Substrate Gas Utilization and C3/C4 Metabolic Analysis of Actinobacillus succinogenes: Integration into a Model for Fermentation Prediction in BES
Abstract
:1. Introduction
1.1. Microbial Succinate Production
1.2. Metabolic Modeling Approaches
1.3. Effects of Redox Potential
1.4. Substrate and Product Inhibition
2. Materials and Methods
2.1. Growth Conditions
2.2. Analytical Procedures
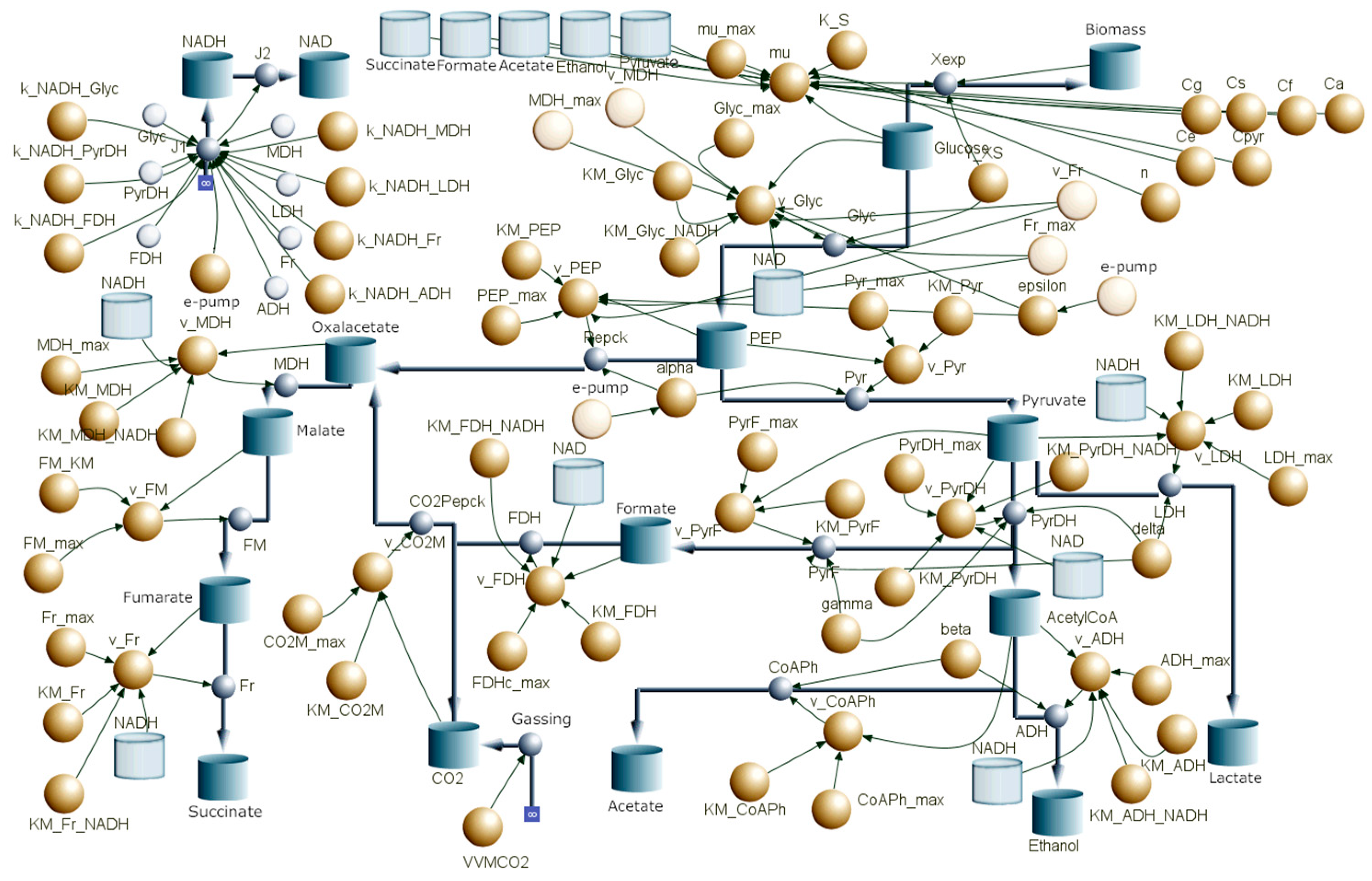
2.3. Modeling
2.4. Calculation
- : Spcific growth rate;
- max: Maximum specific growth rate;
- Substrate concentration;
- : Critical substrate concentration at which inhibition occurs;
- : Monod half-saturation constant.
- : Specific growth rate;
- max: Maximum specific growth rate;
- Substrate concentration;
- : Critical substrate concentration at which inhibition occurs;
- : Monod half-saturation constant;
- Product concentration of product i;
- c*pi: Critical Product concentration of product i at which growth is fully inhibited.
