Two-Step Bio-Based Production of Heme: In Vivo Cell Cultivation Followed by In Vitro Enzymatic Conversion
Abstract
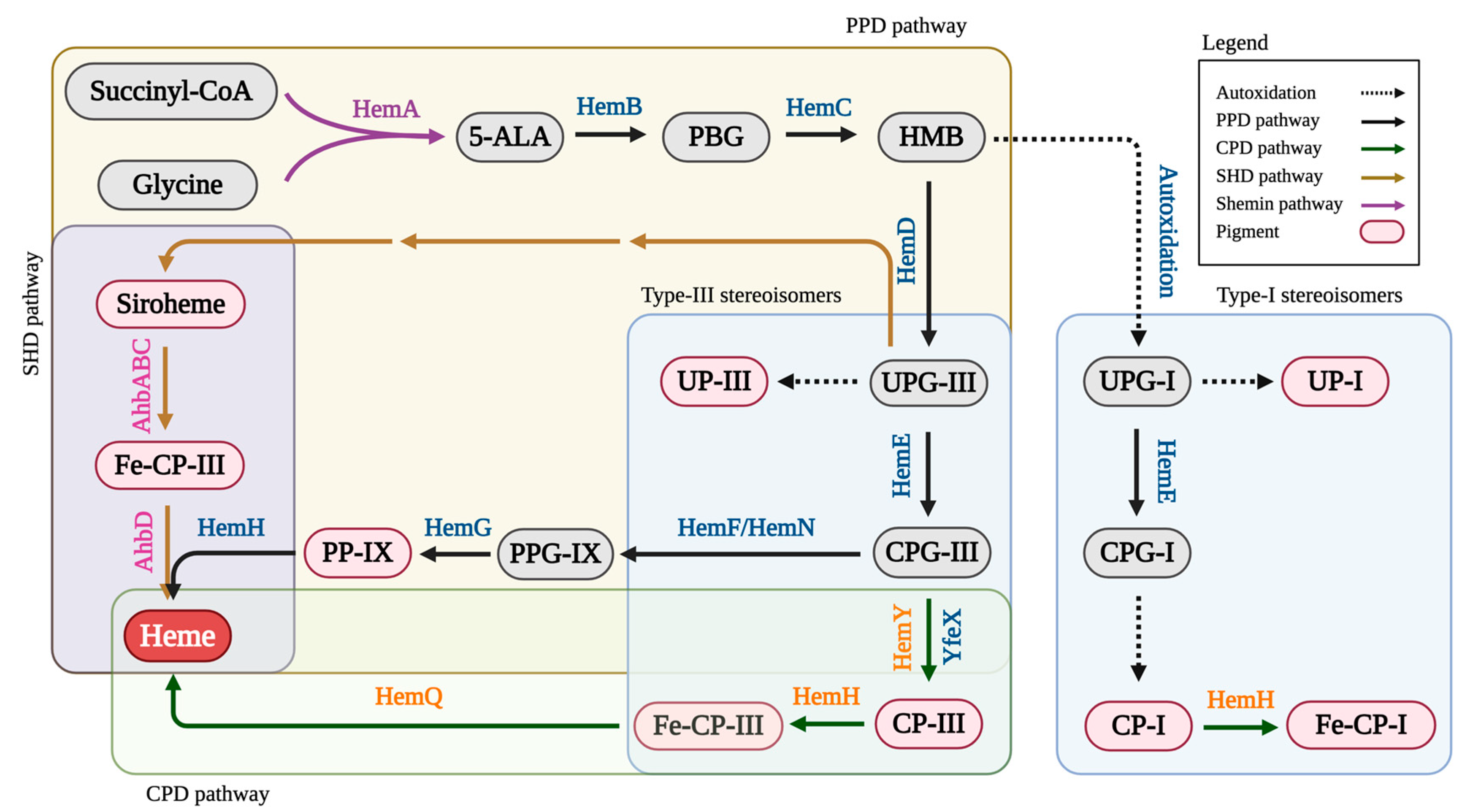
1. Introduction
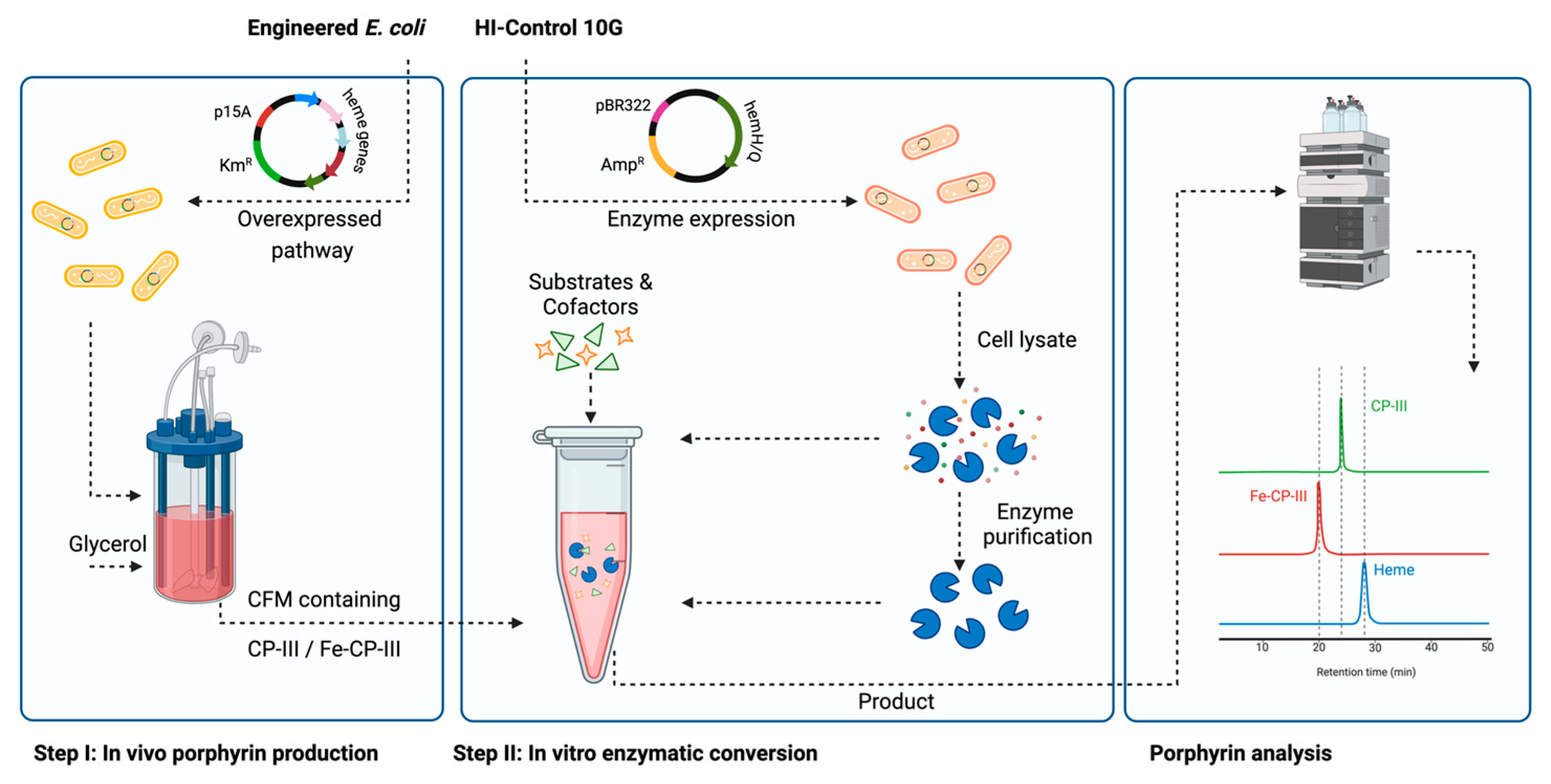
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Media and Bacterial Cell Cultivation
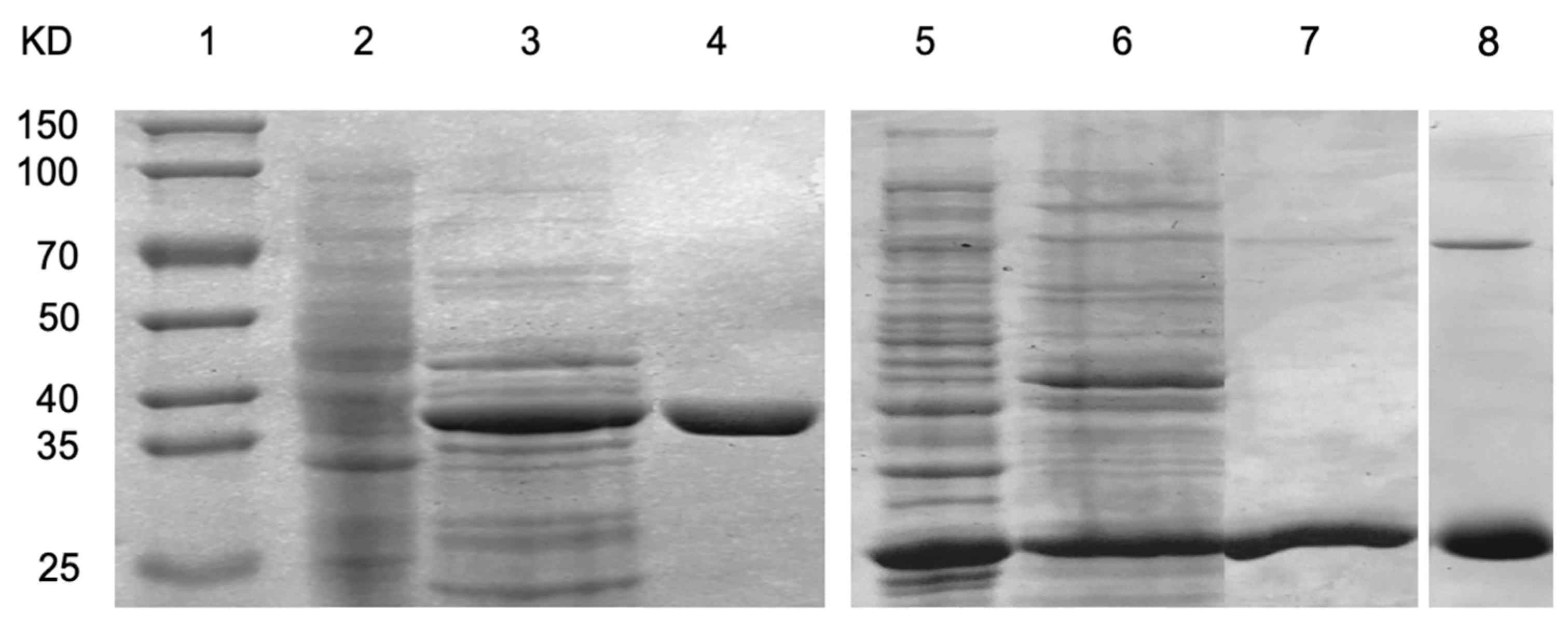
2.3. Enzyme Purification
2.4. In Vitro Experiment Reaction Set-Up
2.5. Intracellular Porphyrin Extraction
2.6. Analysis
3. Results
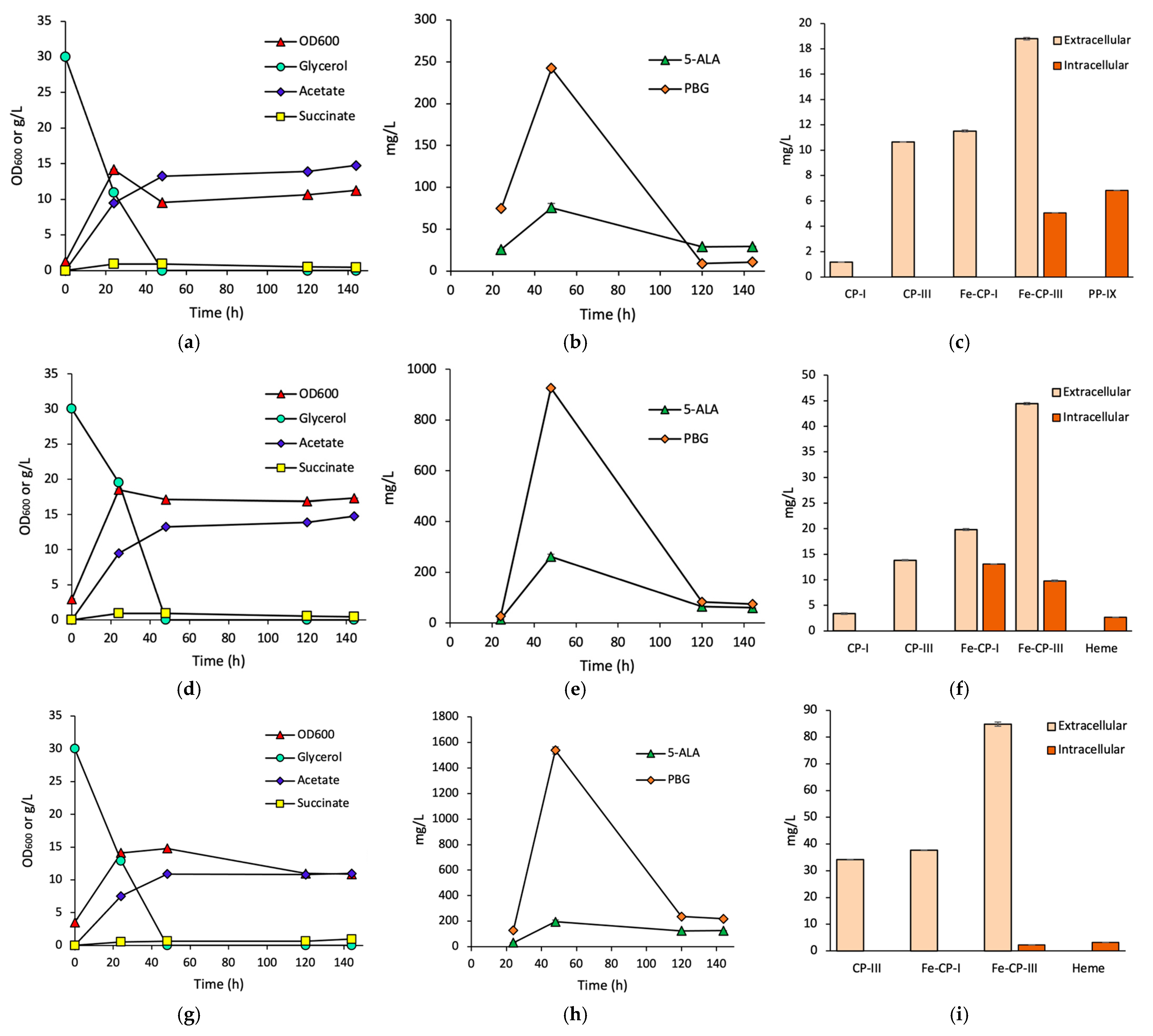
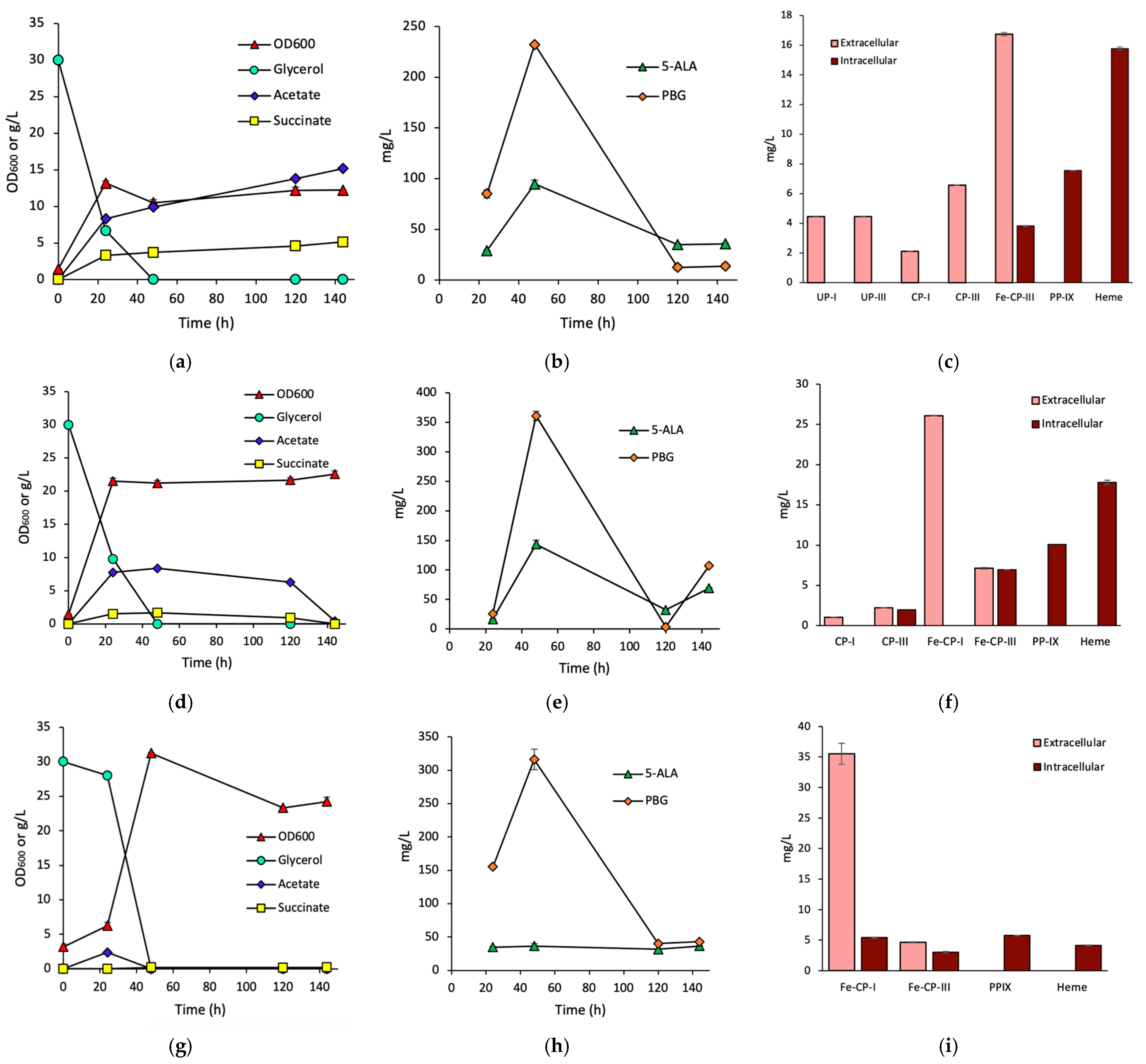
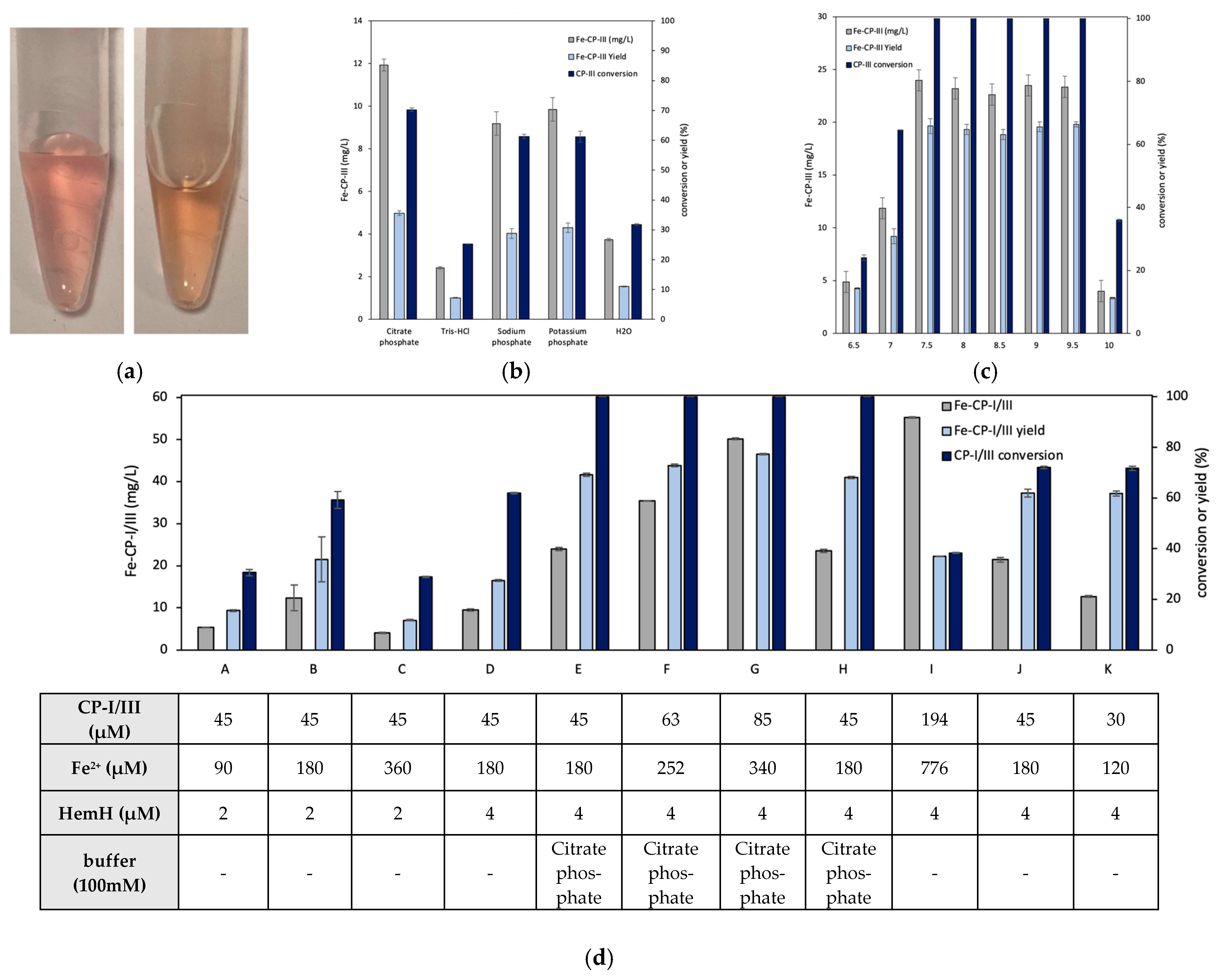
3.1. Effects of HemH on Fe-CP-III Biosynthesis
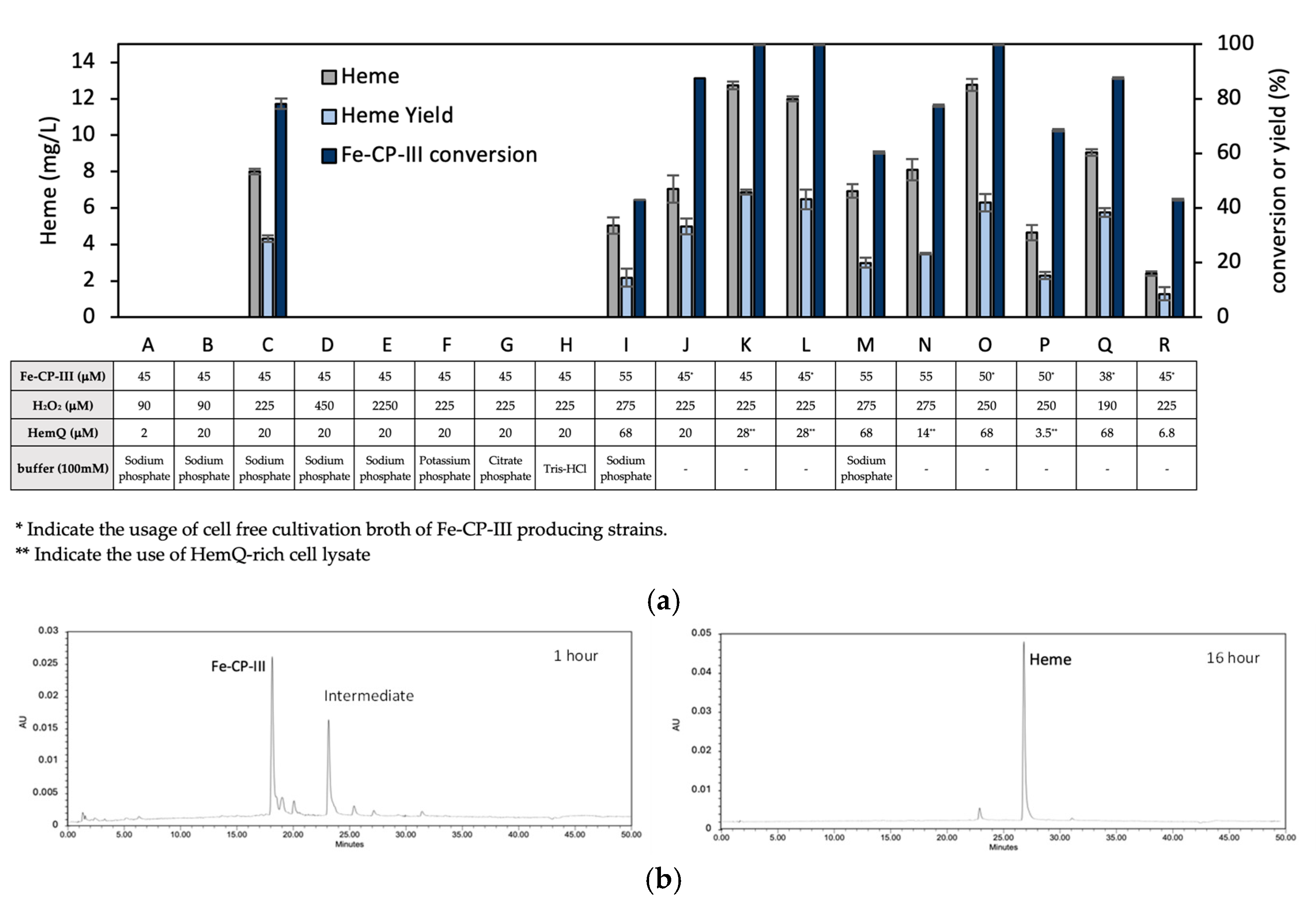
3.2. Effects of HemQ on Heme Biosynthesis
3.3. In Vitro Conversion of CP-III to Fe-CP-III via HemH
3.4. In Vitro Conversion of Fe-CP-III to Heme via HemQ
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ponka, P. Cell biology of heme. Am. J. Med. Sci. 1999, 318, 241–256. [Google Scholar] [PubMed]
- Ajioka, R.S.; Phillips, J.D.; Kushner, J.P. Biosynthesis of heme in mammals. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2006, 1763, 723–736. [Google Scholar]
- Poulos, T.L. Heme enzyme structure and function. Chem. Rev. 2014, 114, 3919–3962. [Google Scholar] [PubMed]
- Xing, Y.; Gao, S.; Zhang, X.; Zang, J. Dietary heme-containing proteins: Structures, applications, and challenges. Foods 2022, 11, 3594. [Google Scholar] [CrossRef]
- Su, H.; Chen, X.; Chen, S.; Guo, M.; Liu, H. Applications of the whole-cell system in the efficient biosynthesis of heme. Int. J. Mol. Sci. 2023, 24, 8384. [Google Scholar] [CrossRef]
- Abdelazim, I.A.; Abu-Faza, M.; Shikanova, S.; Zhurabekova, G.; Maghrabi, M.M. Heme-bound iron in treatment of pregnancy-associated iron deficiency anemia. J. Fam. Med. Prim. Care 2018, 7, 1434–1438. [Google Scholar]
