Improving the Nutritional Value and Safety of Cotton Stalk Feed via Response Surface Methodology and Co-Fermentation Techniques
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Reagents
2.3. Fermentation Preparation
2.4. Screening for Strains with High Cellulase Production
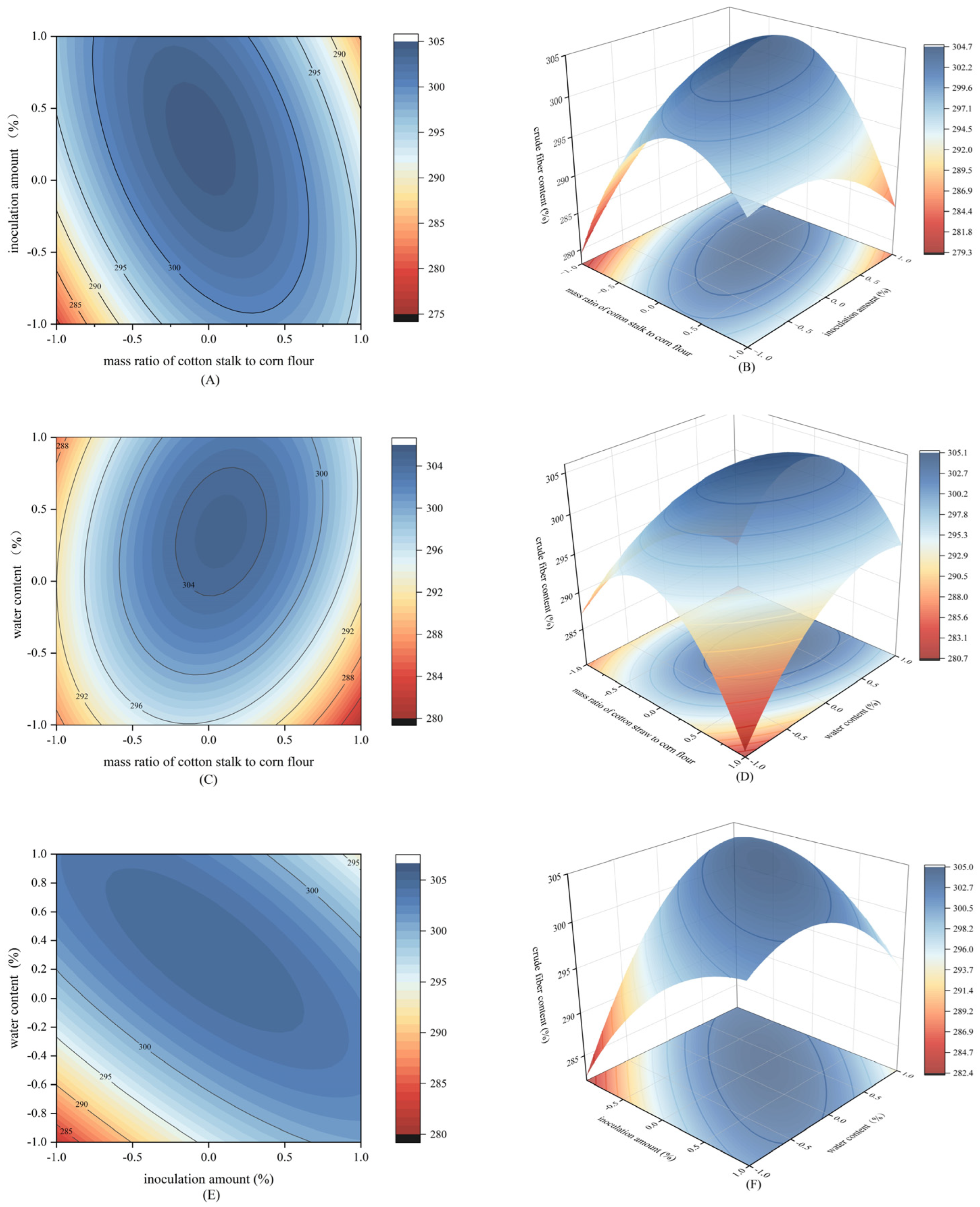
2.5. Experimental Design of Response Surface Methodology
2.6. Determination of Cotton Stalk Composition During Solid-State Fermentation
2.7. Mixed Microbial Solid-State Fermentation Experiment Design
2.8. Feed Quality Analysis
2.9. Statistical Analysis
3. Results
3.1. Determination of High Cellulase-Producing Strains
3.2. Verification Results of the Box–Bhenken Experiment Design
3.3. Degradation of the Cotton Stalk by Aspergillus niger
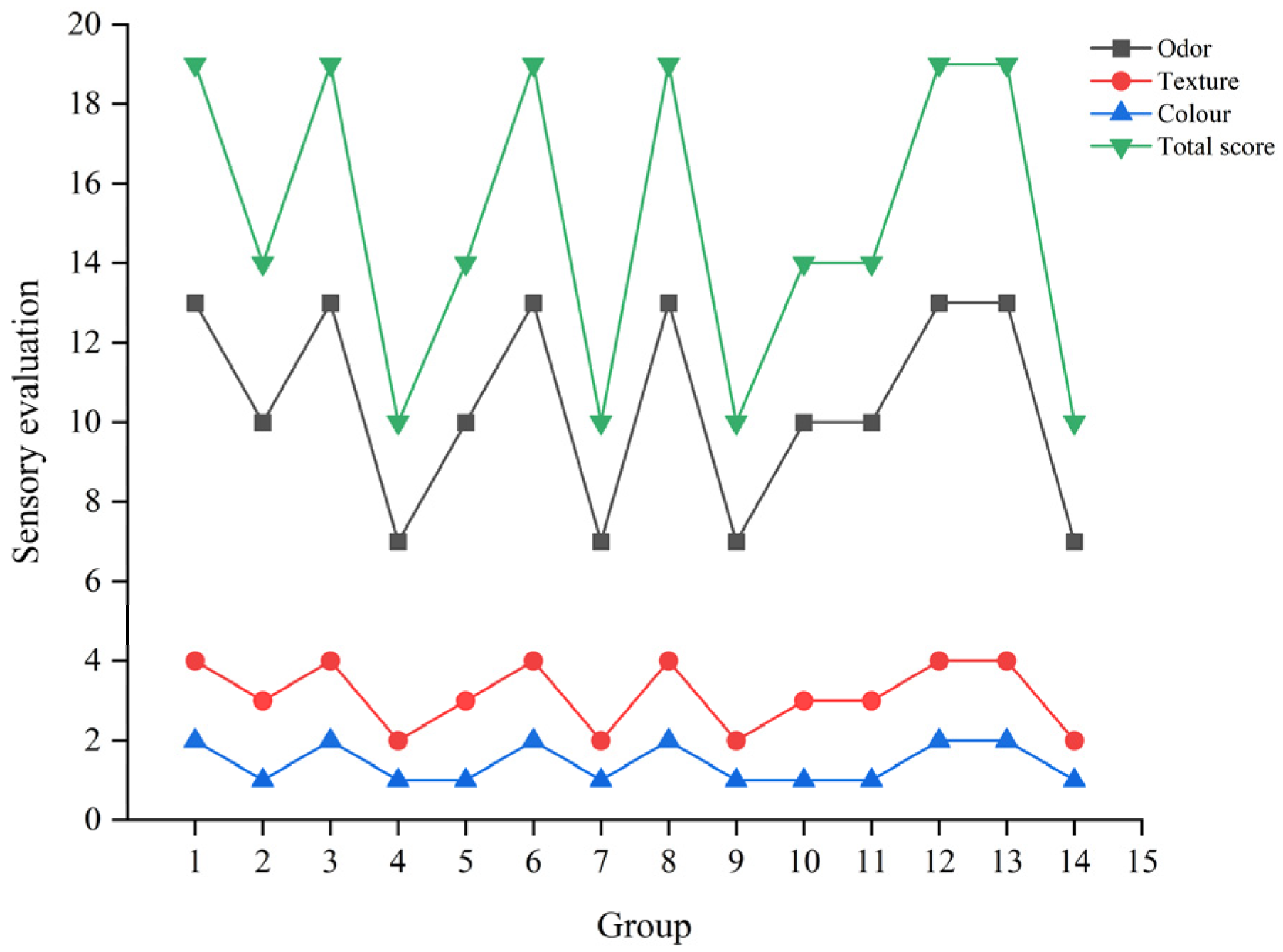
3.4. The Results of Sensory Evaluation
3.5. The Results of Nutritional Value
4. Discussion
4.1. Primary and Re-Screening of Aspergillus niger
4.2. Degradation of Cruder Fiber Content Using Response Surface Methodology
4.3. Fungal and Bacterial Stage Fermentation System
4.4. Prospects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, F.; Bai, J.; Zhang, M.; Zhang, R. Yield estimation of high-density cotton fields using low-altitude UAV imaging and deep learning. Plant Methods 2022, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Singh, A. Physico-chemical pretreatment and enzymatic hydrolysis of cotton stalk for ethanol production by Saccharomyces cerevisiae. Bioresour. Technol. 2017, 244, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Joseph, T.; Baah, K.; Jahanfar, A.; Dubey, B. A comparative life cycle assessment of conventional hand dryer and roll paper towel as hand drying methods. Sci. Total Environ. 2015, 515–516, 109–117. [Google Scholar] [CrossRef]
- Egbuta, M.A.; McIntosh, S.; Waters, D.L.; Vancov, T.; Liu, L. Biological Importance of Cotton By-Products Relative to Chemical Constituents of the Cotton Plant. Molecules 2017, 22, 93. [Google Scholar] [CrossRef] [PubMed]
- Hamawand, I.; Sandell, G.; Pittaway, P.; Chakrabarty, S.; Yusaf, T.; Chen, G.; Seneweera, S.; Al-Lwayzy, S.; Bennett, J.; Hopf, J. Bioenergy from Cotton Industry Wastes: A review and potential. Renew. Sustain. Energy Rev. 2016, 66, 435–448. [Google Scholar] [CrossRef]
- Sadh, P.K.; Duhan, S.; Duhan, J.S. Agro-industrial wastes and their utilization using solid state fermentation: A review. Bioresour. Bioprocess. 2018, 5, 1. [Google Scholar] [CrossRef]