3. Results and Discussion
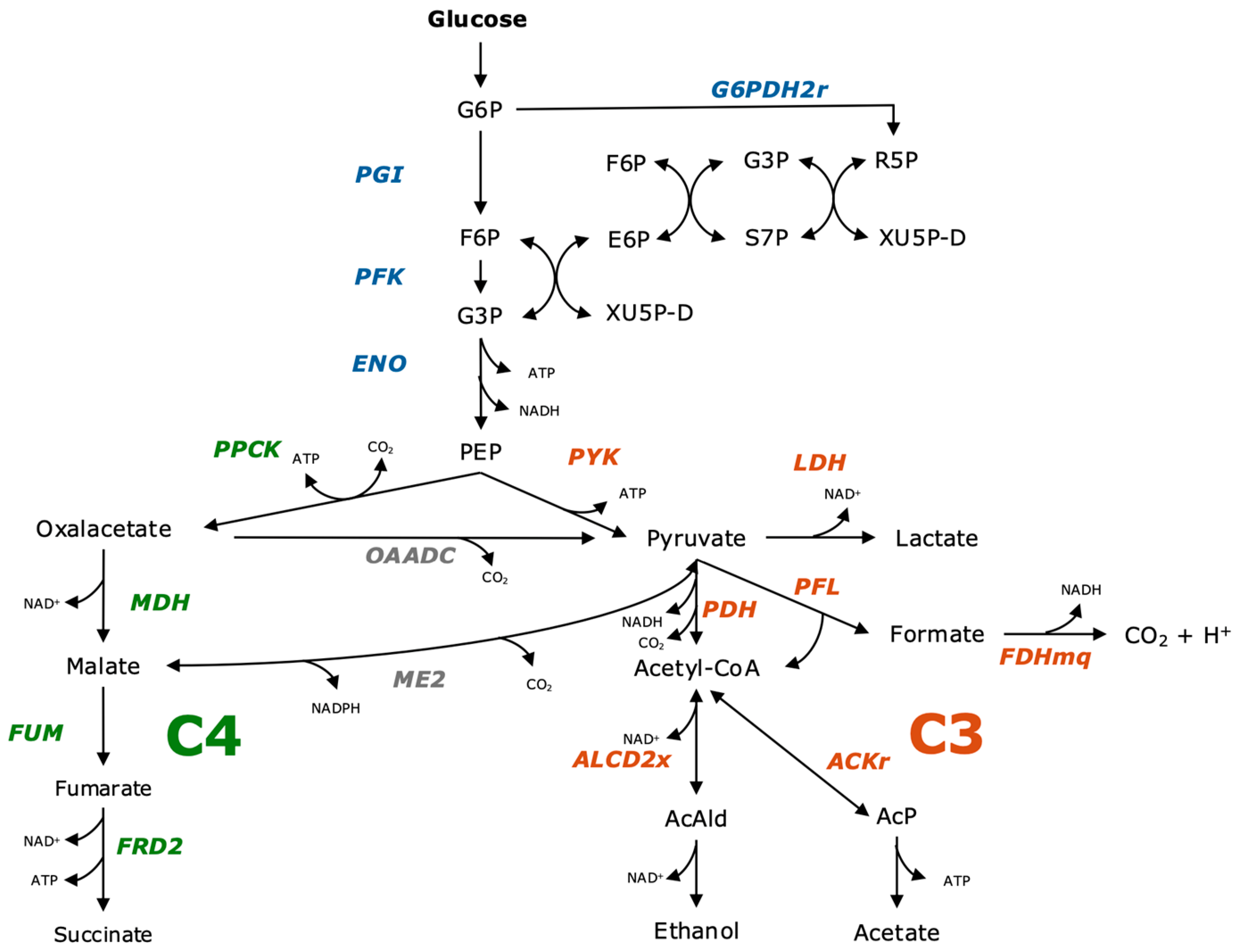
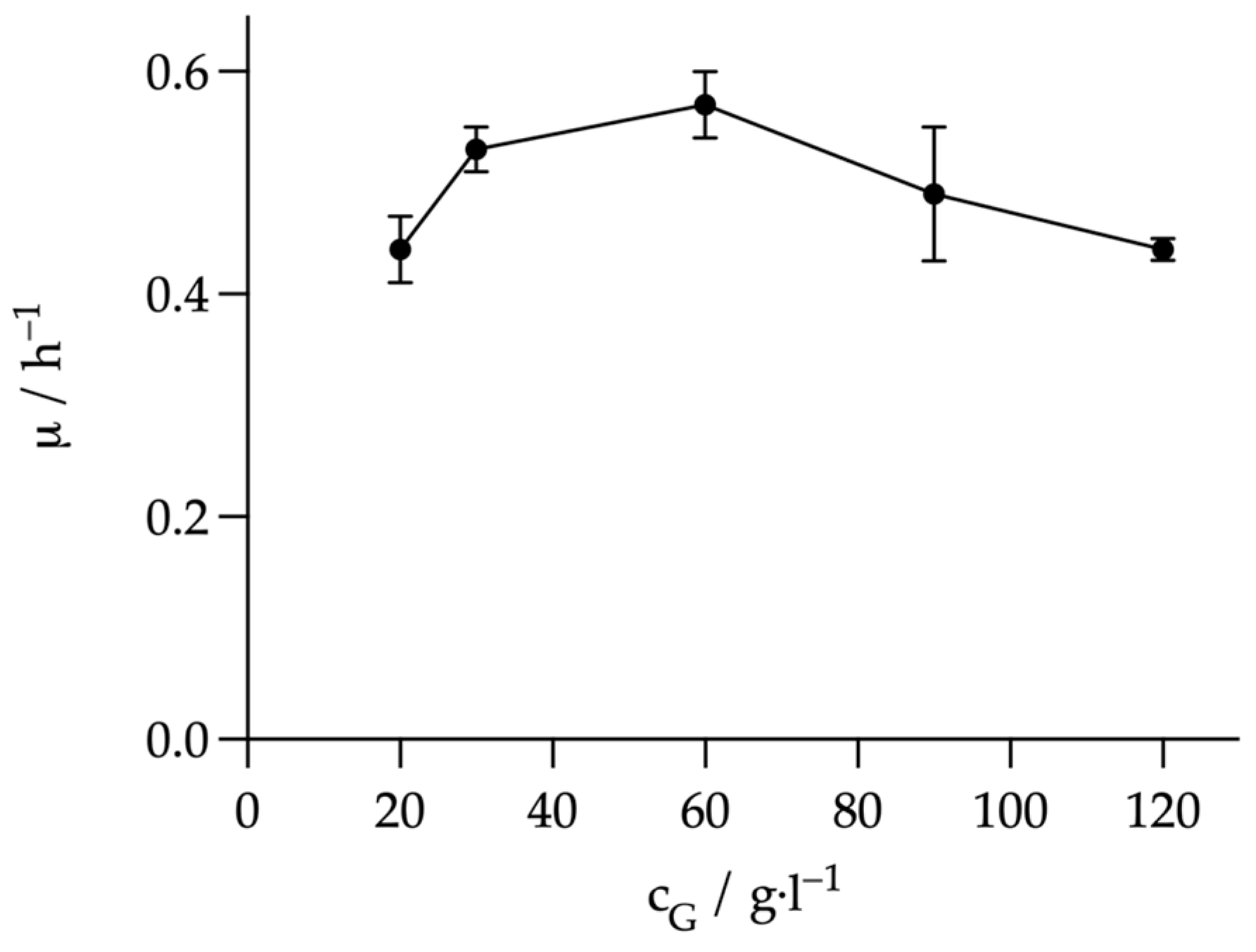
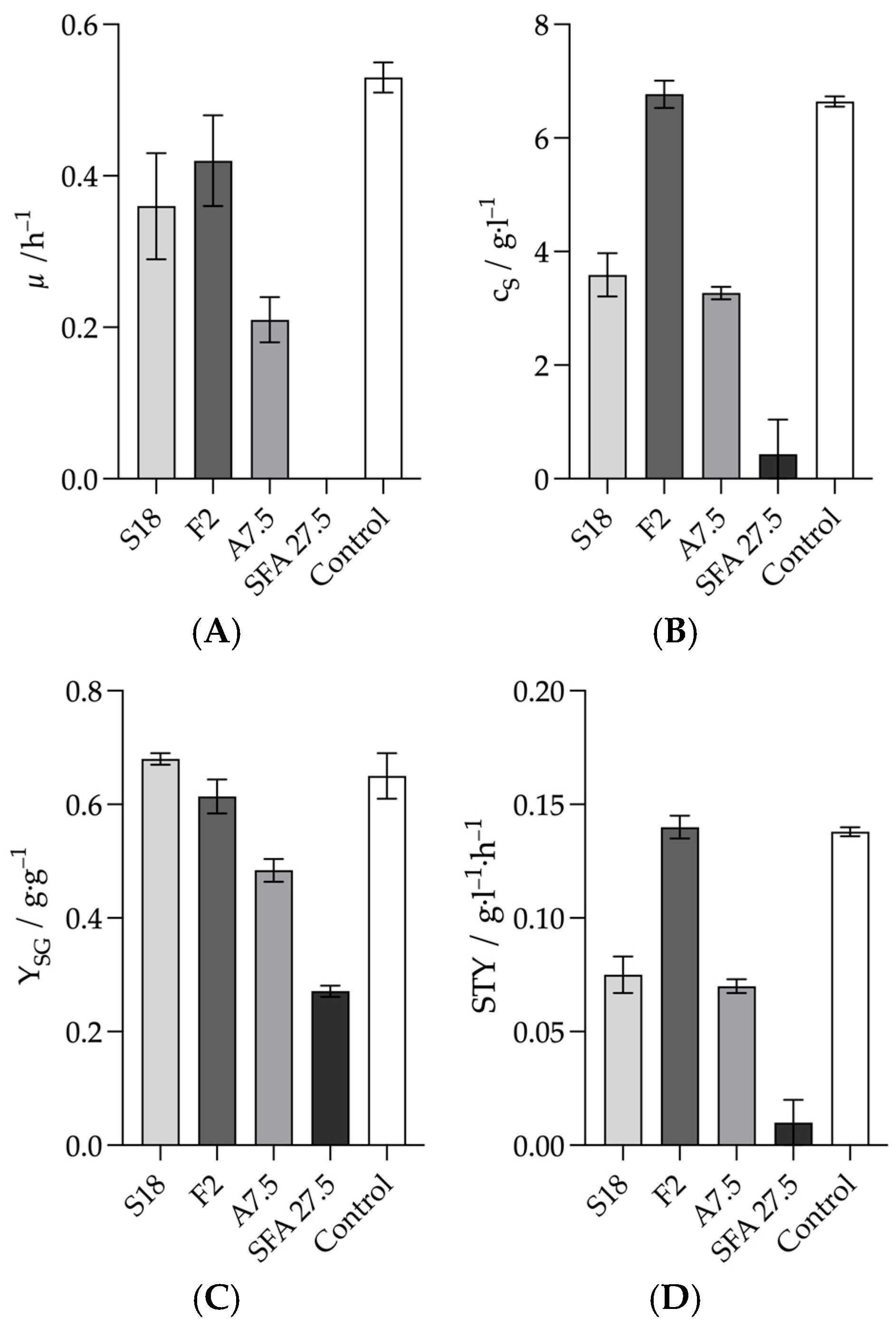
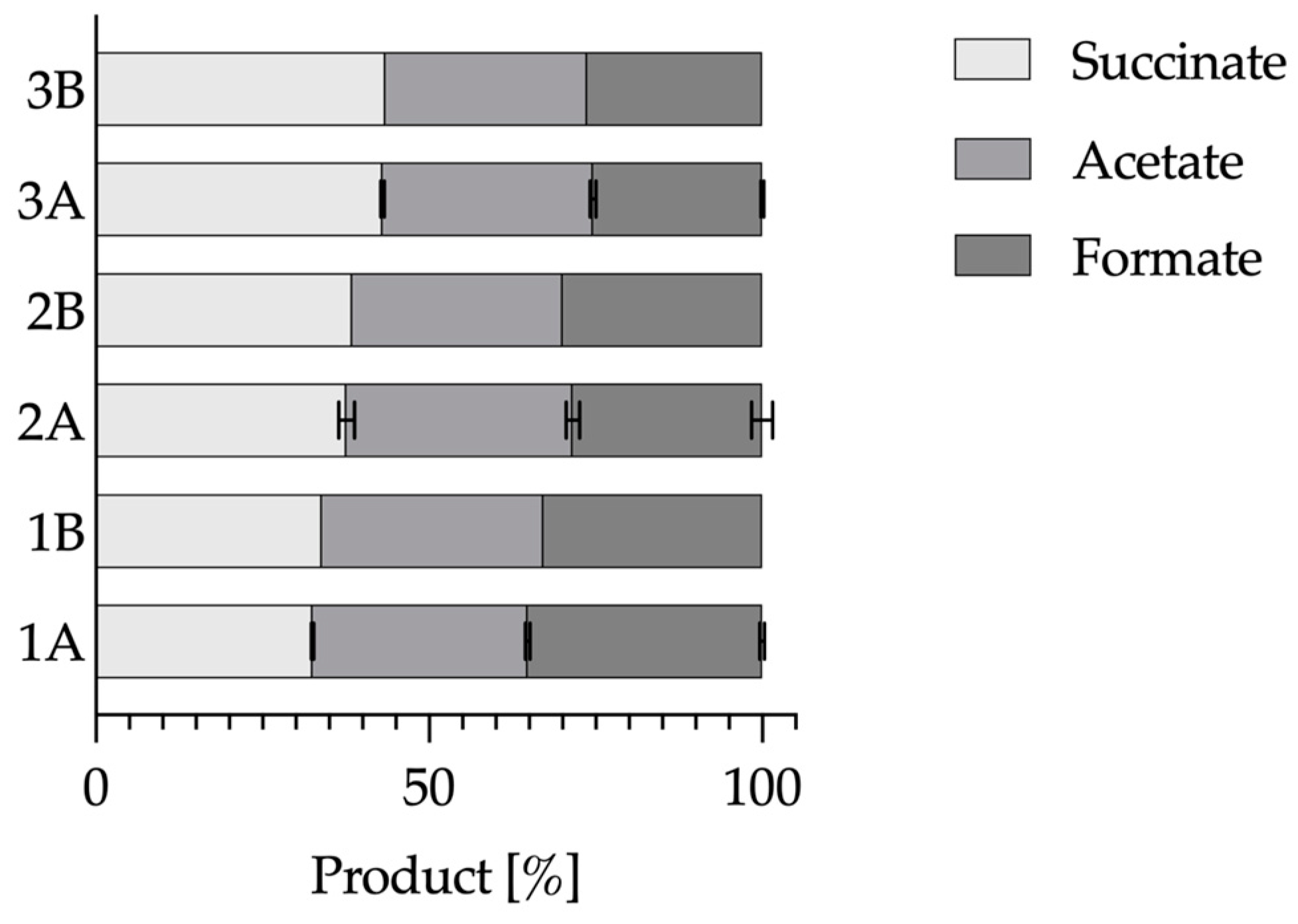
3.1. Influence of Inhibiting Substances on the Metabolism
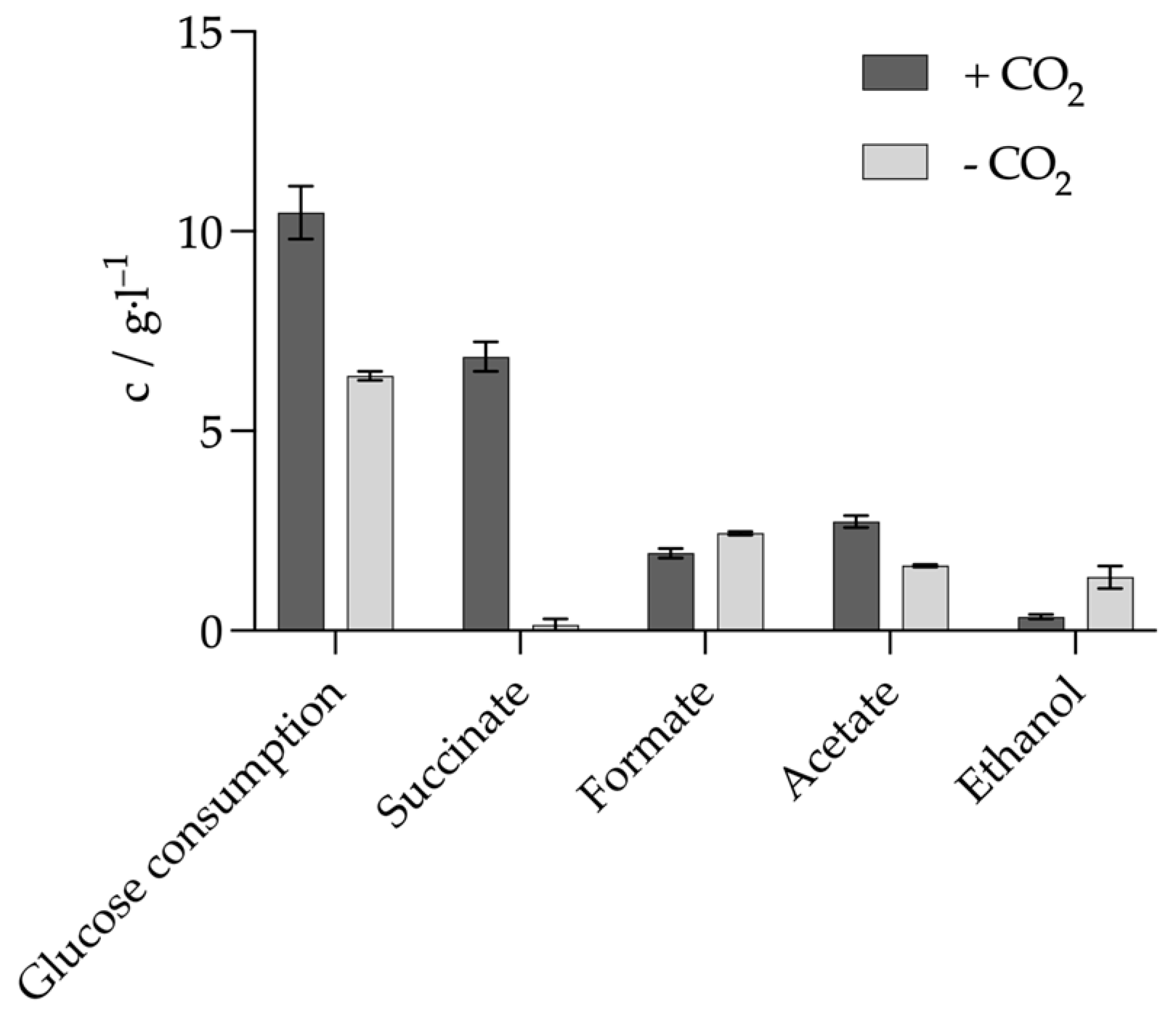
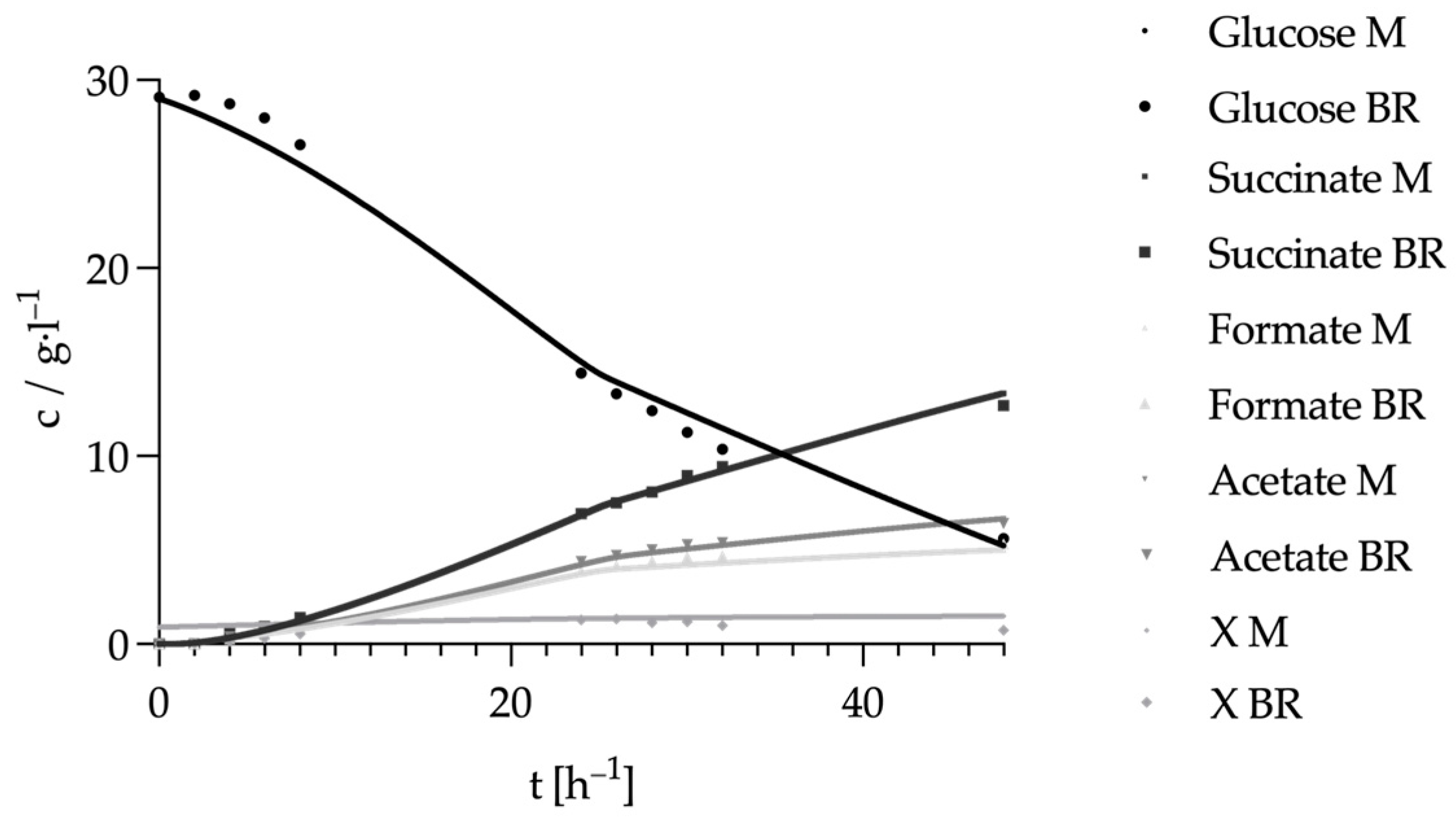
3.2. Alignment of the Metabolism in the Absence of a CO2 Source
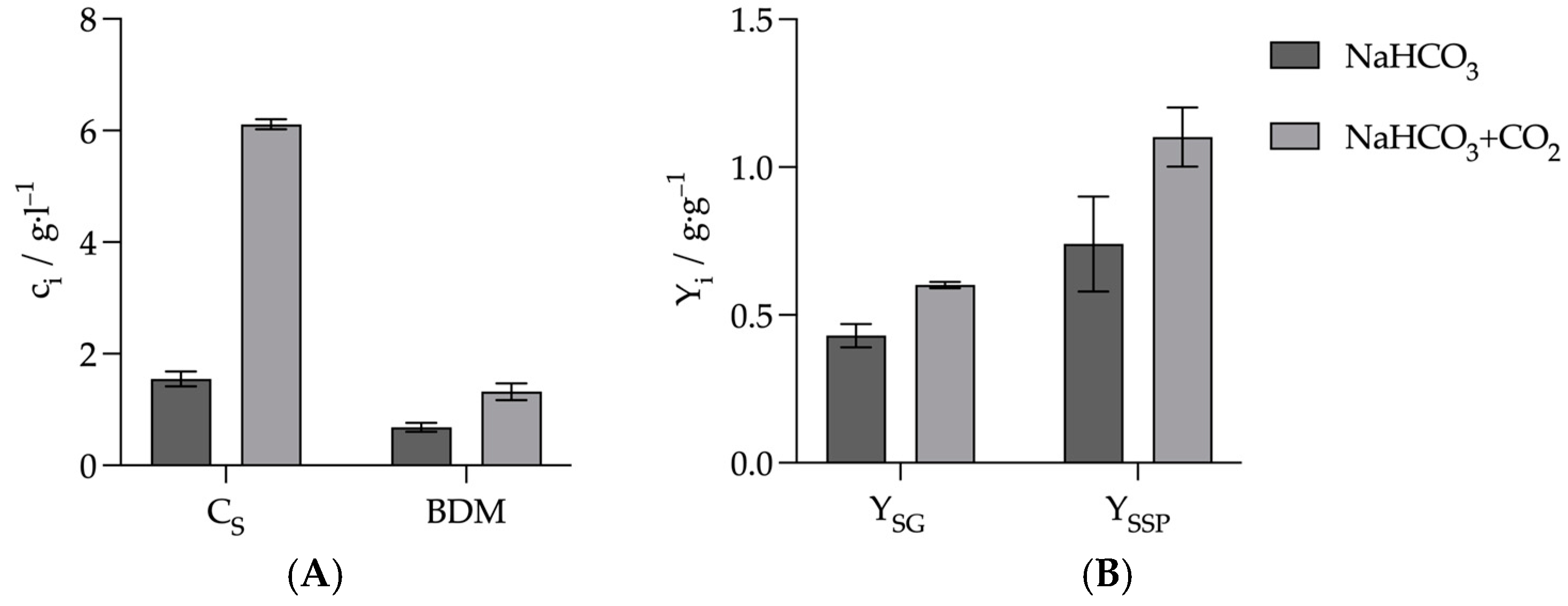
3.3. Optimization of Succinate Production Through the Combination of Sodium Hydrogen Carbonate and Gaseous CO2
3.4. Modeling the Metabolism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meynial-Salles, I.; Dorotyn, S.; Soucaille, P. A New Process for the Continuous Production of Succinic Acid from Glucose at High Yield, Titer, and Productivity. Biotech. Bioeng. 2008, 99, 129–135. [Google Scholar] [CrossRef]
- Dickson, R.; Mancini, E.; Garg, N.; Woodley, J.M.; Gernaey, K.V.; Pinelo, M.; Liu, J.; Mansouri, S.S. Sustainable Bio-Succinic Acid Production: Superstructure Optimization, Techno-Economic, and Lifecycle Assessment. Energy Environ. Sci. 2021, 14, 3542–3558. [Google Scholar] [CrossRef]
- Saxena, R.K.; Saran, S.; Isar, J.; Kaushik, R. Production and Applications of Succinic Acid. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2017; pp. 601–630. ISBN 978-0-444-63662-1. [Google Scholar]
