFU/100 mL | 5.7 | |||||
| Aerobic MBR | 0.4 μm | Municipal wastewater | Culture-based plate count | Fecal coliform | ~107.8 CFU/100 mL | >6.7 | [115] |
| E. coli | ~107.4 CFU/100 mL | >6.1 | |||||
| Enterococci | ~107.5 CFU/100 mL | >6.3 | |||||
| Aerobic MBR | 0.4 μm | Municipal wastewater | Culture-based plate count | E. coli | ~106.9 CFU/100 mL | 6.8 | [128] |
| Enterococci | ~105.9 CFU/100 mL | 5.7 | |||||
| Aerobic MBR | 0.4 μm | Municipal wastewater | Digital PCR | Total bacteria | ~108.5 copies/L | 4.1 | [123] |
| A. Baumannii | ~106.5 copies/L | 2.7 | |||||
| P. aeruginosa | ~103.8 copies/L | 1.0 | |||||
| K. pneumoniae | ~106 copies/L | 3.1 | |||||
| Aerobic MBR | 0.4 μm | Hospital wastewater | Culture-based plate count | Total coliform | ~108.1 CFU/100 mL | 3.2 | [117] |
| Fecal coliform | ~107.3 CFU/100 mL | 3.6 | |||||
| Enterococci | ~106.2 CFU/100 mL | 3.1 | |||||
| Aerobic MBR | 0.2 μm | Hospital wastewater | Culture-based plate count | E. coli | ~105 CFU/100 mL | 3.8 | [129] |
| Enterococci | ~106 CFU/100 mL | 4.5 | |||||
| Aerobic MBR | 0.2 μm | Municipal wastewater | Culture-based plate count | Total coliform | ~108.2 CFU/100 mL | 5.3 | [130] |
| E. coli | ~106.8 CFU/100 mL | 5.1 | |||||
| Enterococci | ~106.0 CFU/100 mL | 5.0 | |||||
| Clostridia | ~105.9 CFU/100 mL | 4.6 | |||||
| Aerobic MBR | 0.1–0.2 μm | Municipal wastewater | Culture-based plate count | Total coliform | ~108 CFU/100 mL | 5.9 | [125] |
| E. coli | ~106.5 CFU/100 mL | 5.1 | |||||
| Clostridia | ~105.5 CFU/100 mL | 4.9 | |||||
| Aerobic MBR | 0.05 μm | High-strength greywater | Culture-based plate count | Total coliform | ~107.4 CFU/100 mL | >7 | [118] |
| E. coli | ~103.8 CFU/100 mL | N.A. | |||||
| Enterococci | ~102.7 CFU/100 mL | 2.6 | |||||
| Clostridia | ~102.9 CFU/100 mL | 2.8 | |||||
| P. aeruginosa | ~107 CFU/100 mL | 6.5 | |||||
| Aerobic MBR | 0.04 μm | Municipal wastewater | Culture-based plate count | Total coliform | N.A. | 5.5 | [131] |
| Fecal coliform | ~107 CFU/100 mL | 6.5 | |||||
| Aerobic MBR | 0.04 μm | Municipal wastewater | Culture-based plate count | Total coliform | ~107 CFU/100 mL | 5.5 | [132] |
| Fecal coliform | ~104 CFU/100 mL | 2.6 | |||||
| Aerobic MBR | 0.04 μm | Municipal wastewater | Culture-based plate count | Total coliform | ~106 to 108 CFU/100 mL | 4.7–5.1 | [133] |
| E. coli | N.A. | N.D. | |||||
| Aerobic MBR | 0.04 μm | Synthetic wastewater | Culture-based plate count | Total coliform | N.A. | 6.5 | [134] |
| E. coli | N.A. | 5.5 | |||||
| C. perfringens | N.A. | 5.0 | |||||
| Aerobic MBR | 0.04 μm | Municipal wastewater | Culture-based plate count | Fecal coliform | ~107 CFU/100 mL | 6.9 | [135] |