- Li, T.; Wang, H.; Cui, J.; Wang, W.; Li, W.; Jiang, M.; Shi, X.; Song, J.; Wang, J.; Lv, X.; et al. Improving the accuracy of cotton seedling emergence rate estimation by fusing UAV-based multispectral vegetation indices. Front. Plant Sci. 2024, 15, 1333089. [Google Scholar] [CrossRef]
- Wei, Y.; Wu, D.; Wei, D.; Zhao, Y.; Wu, J.; Xie, X.; Zhang, R.; Wei, Z. Improved lignocellulose-degrading performance during straw composting from diverse sources with actinomycetes inoculation by regulating the key enzyme activities. Bioresour. Technol. 2019, 271, 66–74. [Google Scholar] [CrossRef]
- Xu, X.; Xu, Z.; Shi, S.; Lin, M. Lignocellulose degradation patterns, structural changes, and enzyme secretion by Inonotus obliquus on straw biomass under submerged fermentation. Bioresour. Technol. 2017, 241, 415–423. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, L.; Sun, Y.; Xu, G.; Wang, W.; Piao, R.; Cui, Z.; Zhao, H. Degradation of lignocelluloses in straw using AC-1, a thermophilic composite microbial system. PeerJ 2021, 9, e12364. [Google Scholar] [CrossRef]
- Liu, H.; Yang, X.; Mai, L.; Lin, J.; Zhang, L.; Wang, D.; Li, Q. Comparative Proteomic Analysis of Bacillus subtilis and Aspergillus niger in Black Soldier Fly Co-Fermentation. Fermentation 2022, 8, 593. [Google Scholar] [CrossRef]
- Zhao, M.; Yu, D.; Liu, Q.; Ma, S.; Xu, J.; Yu, J. Co-fermentation of Bacillus subtilis and Bacillus licheniformis and its application in the feeding of Koi. Aquac. Res. 2022, 53, 6056–6068. [Google Scholar] [CrossRef]
- Zhu, Y.-X.; Zhang, X.; Yang, W.-C.; Li, J.-F. Enhancement of Biomass Conservation and Bioethanol Production of Sweet Sorghum Silage by Constructing Synergistic Microbial Consortia. Microbiol. Spectr. 2023, 11, e03659-22. [Google Scholar] [CrossRef]
- Ualieva, P.S.; Abdieva, G.Z.; Akimbekov, N.S.; Malik, A.M.; Tastambek, K.T. Construction and characterisation of a novel microbial consortium for animal feed enrichment. ES Food Agrofor. 2024, 17, 1243. [Google Scholar] [CrossRef]
- Sharma, M.; Mahajan, C.; Bhatti, M.S.; Chadha, B.S. Profiling and production of hemicellulases by thermophilic fungus Malbranchea flava and the role of xylanases in improved bioconversion of pretreated lignocellulosics to ethanol. 3 Biotech 2016, 6, 30. [Google Scholar] [CrossRef]
- Osiro, K.O.; de Camargo, B.R.; Satomi, R.; Hamann, P.R.; Silva, J.P.; de Sousa, M.V.; Quirino, B.F.; Aquino, E.N.; Felix, C.R.; Murad, A.M.; et al. Characterization of Clostridium thermocellum (B8) secretome and purified cellulosomes for lignocellulosic biomass degradation. Enzym. Microb. Technol. 2017, 97, 43–54. [Google Scholar] [CrossRef]