- Zhu, L.-W.; Wang, C.-C.; Liu, R.-S.; Li, H.-M.; Wan, D.-J.; Tang, Y.-J. Actinobacillus succinogenes ATCC 55618 Fermentation Medium Optimization for the Production of Succinic Acid by Response Surface Methodology. J. Biomed. Biotechnol. 2012, 2012, 626137. [Google Scholar] [CrossRef]
- Stylianou, E.; Pateraki, C.; Ladakis, D.; Vlysidis, A.; Koutinas, A. Optimization of Fermentation Medium for Succinic Acid Production Using Basfia Succiniciproducens. Environ. Technol. Innov. 2021, 24, 101914. [Google Scholar] [CrossRef]
- Song, H.; Lee, J.W.; Choi, S.; You, J.K.; Hong, W.H.; Lee, S.Y. Effects of Dissolved CO2 Levels on the Growth ofMannheimia Succiniciproducens and Succinic Acid Production. Biotechnol. Bioeng. 2007, 98, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Raab, A.M.; Gebhardt, G.; Bolotina, N.; Weuster-Botz, D.; Lang, C. Metabolic Engineering of Saccharomyces Cerevisiae for the Biotechnological Production of Succinic Acid. Metab. Eng. 2010, 12, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhu, J.; Zhu, X.; Song, M.; Zhang, T.; Xin, F.; Dong, W.; Ma, J.; Jiang, M. Expression of Global Regulator IrrE for Improved Succinate Production under High Salt Stress by Escherichia coli. Bioresour. Technol. 2018, 254, 151–156. [Google Scholar] [CrossRef]