- Adam, S.M.; Wijeratne, G.B.; Rogler, P.J.; Diaz, D.E.; Quist, D.A.; Liu, J.J.; Karlin, K.D. Synthetic Fe/Cu complexes: Toward understanding heme-copper oxidase structure and function. Chem. Rev. 2018, 118, 10840–11022. [Google Scholar]
- Hofbauer, S.; Schaffner, I.; Furtmüller, P.G.; Obinger, C. Chlorite dismutases–a heme enzyme family for use in bioremediation and generation of molecular oxygen. Biotechnol. J. 2014, 9, 461–473. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, J.; Zheng, Y.; Chen, T.; Wang, Z. Microbial synthesis of heme b: Biosynthetic pathways, current strategies, detection, and future prospects. Molecules 2023, 28, 3633. [Google Scholar] [CrossRef]
- Yang, S.; Guo, Z.; Sun, J.; Wei, J.; Ma, Q.; Gao, X. Recent advances in microbial synthesis of free heme. Appl. Microbiol. Biotechnol. 2024, 108, 68. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Y.; Wang, W.; Cao, T.; Zhang, L.; Wang, Z.; Chi, X.; Shi, T.; Wang, H.; He, X. High-yield porphyrin production through metabolic engineering and biocatalysis. Nat. Biotechnol. 2024, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Layer, G.; Jahn, M.; Moser, J.; Jahn, D. Radical SAM enzymes involved in tetrapyrrole biosynthesis and insertion. ACS Bio. Med. Chem. Au. 2022, 2, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.; Mingers, T.; Haskamp, V.; Jahn, D.; Jahn, M. Biosynthesis and insertion of heme. In Aerobic Utilization of Hydrocarbons, Oils, and Lipids; Springer: Cham, Switzerland, 2019; pp. 1–28. [Google Scholar]
