1. Introduction
Whole crop corn silage is more nutritious for livestock, has a higher yield and provides more energy at a lower cost than other forages. The fermentation of corn silage is dominated by a variety of microbes that conserve the quality of the corn silage and improve animal performance [
1]. The ensiling process involved in the fermentation of forage materials under anaerobic conditions is predominantly driven by lactic acid bacteria [
2]. LAB can ferment water-soluble carbohydrates (WSCs) into organic acids, particularly lactic acid and marginal levels of acetic acid. The LAB-mediated fermentation process lowers pH via increasing organic acids, especially lactic acid, which inhibits spoilage microorganisms and preserves the nutritional content of forage [
3,
4]. However, undesirable microbes can still proliferate under suboptimal situations, which negatively impacts the silage quality, which leads to reduced animal performance and productivity [
5].
Silage spoilage is primarily caused by undesirable microbial multiplications, which include bacteria, yeast and fungi, which cause nutrient loss, reduced digestibility and mycotoxin contamination, all of which affect silage quality.
Clostridia, enterobacteria, and
bacilli are the main bacterial groups responsible for spoilage.
Clostridium spp. convert lactic acid into butyric acid, leading to an increase in pH, protein degradation, and the production of ammonia, resulting in increased pH and a lower quality of feed and unpalatable silage [
6,
7]. Enterobacteria such as
klebsiella sp. and
Escherichia coli strongly compete with lactic acid bacteria growth in the early stage of fermentation, causing increased pH and protein degradation, whereas
bacillus spp. can contribute to spoilage by producing heat-resistant spores and degrading proteins [
7,
8]. Yeasts, particularly
Candida,
Pichia, and
Saccharomyces spp., are among the first microorganisms to initiate aerobic spoilage and they metabolize lactic acid and residual sugars, leading to increased pH and ethanol production and reduced aerobic stability, which negatively impacts silage palatability [
1,
9]. Molds such as
Aspergillus,
Penicillium, and
Fusarium spp. are major threats when silage is exposed to oxygen, leading to the loss of nutrient content and production of mycotoxins, which affect liver and kidney functions and cause immunosuppression and reproductive disorders in animals [
10,
11]. To mitigate spoilage, effective ensiling practices such as the use of biological inoculants, oxygen barrier films, and proper feedout management are essential to limit microbial growth and preserve silage quality [
2,
12,
13].
Lactic acid bacteria (LAB) have been extensively used in silage production for their potential to accelerate the accumulation of lactic acid, reducing pH, preventing protein degradation, reducing dry matter loss and enhancing animal performance [
5,
14,
15]. Mostly, LAB such as
Lactobacillus plantarum,
Pediococcus pentosaceus,
Lactobacillus buchneri,
Lactococus lactis and
Leuconostoc sp. have been studied for their ability to enhance fermentation, reduce spoilage, and prolong silage stability [
16,
17]. But
Levilactobacillus casei has often not been used in silage production as its effects are not clear. Few studies have shown positive effects of
L. paracasei on silage fermentation such as increased lactic acid and reduced acetic acid levels [
18,
19,
20]. Another study reported that
L. paracasei could not exert beneficial effects on silage fermentation [
21]. This study aims to explore the potential of
Levilactobacillus casei as a biological silage additive in corn silage production, especially in enhancing fermentative metabolites and microbial dynamics. The study objectives include assessing the effects of newly isolated
L. paracasei on fermentative metabolites such as pH, lactic acid, acetic acid, butyric acid, and propionic acid levels, nutrients such as crude protein (CP), acid detergent fiber
(ADF), and bacterial dynamics and to compare the efficiency of
L. paracasei with a commercially available cocktail LAB inoculant to confirm whether
L. paracasei can contribute to improving silage fermentation and prevent nutrient loss. Furthermore, heatmap correlation was used to examine interactions between bacterial communities and treatments or fermentative metabolites.