- Nghiem, N.P.; Davison, B.H.; Suttle, B.E.; Richardson, G.R. Production of Succinic Acid by Anaerobiospirillum succiniciproducens. Appl. Biochem. Biotechnol. 1997, 63–65, 565–576. [Google Scholar] [CrossRef]
- Salma, A.; Djelal, H.; Abdallah, R.; Fourcade, F.; Amrane, A. Well Knowledge of the Physiology of Actinobacillus succinogenes to Improve Succinic Acid Production. Appl. Microbiol. 2021, 1, 304–328. [Google Scholar] [CrossRef]
- Tix, J.; Gotthardt, L.; Bode, J.; Karabacak, B.; Nordmann, J.; Hengsbach, J.-N.; Ulber, R.; Tippkötter, N. Enhancement of Succinic Acid Production by Actinobacillus succinogenes in an Electro-Bioreactor. Fermentation 2024, 10, 504. [Google Scholar] [CrossRef]
- Dessie, W.; Xin, F.; Zhang, W.; Jiang, Y.; Wu, H.; Ma, J.; Jiang, M. Opportunities, Challenges, and Future Perspectives of Succinic Acid Production by Actinobacillus succinogenes. Appl. Microbiol. Biotechnol. 2018, 102, 9893–9910. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Miguel, J.; Vilaça, P.; Soares, S.; Rocha, I.; Carneiro, S. Reconstruction of a Genome-Scale Metabolic Model for Actinobacillus succinogenes 130Z. BMC