| Enterococci | ~106.4 CFU/100 mL | 6.1 | |||||
| Aerobic MBR | 0.03–0.1 μm | Municipal wastewater | Culture-based plate count | Total coliform | ~107 CFU/100 mL | 5.8–6.9 | [136] |
| Fecal coliform | ~106 CFU/100 mL | 5.4–6.0 | |||||
| Aerobic MBR | 0.03–0.2 μm | Municipal wastewater | Culture-based plate count | Total coliform | ~106 to 108 CFU/100 mL | 5.5–6.7 | [137] |
| Fecal coliform | ~106 to 107 CFU/100 mL | 5.3–6.5 | |||||
| AAO MBR | N.A. | Combined blackwater, greywater and kitchen wastewater | Culture-based plate count Quantitative PCR | Fecal coliform | ~104.8 CFU/100 mL | 6.2 | [121] |
| E. coli | ~103.5 CFU/100 mL | 4.7 | |||||
| Salmonella | ~101.5 CFU/100 mL | 2.3 | |||||
| Shigella | ~100.8 CFU/100 mL | 2.6 | |||||
| AAO MBR | 0.45 μm | Greywater | Culture-based plate count | Fecal coliform | ~104 CFU/100 mL | 0.5 | [114] |
| E. coli | ~104 CFU/100 mL | 0.7 | |||||
| Staphylococcus | ~104 CFU/100 mL | 0.1 | |||||
| Salmonella | ~104 CFU/100 mL | 0.1 | |||||
| AAO MBR | 0.4 μm | Municipal wastewater | Culture-based plate count | E. coli | ~106 CFU/100 mL | 6.1 | [122] |
| Clostridia | ~105.6 CFU/100 mL | 4.8 | |||||
| Anaerobic MBR | 0.3 μm | Municipal wastewater | Digital PCR | Total bacteria | ~108.5 copies/L | 3.1 | [123] |
| A. baumannii | ~106.5 copies/L | 4.2 | |||||
| P. aeruginosa | ~103.8 copies/L | 1.2 | |||||
| K. pneumoniae | ~106 copies/L | 3.9 | |||||
| Anaerobic MBR | 0.03 μm | Dairy wastewater | Culture-based plate count | E. coli | ~107 CFU/100 mL | 6.7 | [113] |
| Enterococci | ~107.9 CFU/100 mL | 7.4 | |||||
| C. perfringens | ~106.5 CFU/100 mL | N.D. | |||||
| Anaerobic MBR | 100 kDa MWCO | Municipal wastewater | Culture-based plate count | Total coliform | ~106.9 CFU/100 mL | N.D. (in 1 mL) | [112] |
| Fecal coliform | ~106.6 CFU/100 mL | N.D. | |||||
| Streptococci | ~105.7 CFU/100 mL | N.D. |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harb, M.; Hong, P.-Y. Anaerobic Membrane Bioreactor Effluent Reuse: A Review of Microbial Safety Concerns. Fermentation 2017, 3, 39. https://doi.org/10.3390/fermentation3030039
Harb M, Hong P-Y. Anaerobic Membrane Bioreactor Effluent Reuse: A Review of Microbial Safety Concerns. Fermentation. 2017; 3(3):39. https://doi.org/10.3390/fermentation3030039
Chicago/Turabian StyleHarb, Moustapha, and Pei-Ying Hong. 2017. "Anaerobic Membrane Bioreactor Effluent Reuse: A Review of Microbial Safety Concerns" Fermentation 3, no. 3: 39. https://doi.org/10.3390/fermentation3030039
APA StyleHarb, M., & Hong, P.-Y. (2017). Anaerobic Membrane Bioreactor Effluent Reuse: A Review of Microbial Safety Concerns. Fermentation, 3(3), 39. https://doi.org/10.3390/fermentation3030039