- Bali, S.; Lawrence, A.D.; Lobo, S.A.; Saraiva, L.M.; Golding, B.T.; Palmer, D.J.; Howard, M.J.; Ferguson, S.J.; Warren, M.J. Molecular hijacking of siroheme for the synthesis of heme and d 1 heme. Proc. Natl. Acad. Sci. USA 2011, 108, 18260–18265. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.K.; Dailey, T.A.; Anderle, B.; Warren, M.J.; Bergonia, H.A.; Dailey, H.A.; Phillips, J.D. Exploiting differences in heme biosynthesis between bacterial species to screen for novel antimicrobials. Biomolecules 2023, 13, 1485. [Google Scholar] [CrossRef]
- Celis, A.I.; DuBois, J.L. Making and breaking heme. Curr. Opin. Struct. Biol. 2019, 59, 19–28. [Google Scholar] [CrossRef]
- Ge, J.; Wang, X.; Bai, Y.; Wang, Y.; Wang, Y.; Tu, T.; Qin, X.; Su, X.; Luo, H.; Yao, B. Engineering Escherichia coli for efficient assembly of heme proteins. Microb. Cell Factories 2023, 22, 59. [Google Scholar] [CrossRef]
- Aftab, H.; Donegan, R.K. Regulation of heme biosynthesis via the coproporphyrin dependent pathway in bacteria. Front. Microbiol. 2024, 15, 1345389. [Google Scholar] [CrossRef]
- Beas, J.Z.; Videira, M.A.; Saraiva, L.M. Regulation of bacterial haem biosynthesis. Coord. Chem. Rev. 2022, 452, 214286. [Google Scholar] [CrossRef]
- Zhang, J.; Kang, Z.; Chen, J.; Du, G. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci. Rep. 2015, 5, 8584. [Google Scholar]
- Jiang, M.; Hong, K.; Mao, Y.; Ma, H.; Chen, T.; Wang, Z. Natural 5-aminolevulinic acid: Sources, biosynthesis, detection and applications. Front. Bioeng. Biotechnol. 2022, 10, 841443. [Google Scholar] [CrossRef]
- Choby, J.E.; Skaar, E.P. Heme synthesis and acquisition in bacterial pathogens. J. Mol. Biol. 2016, 428, 3408–3428. [Google Scholar] [PubMed]
- Anzaldi, L.L.; Skaar, E.P. Overcoming the heme paradox: Heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 2010, 78, 4977–4989. [Google Scholar] [CrossRef] [PubMed]