Syst. Biol. 2018, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, D.; Song, Z.; Zhou, W.; Wu, Y.; Xing, J.; Su, Z. Dual-Phase Fermentation Enables Actinobacillus succinogenes 130ZT to Be a Potential Role for High-Level Lactate Production from the Bioresource. Bioresour. Technol. 2010, 101, 7665–7667. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Laivenieks, M.; Schindler, B.D.; McKinlay, A.A.; Siddaramappa, S.; Challacombe, J.F.; Lowry, S.R.; Clum, A.; Lapidus, A.L.; Burkhart, K.B.; et al. A Genomic Perspective on the Potential of Actinobacillus succinogenes for Industrial Succinate Production. BMC Genom. 2010, 11, 680. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Shachar-Hill, Y.; Zeikus, J.G.; Vieille, C. Determining Actinobacillus succinogenes Metabolic Pathways and Fluxes by NMR and GC-MS Analyses of 13C-Labeled Metabolic Product Isotopomers. Metab. Eng. 2007, 9, 177–192. [Google Scholar] [CrossRef]
- Wu, J.; Li, Y.; Yin, J.; Wang, C.; Qi, X.; Zhou, Y.; Liu, H.; Wu, P.; Zhang, J. Mutation Breeding of High-Stress Resistant Strains for Succinic Acid Production from Corn Straw. Appl. Microbiol. Biotechnol. 2024, 108, 278. [Google Scholar] [CrossRef] [PubMed]