4. Discussion
This study demonstrated that lactic acid bacteria (LAB) inhibit fungal and bacterial spoilage organisms in corn silage, improving fermentation quality and microbial populations after two months’ storage. The antibacterial and antifungal activity of the isolated strains, especially K-68, K-56, and K-50, were significant against all tested bacteria and fungi. Isolated LAB effectively suppresses fungi such as
Aspergillus sp.,
Penicillium sp., and bacteria such as
Klebsiella sp. and
E. coli. These results aligned with previous studies indicating that LAB can produce various organic acids, hydrogen peroxide and bacteriocins, which exhibit strong antimicrobial properties against spoilage microorganisms [
17,
29]. The maximum zone of inhibition was observed against all tested spoilage microorganisms with K-68. This confirms that this strain possesses strong antibacterial and antifungal properties than other strains. The maximum inhibition zone was observed for all tested spoilage microorganisms with K-68. This confirms that this strain possesses stronger antibacterial and antifungal properties than others. A 16srRNA sequence analysis of the selected strain revealed that it belongs to
Lacticaseibacillus casei.
Silage fermentation preserves forage nutrients. The ensiling method preserves plant nutrients through spontaneous lactic acid production in an anaerobic condition. The ensiling of forage has gained a great deal of attention as it offers ruminants a consistent, reliable and predictable supply of feed. Lactic acid bacteria (LAB) play a key role in the fermentation of forages. LAB can convert water-soluble carbohydrates (WSCs) in plants into lactic acid and acetic acid, resulting in a lower pH of ensiled forages. LAB typically occurs at low levels in plants, not sufficient to cause rapid fermentation. Thus, adding LAB as a biological additive during ensiling is crucial to induce rapid fermentation that increases lactic acid content and reduces acetic acid and butyric acid levels, as well as reducing proteolysis and increasing dry matter recovery [
30,
31]. In this study, corn silage was developed with
Lacticaseibacillus paracasei- K68 (LABK) or cocktail LAB (LABC) and its fermentation efficiency was evaluated after two months. A reduction in pH and higher levels of lactic acid were observed after LABK treatment compared to the control silages. A reduction in pH and an increase in lactic acid are closely related to the preservation of ensiled silage. A rapid acidification process inhibits microbial growth during ensiling, reducing proteolytic degradation and inhibiting undesirable growth [
12]. In general, pH is used to monitor silage quality; 3.8–4.2 is usually considered ideal for ensiling [
32]. For silage with a high moisture content, pH 4.2 is also considered a benchmark for well-conserved silage [
33]. In the present study, the pH was reduced below the desired pH (pH < 4) in all experimental silages. However, silage developed with LABK or LABC significantly lowered the pH of corn silage compared to control silage, suggesting an addition of LAB could have significant impacts on pH reduction.
In silage, lactic acid, and acetic are the main acids, and they are the most common acids produced by
Lactobacillus from WSCs [
9]. Lactic acid is present at high concentrations during fermentation, and it is 10–20 times stronger than other acids [
9] and it reduces silage pH. In this study, silage treated with
Lacticaseibacillus paracasei- K68 (LABK) showed a marked decrease in pH and an increase in lactic acid levels, indicating enhanced lactic acid fermentation. Additionally, cocktail LAB (LABC) also increased fermentation characteristics, though slightly less than LABK and greater than the control silage. The findings demonstrate the effectiveness of LABK in driving the production of lactic acid, which lowers the pH of the surrounding environment and creates unfavorable conditions for spoilage microorganism’s growth. In the control silage, lactic acid production was lower due to a lower number of LABs and a higher number of yeast and mold counts. The lack of LAB dominance over fermentation for corn silage was supported by the microbial enumeration, and silages produced with either LABK or LABC had significantly higher numbers of LABs and lower numbers of yeasts and molds counts compared to the control silage. Notably, LABK-treated silages had the lowest counts of yeast and mold, suggesting its dominant ability to reduce yeast and fungal multiplications in corn silages. The reduction in undesirable microbial activity is essential for extending the shelf life of fermented silage and maintaining its nutritional value for a long time. A higher LAB count in LAB-treated silages also indicates a healthier microbial environment dominated by beneficial bacteria. It has been shown that high acetic acid and butyric acid contents are the primary negative indicators of fermentation quality and reduce dry matter content and energy production during fermentation [
31]. All experimental corn silages contained noticeable levels of acetic acid, but there were no significant differences between treatments. In terms of trend, LABK-treated silages had the lowest level of acetic acid, followed by LABC-treated silages. This indicates that the presence of LAB treatment modulated the level of acetic acid slightly. The negative indicator acid butyric acid was absent in all experimental silages. The levels of propionic acid and isobutyric acid were not significantly changed between treatments (
p < 0.05). LABK- and LABC-treated silage did not differ in the amount of ADF, NDF, and CP compared to the control silage, indicating that neither LABK nor LABC had the ability to degrade the fiber content of corn silage.