- Miscevic, D.; Mao, J.Y.; Kefale, T.; Abedi, D.; Moo-Young, M.; Perry Chou, C. Strain engineering for high-level 5-aminolevulinic acid production in Escherichia coli. Biotechnol. Bioeng. 2021, 118, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Lall, D.; Miscevic, D.; Bruder, M.; Westbrook, A.; Aucoin, M.; Moo-Young, M.; Perry Chou, C. Strain engineering and bioprocessing strategies for biobased production of porphobilinogen in Escherichia coli. Bioresour. Bioprocess. 2021, 8, 122. [Google Scholar]
- Arab, B.; Westbrook, A.W.; Moo-Young, M.; Liu, Y.; Chou, C.P. Bio-Based Production of Uroporphyrin in Escherichia coli. Synth. Biol. Eng. 2024, 2, 10002. [Google Scholar]
- Arab, B.; Moo-Young, M.; Liu, Y.; Chou, C.P. Manipulating Intracellular Oxidative Conditions to Enhance Porphyrin Production in Escherichia coli. Bioengineering 2025, 12, 83. [Google Scholar] [CrossRef]
- Arab, B.; Westbrook, A.; Moo-Young, M.; Liu, Y.; Chou, C.P. High-Level Bio-Based Production of Coproporphyrin in Escherichia coli. Fermentation 2024, 10, 250. [Google Scholar] [CrossRef]
- Tinafar, A.; Jaenes, K.; Pardee, K. Synthetic biology goes cell-free. BMC Biol. 2019, 17, 64. [Google Scholar]
- Hodgman, C.E.; Jewett, M.C. Cell-free synthetic biology: Thinking outside the cell. Metab. Eng. 2012, 14, 261–269. [Google Scholar] [CrossRef]
- Guterl, J.K.; Sieber, V. Biosynthesis “debugged”: Novel bioproduction strategies. Eng. Life Sci. 2013, 13, 4–18. [Google Scholar]
- Dudley, Q.M.; Karim, A.S.; Jewett, M.C. Cell-free metabolic engineering: Biomanufacturing beyond the cell. Biotechnol. J. 2015, 10, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Miscevic, D.; Mao, J.Y.; Moo-Young, M.; Chou, C.H.P. High-level heterologous production of propionate in engineered Escherichia coli. Biotechnol. Bioeng. 