- Santos, G.B.; de Sousa Francisco Filho, Á.; Rêgo da Silva Rodrigues, J.; Rodrigues de Souza, R. Cellulase production by Aspergillus niger using urban lignocellulosic waste as substrate: Evaluation of different cultivation strategies. J. Environ. Manag. 2022, 305, 114431. [Google Scholar] [CrossRef]
- Guo, T.-R.; Zeng, Q.; Yang, G.; Ye, S.-S.; Chen, Z.-Y.; Xie, S.-Y.; Wang, H.; Mo, Y.-W. Isolation, identification, biological characteristics, and antifungal efficacy of sodium bicarbonate combined with natamycin on Aspergillus niger from Shengzhou nane (Prunus salicina var. taoxingli) fruit. Front. Microbiol. 2023, 13, 1075033. [Google Scholar] [CrossRef]
- Bampidis, V.; Azimonti, G.; Bastos, M.L.; Christensen, H.; Dusemund, B.; Durjava, M.; Kouba, M.; López-Alonso, M.; López Puente, S.; Marcon, F.; et al. Safety and efficacy of a feed additive consisting of β-mannanase produced by Aspergillus nigerCBS 120604 (Nutrixtend Optim) for use in all poultry for fattening (Kerry Ingredients & Flavours Ltd.). EFSA J. Eur. Food Saf. Auth. 2023, 21, e08045. [Google Scholar] [CrossRef]
- Meng, J.; Chroumpi, T.; Mäkelä, M.R.; de Vries, R.P. Xylitol production from plant biomass by Aspergillus niger through metabolic engineering. Bioresour. Technol. 2022, 344, 126199. [Google Scholar] [CrossRef]
- Li, H.; Zhang, M.; Zhang, Y.; Xu, X.; Zhao, Y.; Jiang, X.; Zhang, R.; Gui, Z. Characterization of Cellulose-Degrading Bacteria Isolated from Silkworm Excrement and Optimization of Its Cellulase Production. Polymers 2023, 15, 4142. [Google Scholar] [CrossRef] [PubMed]
- Dashtban, M.; Maki, M.; Leung, K.T.; Mao, C.; Qin, W. Cellulase activities in biomass conversion: Measurement methods and comparison. Crit. Rev. Biotechnol. 2010, 30, 302–309. [Google Scholar] [CrossRef]
- Li, J.; Wu, K.; Wu, J.; Yang, C.; Sun, B.; Deng, M.; Liu, D.; Li, Y.; Liu, G.; Guo, Y. Effects of Fresh Corn Stover to Corn Flour Ratio on Fermentation Quality and Bacterial Community of Mixed Silage. Fermentation 2024, 10, 654. [Google Scholar] [CrossRef]
- Marques, N.P.; de Cassia Pereira, J.; Gomes, E.; da Silva, R.; Araújo, A.R.; Ferreira, H.; Rodrigues, A.; Dussán, K.J.; Bocchini, D.A. Cellulases and xylanases production by endophytic fungi by solid state fermentation using lignocellulosic substrates and enzymatic saccharification of pretreated sugarcane bagasse. Ind. Crop. Prod. 2018, 122, 66–75. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, K.; Lin, Y.; Li, M.; Wang, X.; Yu, Q.; Sun, H.; Cheng, Q.; Xie, Y.; Wang, C. Effect of cellulase and lactic acid bacteria on the fermentation quality, carbohydrate conversion, and microbial community of ensiling oat with different moisture contents. Front. Microbiol. 2022, 13, 1013258. [Google Scholar] [CrossRef]