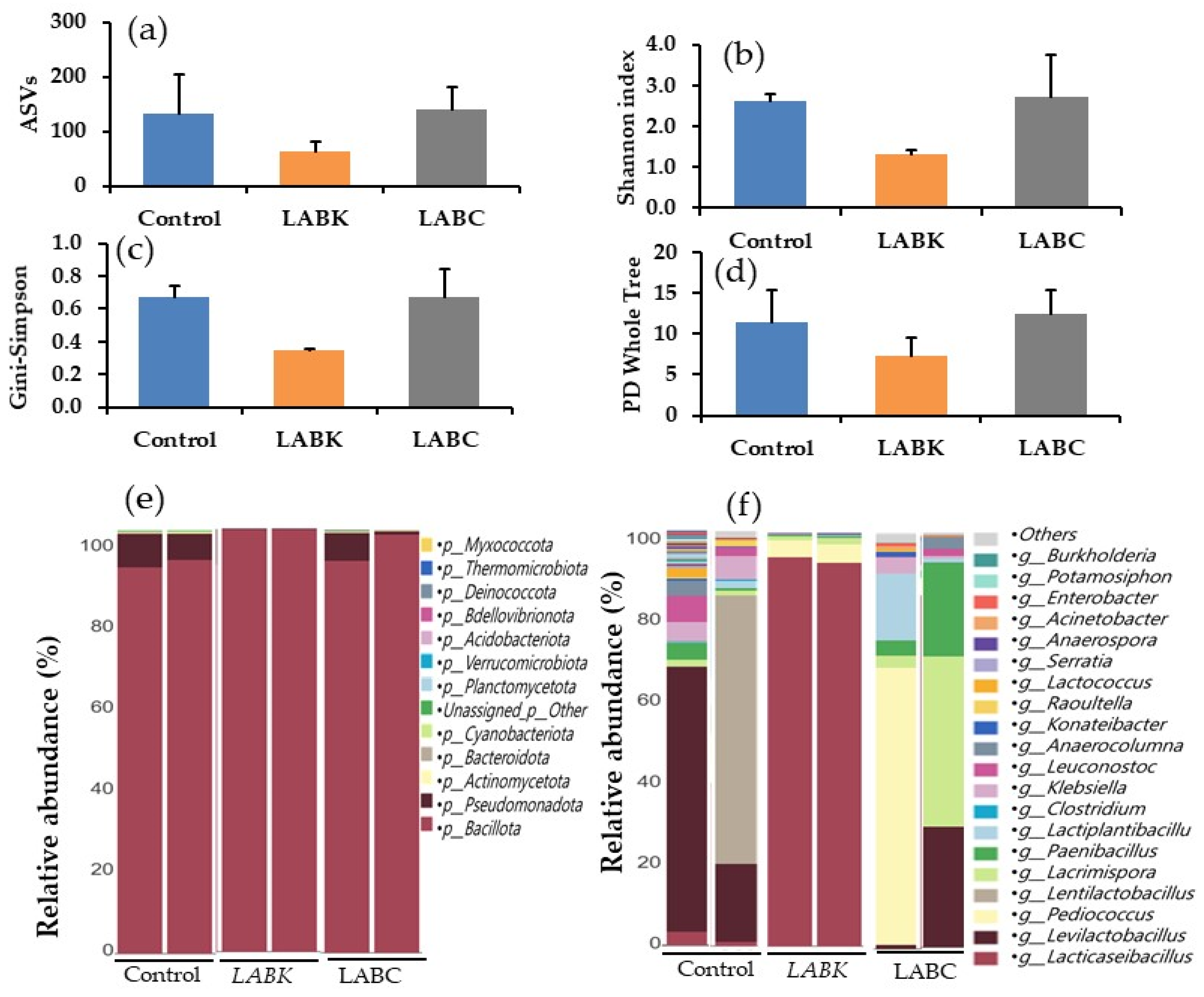
The relative abundance and diversity of bacteria in experimental corn silages were investigated by the Illumina sequencing platform. It is a powerful method used to understand microbial diversity and composition in samples [
34]. Corn silage after 2 months of fermentation under different treatments such as control,
L. paracasei-K68 (LABK), and cocktail LAB (LABC) shows significant differences in microbial diversity and composition that affect silage quality, fermentation stability, and microbial resilience. Alpha diversity indices reveal that the LABC treatment maintained the highest microbial richness and evenness, as shown by the Shannon index, Gini–Simpson index, and PD whole tree value. This suggests that LABC promotes a more diverse and stable microbial community in fermented corn silage. In contrast, LABK significantly reduced microbial diversity (63 ASVs) compared to LABC and the control. The LABK group had the lowest Shannon index. This reduction in diversity indicates that LABK dominates other microbial growth and favors positive fermentation. The majority of bacteria involved in lactic acid fermentation in silage belong to the phylum
Bacillota [
35] and include
Lactobacillus,
Pediococcus,
Lactococcus,
Weissella and
Leuconostoc [
36]. The fermentation quality and microbial composition of corn silage are profoundly affected by lactic acid bacteria inoculants. A phylum-level analysis of bacterial populations revealed significant changes in dominant microbial communities under different treatment conditions such as control, LABK, and cocktail LAB (LABC). These shifts provide insight into the role that inoculation plays in silage fermentation and other bacterial growth suppression.