- Almqvist, H.; Pateraki, C.; Alexandri, M.; Koutinas, A.; Lidén, G. Succinic Acid Production by Actinobacillus succinogenes from Batch Fermentation of Mixed Sugars. J. Ind. Microbiol. Biotechnol. 2016, 43, 1117–1130. [Google Scholar] [CrossRef]
- Kádár, Z.; Fonseca, C. Bio-Products from Sugar-Based Fermentation Processes. In Biorefinery; Bastidas-Oyanedel, J.-R., Schmidt, J.E., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 281–312. ISBN 978-3-030-10960-8. [Google Scholar]
- Bradfield, M.F.A.; Nicol, W. Continuous Succinic Acid Production by Actinobacillus succinogenes in a Biofilm Reactor: Steady-State Metabolic Flux Variation. Biochem. Eng. J. 2014, 85, 1–7. [Google Scholar] [CrossRef]
- Li, J.; Jiang, M.; Chen, K.; Shang, L.; Wei, P.; Ying, H.; Ye, Q.; Ouyang, P.; Chang, H. Enhanced Production of Succinic Acid by Actinobacillus succinogenes with Reductive Carbon Source. Process Biochem. 2010, 45, 980–985. [Google Scholar] [CrossRef]
- Quraishi, M.; Wani, K.; Pandit, S.; Gupta, P.K.; Rai, A.K.; Lahiri, D.; Jadhav, D.A.; Ray, R.R.; Jung, S.P.; Thakur, V.K.; et al. Valorisation of CO2 into Value-Added Products via Microbial Electrosynthesis (MES) and Electro-Fermentation Technology. Fermentation 2021, 7, 291. [Google Scholar] [CrossRef]