- Quirino, D.F.; Lima, N.S.A.; Palma, M.N.N.; Franco, M.O.; Detmann, E. Evaluation of Heating Times for Loss on Drying at 105 °C for Estimation of Laboratory Dry Matter in Animal Feeds. J. AOAC Int. 2023, 106, 261–266. [Google Scholar] [CrossRef]
- Mei, J.; Shen, X.; Gang, L.; Xu, H.; Wu, F.; Sheng, L. A novel lignin degradation bacteria-Bacillus amyloliquefaciens SL-7 used to degrade straw lignin efficiently. Bioresour. Technol. 2020, 310, 123445. [Google Scholar] [CrossRef]
- Martin, E.; Dubessay, P.; Record, E.; Audonnet, F.; Michaud, P. Recent advances in laccase activity assays: A crucial challenge for applications on complex substrates. Enzym. Microb. Technol. 2024, 173, 110373. [Google Scholar] [CrossRef]
- Chavan, S.; Shete, A.; Dharne, M.S. Strain improvement for cellulolytic enzymes for effective saccharification of lignocellulosic biomass by mutant of Penicillium funiculosum NCIM 1228. Syst. Microbiol. Biomanufacturing 2024, 4, 716–730. [Google Scholar] [CrossRef]
- Sousa, D.; Salgado, J.M.; Cambra-López, M.; Dias, A.C.; Belo, I. Degradation of lignocellulosic matrix of oilseed cakes by solid-state fermentation: Fungi screening for enzymes production and antioxidants release. J. Sci. Food Agric. 2022, 102, 1550–1560. [Google Scholar] [CrossRef]
- Haokok, C.; Lunprom, S.; Reungsang, A.; Salakkam, A. Efficient production of lactic acid from cellulose and xylan in sugarcane bagasse by newly isolated Lactiplantibacillus plantarum and Levilactobacillus brevis through simultaneous saccharification and co-fermentation process. Heliyon 2023, 9, e17935. [Google Scholar] [CrossRef] [PubMed]
- Kargar, S.; Kanani, M. Reconstituted versus dry alfalfa hay in starter feed diets of Holstein dairy calves: Effects on growth performance, nutrient digestibility, and metabolic indications of rumen development. J. Dairy Sci. 2019, 102, 4051–4060. [Google Scholar] [CrossRef]
- Muhandiram, N.P.K.; Humphreys, M.W.; Fychan, R.; Davies, J.W.; Sanderson, R.; Marley, C.L. Designing agricultural grasses to help mitigate proteolysis during ensiling to optimize protein feed provisions for livestock. Food Energy Secur. 2023, 12, e475. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xie, H.; Luo, Q.; Yang, K.; Xia, C.; Guan, J.; Zhou, W.; Sun, B.; Wang, Z.; Cui, S. Mode of innovative green production for concrete engineering: Life cycle assessment of accelerators prepared from aluminum mud wastes. Environ. Sci. Pollut. Res. Int. 2023, 30, 79106–79119. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Sun, J.; Chu, J. High-content screening of Aspergillus niger with both increased production and high secretion rate of glucose oxidase. Biotechnol. Lett. 2018, 40, 103–110. [Google Scholar] [CrossRef]