2020, 117, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Srirangan, K.; Liu, X.; Westbrook, A.; Akawi, L.; Pyne, M.E.; Moo-Young, M.; Chou, C.P. Biochemical, genetic, and metabolic engineering strategies to enhance coproduction of 1-propanol and ethanol in engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2014, 98, 9499–9515. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Neidhardt, F.C.; Bloch, P.L.; Smith, D.F. Culture medium for Enterobacteria. J. Bacteriol. 1974, 119, 736–747. [Google Scholar]
- Hofbauer, S.; Mlynek, G.; Milazzo, L.; Pühringer, D.; Maresch, D.; Schaffner, I.; Furtmüller, P.G.; Smulevich, G.; Djinović-Carugo, K.; Obinger, C. Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ—Lessons from the first crystal structure and kinetic studies. FEBS J. 2016, 283, 4386–4401. [Google Scholar]
- Dailey, T.A.; Boynton, T.O.; Albetel, A.-N.; Gerdes, S.; Johnson, M.K.; Dailey, H.A. Discovery and characterization of HemQ: An essential heme biosynthetic pathway component. J. Biol. Chem. 2010, 285, 25978–25986. [Google Scholar]
- Fyrestam, J.; Östman, C. Determination of heme in microorganisms using HPLC-MS/MS and cobalt (III) protoporphyrin IX inhibition of heme acquisition in Escherichia coli. Anal. Bioanal. Chem. 2017, 409, 6999–7010. [Google Scholar]
- Mauzerall, D.; Granick, S. The Occurence and Determination og δ-Aminolevulinic Acid and Porphobilinogen in Urine. J. Biol. Chem. 1956, 219, 435–446. [Google Scholar] [CrossRef]
- Kwon, S.J.; De Boer, A.L.; Petri, R.; Schmidt-Dannert, C. High-Level Production of Porphyrins in Metabolically Engineered Escherichia coli: Systematic Extension of a Pathway Assembled from Overexpressed Genes Involved in Heme Biosynthesis. Appl. Environ. Microbiol. 2003, 69, 4875–4883. [Google Scholar] [CrossRef]