- Li, J.; Cheng, G.; Dong, S. Direct Electron Transfer to Cytochrome c Oxidase in Self-Assembled Monolayers on Gold Electrodes. J. Electroanal. Chem. 1996, 416, 97–104. [Google Scholar] [CrossRef]
- Gong, Z.; Yu, H.; Zhang, J.; Li, F.; Song, H. Microbial Electro-Fermentation for Synthesis of Chemicals and Biofuels Driven by Bi-Directional Extracellular Electron Transfer. Synth. Syst. Biotechnol. 2020, 5, 304–313. [Google Scholar] [CrossRef]
- Lin, S.K.C.; Du, C.; Koutinas, A.; Wang, R.; Webb, C. Substrate and Product Inhibition Kinetics in Succinic Acid Production by Actinobacillus succinogenes. Biochem. Eng. J. 2008, 41, 128–135. [Google Scholar] [CrossRef]
- Li, Q.; Wang, D.; Wu, Y.; Yang, M.; Li, W.; Xing, J.; Su, Z. Kinetic Evaluation of Products Inhibition to Succinic Acid Producers Escherichia Coli NZN111, AFP111, BL21, and Actinobacillus succinogenes 130ZT. J. Microbiol. 2010, 48, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Corona-González, R.I.; Bories, A.; González-Álvarez, V.; Pelayo-Ortiz, C. Kinetic Study of Succinic Acid Production by Actinobacillus succinogenes ZT-130. Process Biochem. 2008, 43, 1047–1053. [Google Scholar] [CrossRef]
- 545: TRYPTONE SOYA BROTH (TSB). Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium545.pdf (accessed on 3 July 2024).