- Detain, J.; Besaury, L. Degradation of lignocellulose by different bacterial and fungal co-cultures. Curr. Res. Microb. Sci. 2024, 7, 100271. [Google Scholar] [CrossRef]
- Cui, T.; Yuan, B.; Guo, H.; Tian, H.; Wang, W.; Ma, Y.; Li, C.; Fei, Q. Enhanced lignin biodegradation by consortium of white rot fungi: Microbial synergistic effects and product mapping. Biotechnol. Biofuels 2021, 14, 162. [Google Scholar] [CrossRef]
- Sun, S.; Li, F.; Li, M.; Zhang, W.; Jiang, Z.; Zhao, H.; Pu, Y.; Ragauskas, A.J.; Dai, S.Y.; Zhang, X. Lytic polysaccharide monooxygenase synergized with lignin-degrading enzymes for efficient lignin degradation. iScience 2023, 26, 107870. [Google Scholar] [CrossRef]
- Malik, K.; Sharma, P.; Yang, Y.; Zhang, P.; Zhang, L.; Xing, X.; Yue, J.; Song, Z.; Nan, L.; Yujun, S.; et al. Lignocellulosic biomass for bioethanol: Insight into the advanced pretreatment and fermentation approaches. Ind. Crops Prod. 2022, 188, 115569. [Google Scholar] [CrossRef]
- Asina, F.; Brzonova, I.; Voeller, K.; Kozliak, E.; Kubátová, A.; Yao, B.; Ji, Y. Biodegradation of lignin by fungi, bacteria and laccases. Bioresour. Technol. 2016, 220, 414–424. [Google Scholar] [CrossRef]
- Šuchová, K.; Fehér, C.; Ravn, J.L.; Bedő, S.; Biely, P.; Geijer, C. Cellulose- and xylan-degrading yeasts: Enzymes, applications and biotechnological potential. Biotechnol. Adv. 2022, 59, 107981. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bao, F.; Wei, H.; Zhang, Y. Screening of cellulose-degrading bacteria and optimization of cellulase production from Bacillus cereus A49 through response surface methodology. Sci. Rep. 2024, 14, 7755. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Wang, B.; Sun, Y.; Yang, J.; Zhou, J.; Wang, T.; Zhang, W.; Qi, C.; Guo, Y. Effects of Aspergillus niger on cyanogenic glycosides removal and fermentation qualities of ratooning sorghum. Front. Microbiol. 2023, 14, 1128057. [Google Scholar] [CrossRef] [PubMed]
- Pongsub, S.; Suntara, C.; Khota, W.; Boontiam, W.; Cherdthong, A. The Chemical Composition, Fermentation End-Product of Silage, and Aerobic Stability of Cassava Pulp Fermented with Lactobacillus casei TH14 and Additives. Vet. Sci. 2022, 9, 617. [Google Scholar] [CrossRef]