- Hansson, M.; Hederstedt, L. Purification and characterisation of a water-soluble ferrochelatase from Bacillus subtilis. Eur. J. Biochem. 1994, 220, 201–208. [Google Scholar] [PubMed]
- Dailey, H.A.; Dailey, T.A.; Gerdes, S.; Jahn, D.; Jahn, M.; O’Brian, M.R.; Warren, M.J. Prokaryotic heme biosynthesis: Multiple pathways to a common essential product. Microbiol. Mol. Biol. Rev. 2017, 81, e00048-16. [Google Scholar] [CrossRef] [PubMed]
- Dailey, H.A.; Gerdes, S. HemQ: An iron-coproporphyrin oxidative decarboxylase for protoheme synthesis in Firmicutes and Actinobacteria. Arch. Biochem. Biophys. 2015, 574, 27–35. [Google Scholar]
- Santos, C.N.S.; Stephanopoulos, G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr. Opin. Chem. Biol. 2008, 12, 168–176. [Google Scholar]
- Ko, Y.J.; Lee, M.-E.; Cho, B.-H.; Kim, M.; Hyeon, J.E.; Han, J.H.; Han, S.O. Bioproduction of porphyrins, phycobilins, and their proteins using microbial cell factories: Engineering, metabolic regulations, challenges, and perspectives. Crit. Rev. Biotechnol. 2024, 44, 373–387. [Google Scholar] [CrossRef]
- Khan, A.A.; Quigley, J.G. Control of intracellular heme levels: Heme transporters and heme oxygenases. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2011, 1813, 668–682. [Google Scholar]
- Sirithanakorn, C.; Imlay, J.A. Evidence for endogenous hydrogen peroxide production by E. coli fatty acyl-CoA dehydrogenase. PLoS ONE 2024, 19, e0309988. [Google Scholar]
- Dailey, H.A.; Gerdes, S.; Dailey, T.A.; Burch, J.S.; Phillips, J.D. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc. Natl. Acad. Sci. USA 2015, 112, 2210–2215. [Google Scholar] [CrossRef]
- Hao, G.-F.; Tan, Y.; Yang, S.-G.; Wang, Z.-F.; Zhan, C.-G.; Xi, Z.; Yang, G.-F. Computational and experimental insights into the mechanism of substrate recognition and feedback inhibition of protoporphyrinogen oxidase. PLoS ONE 2013, 8, e69198. [Google Scholar] [CrossRef]