- Wang, Z.; Li, H.; Feng, J.; Zhang, A.; Ying, H.; He, X.; Jiang, M.; Chen, K.; Ouyang, P. Enhanced Succinic Acid Production from Polyacrylamide-pretreated Cane Molasses in Microbial Electrolysis Cells. J. Chem. Tech. Biotech. 2018, 93, 855–860. [Google Scholar] [CrossRef]
- Tix, J.; Moll, F.; Krafft, S.; Betsch, M.; Tippkötter, N. Hydrogen Production from Enzymatic Pretreated Organic Waste with Thermotoga Neapolitana. Energies 2024, 17, 2938. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, Q.; Wu, M.; Liu, H.; Zhou, J.; Dong, W.; Ma, J.; Jiang, M.; Xin, F. Metabolic Regulation of Organic Acid Biosynthesis in Actinobacillus succinogenes. Front. Bioeng. Biotechnol. 2019, 7, 216. [Google Scholar] [CrossRef]
- Guarnieri, M.T.; Chou, Y.-C.; Salvachúa, D.; Mohagheghi, A.; St. John, P.C.; Peterson, D.J.; Bomble, Y.J.; Beckham, G.T. Metabolic Engineering of Actinobacillus succinogenes Provides Insights into Succinic Acid Biosynthesis. Appl. Environ. Microbiol. 2017, 83, e00996-17. [Google Scholar] [CrossRef]
- Zou, W.; Zhu, L.-W.; Li, H.-M.; Tang, Y.-J. Significance of CO2 Donor on the Production of Succinic Acid by Actinobacillus succinogenes ATCC 55618. Microb. Cell Fact. 2011, 10, 87. [Google Scholar] [CrossRef]
- Chassagnole, C.; Noisommit-Rizzi, N.; Schmid, J.W.; Mauch, K.; Reuss, M. Dynamic Modeling of the Central Carbon Metabolism of Escherichia coli. Biotech. Bioeng. 2002, 79, 53–73. [Google Scholar] [CrossRef] [PubMed]
- Mokwatlo, S.C.; Nicol, W. Structure and Cell Viability Analysis of Actinobacillus succinogenes Biofilms as Biocatalysts for Succinic Acid Production. Biochem. Eng. J. 2017, 128, 134–140. [Google Scholar] [CrossRef]
- Van Eunen, K.; Rossell, S.; Bouwman, J.; Westerhoff, H.V.; Bakker, B.M. Quantitative Analysis of Flux Regulation Through Hierarchical Regulation Analysis. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2011; Volume 500, pp. 571–595. ISBN 978-0-12-385118-5. [Google Scholar]
- Long, C.P.; Antoniewicz, M.R. High-Resolution 13C Metabolic Flux Analysis. Nat. Protoc. 2019, 14, 2856–2877. [Google Scholar] [CrossRef] [PubMed]

| Organism | Glucose Concentration | Titer | Productivity | Yield | Source |
|---|---|---|---|---|---|
| Basfia succiniciproducens JF4016 (WT) | 30 | 19 | 1.90 | 0.62 | [5] |
| Mannheimia succiniciproducens MBEL55E (WT) | 17.76 | 10.51 | - | 0.59 | [6] |
| Saccharomyces cerevisiae (GM) | 50 | 3.62 | - | 0.11 | [7] |
| Escherichia coli (GM) | 30 | 24.5 | 0.68 | 0.88 | [8] |
| Anaerobiospirillum succiniciproducens ATCC 53488 (WT) | 50 | 32.2 | 1.2 | 0.99 | [9] |
| A. succinogenes Z130 (WT) | 84.6/70 | 52.7 | 1.0 | 0.62 | [4] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tix, J.; Bode, J.; Gotthardt, L.; Tippkötter, N. Substrate Gas Utilization and C3/C4 Metabolic Analysis of Actinobacillus succinogenes: Integration into a Model for Fermentation Prediction in BES. Fermentation 2025, 11, 263. https://doi.org/10.3390/fermentation11050263
Tix J, Bode J, Gotthardt L, Tippkötter N. Substrate Gas Utilization and C3/C4 Metabolic Analysis of Actinobacillus succinogenes: Integration into a Model for Fermentation Prediction in BES. Fermentation. 2025; 11(5):263. https://doi.org/10.3390/fermentation11050263
Chicago/Turabian StyleTix, Julian, Joshua Bode, Leon Gotthardt, and Nils Tippkötter. 2025. "Substrate Gas Utilization and C3/C4 Metabolic Analysis of Actinobacillus succinogenes: Integration into a Model for Fermentation Prediction in BES" Fermentation 11, no. 5: 263. https://doi.org/10.3390/fermentation11050263
APA StyleTix, J., Bode, J., Gotthardt, L., & Tippkötter, N. (2025). Substrate Gas Utilization and C3/C4 Metabolic Analysis of Actinobacillus succinogenes: Integration into a Model for Fermentation Prediction in BES. Fermentation, 11(5), 263. https://doi.org/10.3390/fermentation11050263