- Mirmohammadi, R.; Zamindar, N.; Razavi, S.H.; Mirmohammadi, M.; Paidari, S. Investigation of the possibility of fermentation of red grape juice and rice flour by Lactobacillus plantarum and Lactobacillus casei. Food Sci. Nutr. 2021, 9, 5370–5378. [Google Scholar] [CrossRef]
- Spacova, I.; Allonsius, C.N.; De Boeck, I.; Oerlemans, E.; Tuyaerts, I.; Van de Vliet, N.; van den Broek, M.F.L.; Jimenez, L.; Boyer, M.; Rodriguez, B.; et al. Multifactorial inhibition of Candida albicans by combinations of lactobacilli and probiotic Saccharomyces cerevisiae CNCM I-3856. Sci. Rep. 2024, 14, 9365. [Google Scholar] [CrossRef]
- Vadopalas, L.; Ruzauskas, M.; Lele, V.; Starkute, V.; Zavistanaviciute, P.; Zokaityte, E.; Bartkevics, V.; Pugajeva, I.; Reinolds, I.; Badaras, S.; et al. Combination of Antimicrobial Starters for Feed Fermentation: Influence on Piglet Feces Microbiota and Health and Growth Performance, Including Mycotoxin Biotransformation in vivo. Front. Vet. Sci. 2020, 7, 528990. [Google Scholar] [CrossRef]
- Jiang, Y.; Du, X.; Xu, Q.; Yin, C.; Zhang, H.; Liu, Y.; Liu, X.; Yan, H. Biodegradation of Gossypol by Aspergillus terreus-YJ01. Microorganisms 2023, 11, 2148. [Google Scholar] [CrossRef]
- Yakupova, E.I.; Bobyleva, L.G.; Vikhlyantsev, I.M.; Bobylev, A.G. Congo Red and amyloids: History and relationship. Biosci. Rep. 2019, 39, BSR20181415. [Google Scholar] [CrossRef]
- Zhang, G.; Dong, Y. Design and application of an efficient cellulose-degrading microbial consortium and carboxymethyl cellulase production optimization. Front. Microbiol. 2022, 13, 957444. [Google Scholar] [CrossRef]
- Wei, T.; Chen, H.; Wu, D.; Gao, D.; Cai, Y.; Cao, X.; Xu, H.; Yang, J.; Guo, P. Response surface methodology for the mixed fungal fermentation of Codonopsis pilosula straw using Trichoderma reesei and Coprinus comatus. PeerJ 2023, 11, e15757. [Google Scholar] [CrossRef] [PubMed]
- An, N.-n.; Li, D.; Wang, L.-j.; Wang, Y. Microwave irradiation of corn kernels: Effects on structural, thermal, functional and rheological properties of corn flour. Food Hydrocoll. 2023, 143, 108939. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, Q.; Teng, C.; Zhou, M.; Fan, G.; Qu, P. Preparation and improvement of physicochemical and functional properties of dietary fiber from corn cob fermented by Aspergillus Niger. J. Microbiol. Biotechnol. 2023, 34, 330. [Google Scholar] [CrossRef] [PubMed]
- Vignali, E.; Gigli, M.; Cailotto, S.; Pollegioni, L.; Rosini, E.; Crestini, C. The Laccase-Lig Multienzymatic Multistep System in Lignin Valorization. ChemSusChem 2022, 15, e202201147. [Google Scholar] [CrossRef]