- Yu, F.; Zhou, Y.J. Sustainable production of porphyrins through synthetic biology. Trends Biotechnol. 2024. online. [Google Scholar] [CrossRef]
| Name | Description or Relevant Genotype | Source |
|---|---|---|
| Host strains | ||
| HI-Control 10G | mcrA, ∆(mrr-hsdRMS-mcrBC), endA1, recA1, ϕ80dlacZ∆M15, ∆lacX74, araD139, ∆(ara leu)7697, galU, galK, rpsL (StrR), nupG, λ−, tonA, and Mini-F lacIq1 (GentR) | Lucigen |
| MG1655 | K-12; F−, λ−, and rph-1 | Lab stock |
| Bacillus subtilis 168 | Wild type | Lab stock |
| Staphylococcus aureus | Wild type | Lab stock |
| CPC-Sbm∆iclR∆sdhA | F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514, ∆ldhA, Ptrc::sbm (i.e., with the FRT-Ptrc cassette replacing the 204 bp upstream of the Sbm operon), ∆iclR, and ∆sdhA | [33] |
| FECP1 | CPC-Sbm∆iclR∆sdhA/pK-hemABD-EH | This study |
| FECP2 | CPC-Sbm∆iclR∆sdhA/pK-hemAB-EH | This study |
| FECP3 | CPC-Sbm∆iclR∆sdhA/pK-hemABE-H | This study |
| HEM1 | CPC-Sbm∆iclR∆sdhA/pK-hemABD-EHQ | This study |
| HEM2 | CPC-Sbm∆iclR∆sdhA/pK-hemAB-EHQ | This study |
| HEM3 | CPC-Sbm∆iclR∆sdhA/pK-hemABE-HQS.a | This study |
| Plasmids | ||
| pK-hemABCD-E | p15A ori, KmR, Ptrc::hemABCD-Pgracmax::hemE | [28] |
| pK-hemABD-EH | p15A ori, KmR, Ptrc::hemABD-Pgracmax::hemEH -hemH from B. subtilis 168- | This study |
| pK-hemAB-EH | p15A ori, KmR, Ptrc::hemAB-Pgracmax::hemEH -hemH from B. subtilis 168- | This study |
| pK-hemABE-H | p15A ori, KmR, Ptrc::hemABE-Pgracmax::hemH -hemH from B. subtilis 168- | This study |
| pK-hemABD-EHQ | p15A ori, KmR, Ptrc::hemABD-Pgracmax::hemEHQ -hemH and hemQ from B. subtilis 168- | This study |