Bacillota constituted 91.90% of the bacterial population in the control group, while Pseudomonadota, which includes spoilage-associated and opportunistic pathogens, represented 7.55%. In addition, it had significant proportions of Actinomycetota, Bacteroidota, and Cyanobacteriota, which may reflect an undesirable fermentation process. Silage quality could be reduced due to the relatively high percent of Pseudomonadota in the control silages. However, silage treated with LABK or LABC showed higher bacillota dominance, increasing to 99.77% and 96.76%, respectively. Pseudomonadota abundance in silages treated with LABK or LABC dropped sharply to just 0.7% and 2.83%, respectively, compared to the control silage. The results suggest that LABK treatment effectively promotes beneficial fermentative bacteria growth in ensiled corn. The dominance of Bacillota, particularly members of the Lacticaseibacillus genus, is a desirable outcome of because these bacteria are key producers of lactic acid and play a central role in reducing the pH of silage, which inhibits pathogenic and spoilage organisms. Minor phyla such as Actinomycetota, Bacteroidota, and Cyanobacteriota remain present in all treatments but at much lower levels in LAB-treated silages.
Several studies have reported differences in the microbial dynamics in corn silage produced with LAB or without LAB. Xu et al. reported that the addition of
L. buchneri combined with
Saccharomyces cerevisiae did not affect total numbers of yeast and mold communities in corn silage during ensiling and aerobic exposure, whereas lactobacilli abundance in silage treated with
L. acidophilus or
L. plantarum was increased after 45 and 90 days of ensiling [
37]. Another report claimed that silage produced with
L. salivarius and
L. rhamnosus could affect the microbial dynamics throughout the ensiling periods, resulting in a gradual shift in dominant bacterial genera from
Pediococcus to
Lactobacilli [
38].