- Nurul-Aliyaa, Y.A.; Awang, N.A.; Mohd, M.H. Characterization of white rot fungi from wood decayed for lignin degradation. Lett. Appl. Microbiol. 2023, 76, ovad118. [Google Scholar] [CrossRef]
- Fadel, M.; Hamed, A.A.; Abd-Elaziz, A.M.; Ghanem, M.M.; Roshdy, A.M. Cellulases and animal feed production by solid-state fermentation by Aspergillus fumigatus NRCF-122 mutant. Egypt. J. Chem. 2021, 64, 3511–3520. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, C.; Ai, C.; Song, W.; Zhao, J. Development of probiotic complex based on the synthetic microbial community and probiotic effects in farming Yunlong grouper. Aquaculture 2024, 586, 740708. [Google Scholar] [CrossRef]
- Sheng, F.; Hu, X.; Zeng, J.; Tian, X.; Wu, Z. Citrus pomace co-fermentation improved its protein and amino acids by Bacillus amyloliquefaciens and Candida utilis. Process. Biochem. 2023, 130, 545–554. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, D.; Liu, L.; Chang, Z.; Peng, N. Effective gossypol removal from cottonseed meal through optimized solid-state fermentation by Bacillus coagulans. Microb. Cell Factories 2022, 21, 252. [Google Scholar] [CrossRef]
- Van der Poel, A.; Abdollahi, M.; Cheng, H.; Colovic, R.; Den Hartog, L.; Miladinovic, D.; Page, G.; Sijssens, K.; Smillie, J.; Thomas, M.; et al. Future directions of animal feed technology research to meet the challenges of a changing world. Anim. Feed. Sci. Technol. 2020, 270, 114692. [Google Scholar] [CrossRef]
- Korombé, H.; Bado, V.; Abdou, N.; Umutoni, C.; Ibrahima, A.; Gouro, A. Evaluation of fermentation progress during storage of millet stovers silage based on pH-indicators. Online J. Anim. Feed. Res. 2023, 13, 116–126. [Google Scholar] [CrossRef]
- T.sriwong, K.; Matsuda, T. Recent advances in enzyme immobilization utilizing nanotechnology for biocatalysis. Org. Process Res. Dev. 2022, 26, 1857–1877. [Google Scholar] [CrossRef]
- Tan, Z.; Cheng, H.; Chen, G.; Ju, F.; Fernández-Lucas, J.; Zdarta, J.; Jesionowski, T.; Bilal, M. Designing multifunctional biocatalytic cascade system by multi-enzyme co-immobilization on biopolymers and nanostructured materials. Int. J. Biol. Macromol. 2023, 227, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Guo, Y.; Yang, F.; Zhou, C.; Tang, C.; Zhou, G. Combined application of high-throughput sequencing and UHPLC-Q/TOF-MS-based metabolomics in the evaluation of microorganisms and metabolites of dry-cured ham of different origins. Int. J. Food Microbiol. 2021, 359, 109422. [Google Scholar] [CrossRef]
- Chen, W. Demystification of fermented foods by omics technologies. Curr. Opin. Food Sci. 2022, 46, 100845. [Google Scholar] [CrossRef]