| pK-hemAB-EHQ | p15A ori, KmR, Ptrc::hemAB-Pgracmax::hemEHQ -hemH and hemQ from B. subtilis 168- | This study |
| pK-hemABE-HQS.a | p15A ori, KmR, Ptrc::hemABE-Pgracmax::hemHQ -hemH from B. subtilis 168 and hemQ from S. aureus- | This study |
| pK-hemH-His6 | pBR322 ori, AmpR, Ptrc::hemH:His6 -hemH from B. subtilis 168- | This study |
| pK-His6-hemH | pBR322 ori, AmpR, Ptrc::His6::hemH -hemH from B. subtilis 168- | This study |
| pK-hemQ-His6 | pBR322 ori, AmpR, Ptrc::hemQ:His6 -hemQ from B. subtilis 168- | This study |
| pK-His6-hemQ | pBR322 ori, AmpR, Ptrc::His6::hemQ -hemQ from B. subtilis 168- | This study |
| pK-hemQS.a-His6 | pBR322 ori, AmpR, Ptrc::hemQ:His6 -hemQ from S. aureus- | This study |
| pK-His6-hemQS.a | pBR322 ori, AmpR, Ptrc::His6::hemQ -hemQ from S. aureus- | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arab, B.; Moo-Young, M.; Liu, Y.; Chou, C.-H.P. Two-Step Bio-Based Production of Heme: In Vivo Cell Cultivation Followed by In Vitro Enzymatic Conversion. Fermentation 2025, 11, 198. https://doi.org/10.3390/fermentation11040198
Arab B, Moo-Young M, Liu Y, Chou C-HP. Two-Step Bio-Based Production of Heme: In Vivo Cell Cultivation Followed by In Vitro Enzymatic Conversion. Fermentation. 2025; 11(4):198. https://doi.org/10.3390/fermentation11040198
Chicago/Turabian StyleArab, Bahareh, Murray Moo-Young, Yilan Liu, and Chih-Hsiung Perry Chou. 2025. "Two-Step Bio-Based Production of Heme: In Vivo Cell Cultivation Followed by In Vitro Enzymatic Conversion" Fermentation 11, no. 4: 198. https://doi.org/10.3390/fermentation11040198
APA StyleArab, B., Moo-Young, M., Liu, Y., & Chou, C.-H. P. (2025). Two-Step Bio-Based Production of Heme: In Vivo Cell Cultivation Followed by In Vitro Enzymatic Conversion. Fermentation, 11(4), 198. https://doi.org/10.3390/fermentation11040198