L. Plantarum treatment modulated the bacterial communities at the early stage of fermentation, but silage produced with
L. buchneri modulated the bacterial dynamics at the later stage of fermentation [
39,
40]. In the present study,
Lacticaseibacillus dominated at the genus level (93.26%), almost completely eliminating other bacterial genera in corn silage treated with
L. paracasei K-68. Hence,
L. paracasei K-68 is highly competitive, outcompeting other microorganisms rapidly and ensuring rapid lactic acid fermentation. The LABC treatment maintained a more balanced bacterial composition, with substantial proportions of
Pediococcus (23.47%),
Lacrimispora (14.81%),
Paenibacillus (9.45%), and
Lacticaseibacillus (28.14%). Control silage had a wider distribution of bacteria, including
Levilactobacillus (28.22%),
Lentilactobacillus (21.60%),
Clostridium (8.00%) and
Klebsiella (5.07%).
Clostridium and
Klebsiella genera include spoilage-associated species that cause butyric acid fermentation, leading to poor silage quality. According to these data,
L. paracasei K-68 inoculum modulated bacterial communities after 2-month fermentation, resulting in rapid shifts in
Levilactobacillus,
Lentilactobacillus,
Clostridium, and
Klebsiella abundances into
Lacticaseibacillus. In addition, LABC treatment also significantly modulates bacterial communities, and this results in shifts in
Levilactobacillus,
Lentilactobacillus,
Clostridium and
Klebsiella into
Lacticaseibacillus Pediococcus and
Lactiplantibacillus.
Species-level distributions provide insight into microbial dynamics. The
L. paracasei K-68 (LABK)-treated silage was almost entirely dominated by
Lacticaseibacillus paracasei (93.25%), demonstrating its ability to outcompete other bacterial species and drive fermentation. On the other hand, LABC treatment showed a more heterogeneous bacterial community, with high proportions of
Pediococcus pentosaceus (23.47%),
Lacrimispora xylanolytica (14.63%), and
Lacticaseibacillus paracasei (28.41%). LABC treatment maintained a balance between efficient lactic acid fermentation and microbial diversity, which may improve fermentation stability and resilience. In the control silage,
Levilactobacillus brevis (28.22%),
Lentilactobacillus hilgardii (21.58%), and
Clostridium beijerinckii (7.87%) were the most abundant bacteria. There is a noticeable presence of
Clostridium beijerinckii, which is linked to butyric acid production, and it negatively affects silage quality and preservation. This highlights that LABK exhibits a more significant effect on the fermentation of corn silage and its bacterial dynamics than cocktail LAB and the control, which prevent the multiplication of spoilage microorganisms. Particularly, most of the 16srRNA sequences belong to
Lacticaseibacillus paracasei (93.25%), evidence that
L. paracasei K-68 outcompetes other bacterial growth and induces positive lactic acid fermentation within a short period of time. The application of
L paracasei K-68 in corn silage production is similar to that of
L Plantarum and differs from
L. Buchneri [
39,
40].
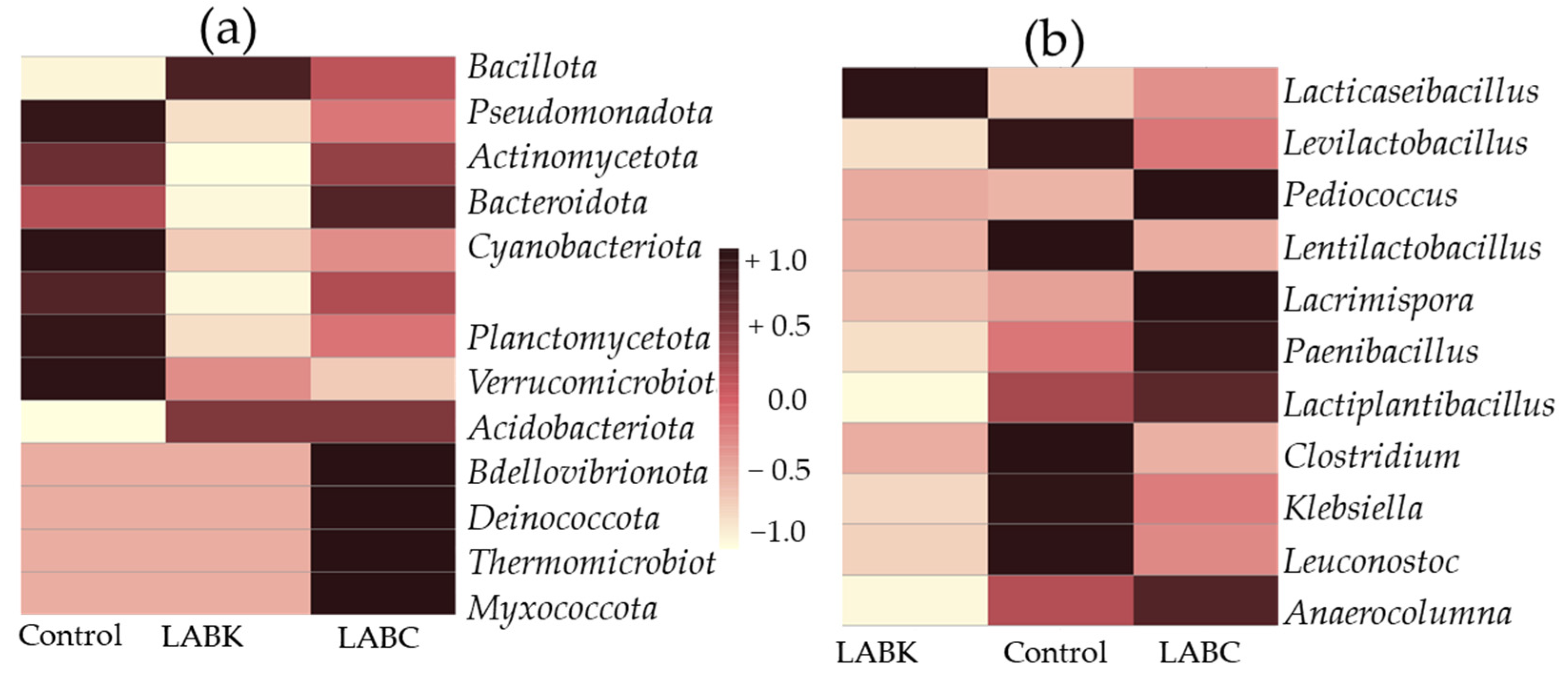
The heatmap analysis provides valuable insights into the relationships between microbial communities and different treatments. The control silage showed a positive correlation with phyla such as Pseudomonadota, Planctomycetota, Verrucomicrobiota, and Actinomycetota and a negative correlation with Bacillota, which includes beneficial lactic acid bacteria. This pattern reflects an unfavorable environment for corn silage fermentation. In contrast, silage inoculated with L. paracasei K-68 showed a strong positive correlation with Bacillota, and a negative correlation with Pseudomonadota, Bacteroidota, and other spoilage-associated phyla. In LABC, there were positive associations with not only Bacillota but also Bdellovibrionota, Deinococcota, and Myxococcota, suggesting complex microbial interactions. At the genus level, control silage was positively associated with spoilage and undesirable microbes, while L. paracasei K-68 showed a strong association with Lacticaseibacillus, a key genus associated with lactic acid production. Pediococcus, Paenibacillus, Lactiplantibacillus, and Lacrimispora were positively associated with the LABC-treated group, indicating a broader range of genera, but still, these are favorable microbial compositions.
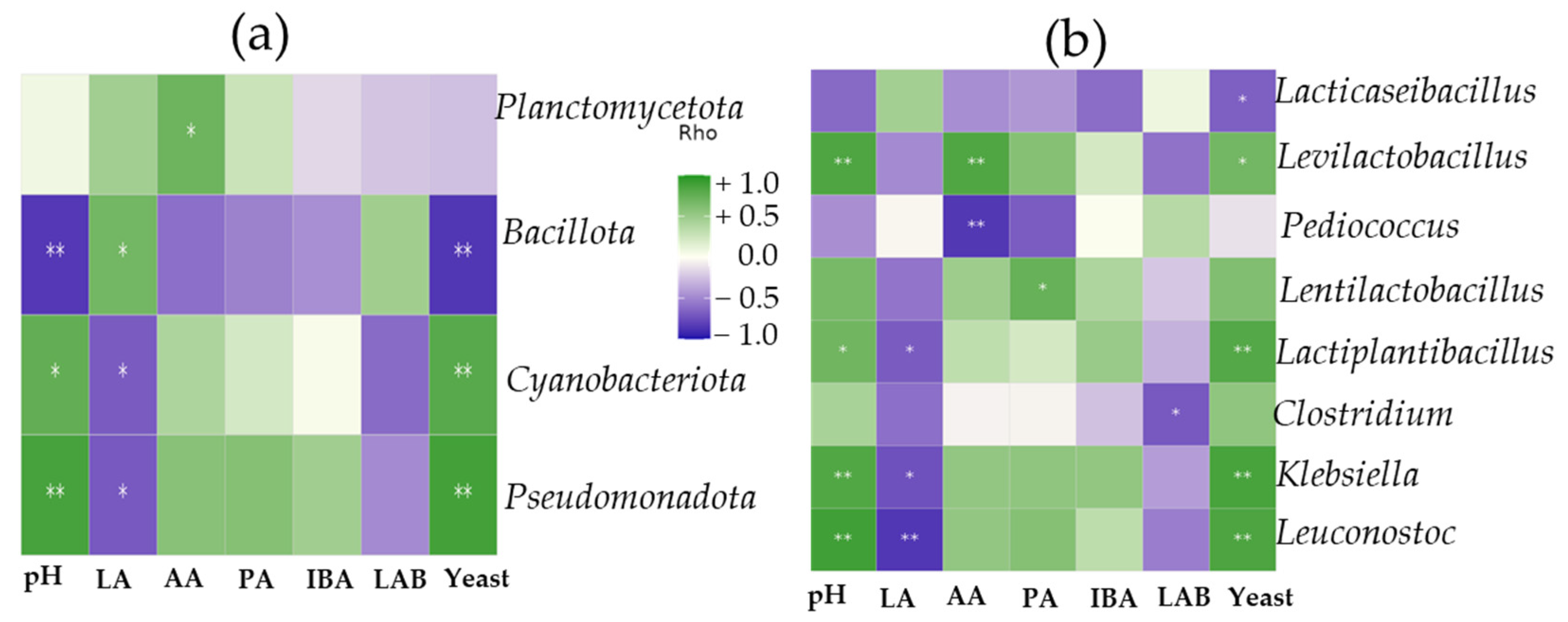
Heatmap correlations between bacterial dynamics and fermentation products further emphasized the role of Bacillota, which was negatively correlated with pH and yeast and positively correlated with lactic acid production. Contrary to this, Pseudomonadota and Cyanobacteriota were negatively associated with lactic acid and positively associated with pH. At the genus level, Lacticaseibacillus showed a strong positive relationship with lactic acid and a negative correlation with pH and yeast growth, which affirms its crucial role in effective silage fermentation and prevents an undesirable fermentation process. Interestingly, Levilactobacillus had a strong positive correlation with acetic acid, while Pediococcus was negatively associated with acetic acid production, indicating functional differences even among beneficial LAB. Importantly, LAB populations in silages were negatively correlated with Clostridium, a harmful genus associated with butyric acid production and silage spoilage. Overall, these findings highlight that targeted inoculation with L. paracasei K-68 not only enhances silage fermentation but also reshapes microbial interactions to favor preservation and feed quality compared to commercial cocktail LAB.