| Factors | Level | ||
|---|---|---|---|
| −1 | 1 | 0 | |
| A: Cotton stalk to corn flour mass ratio | 5:1 | 4:1 | 3:1 |
| B: Inoculation amount | 8% | 10% | 12% |
| C: Moisture content | 65% | 70% | 75% |
| Group | Microbial Strain Combination | Inoculation Amount (%) | Fermentation Time (d) |
|---|---|---|---|
| 1 | A + B + L | 10 | 20 |
| 2 | A + B + C + L | 10 | 10 |
| 3 | A + B + L | 5 | 10 |
| 4 | A + C | 5 | 20 |
| 5 | A + B + C + L | 15 | 20 |
| 6 | A + B + L | 5 | 30 |
| 7 | A + C | 10 | 10 |
| 8 | A + B + L | 15 | 30 |
| 9 | A + C | 15 | 20 |
| 10 | A + B + C + L | 5 | 20 |
| 11 | A + B + C + L | 10 | 30 |
| 12 | A + B + L | 15 | 10 |
| 13 | A + B + L | 10 | 30 |
| 14 | A | 5 | 30 |
| Strain | D (mm) | d (mm) | D/d | A |
|---|---|---|---|---|
| L3 | 2.59 ± 0.40 | 2.40 ± 0.37 | 1.08 | 0.518 |
| L4 | 8.14 ± 0.76 | 3.42 ± 0.85 | 2.38 | 1.628 |
| HQZD | 12.50 ± 3.12 | 4.38 ± 0.98 | 2.85 | 2.500 |
| HQXY | 16.23 ± 2.39 | 3.46 ± 0.68 | 4.69 | 3.246 |
| Z3 | 10.36 ± 1.66 | 4.24 ± 0.82 | 2.44 | 2.072 |
| Z4 | 9.24 ± 2.50 | 5.22 ± 1.02 | 1.77 | 1.848 |
| Strain | Weight Loss Rate | ||
|---|---|---|---|
| 3 d | 5 d | 7 d | |
| L3 | 10.72% ± 0.10 a | 12.56% ± 0.43 a | 15.67% ± 1.64 a |
| L4 | 9.94% ± 0.24 a | 34.85% ± 0.78 c | 60.68% ± 1.14 c |
| HQZD | 25.27% ± 0.92 b | 46.28% ± 1.17 d | 72.36% ± 1.51 d |
| HQXY | 26.35% ± 0.94 b | 51.82% ± 0.81 d | 82.49% ± 1.87 e |
| Z3 | 11.56% ± 0.73 a | 23.34% ± 1.06 b | 40.34% ± 1.18 b |
| Z4 | 8.45% ± 1.20 a | 29.56% ± 0.85 b | 44.78% ± 2.12 b |
| Strains | CMCase (U.mL−1) | FPase (U.mL−1) |
|---|---|---|
| L3 | 150.66 ± 8.19 a | 80.30 ± 4.71 a |
| L4 | 203.29 ± 6.40 b | 114.29 ± 8.50 b |
| HQZD | 243.74 ± 21.78 c | 133.29 ± 11.26 c |
| HQYX | 255.35 ± 9.94 c | 151.69 ± 8.24 d |
| Z3 | 144.83 ± 31.51 a | 70.39 ± 8.52 a |
| Z4 | 126.99 ± 8.71 a | 50.23 ± 6.95 a |
| Group | pH | Crude Protein Content (%) | Dry Matter Content (%) | Gossypol Content (mg·kg−1) |
|---|---|---|---|---|
| 1 | 4.21 ± 0.010 | 24.9 ± 0.073 | 26.89 ± 0.040 | 23.19 ± 0.015 |
| 2 | 4.89 ± 0.058 | 23.1 ± 0.065 | 26.08 ± 0.035 | 48.18 ± 0.030 |
| 3 | 4.52 ± 0.058 | 22.8 ± 0.047 | 26.31 ± 0.032 | 25.31 ± 0.035 |
| 4 | 5.85 ± 0.058 | 22.1 ± 0.030 | 25.84 ± 0.055 | 66.18 ± 0.361 |
| 5 | 4.77 ± 0.021 | 23.5 ± 0.033 | 26.08 ± 0.030 | 47.46 ± 0.076 |
| 6 | 3.92 ± 0.015 | 25.3 ± 0.055 | 26.83 ± 0.070 | 22.65 ± 0.153 |
| 7 | 4.89 ± 0.021 | 22.6 ± 0.021 | 25.69 ± 0.025 | 61.69 ± 0.038 |
| 8 | 3.97 ± 0.012 | 25.5 ± 0.046 | 26.74 ± 0.020 | 17.68 ± 0.026 |
| 9 | 4.94 ± 0.01 | 22.3 ± 0.053 | 25.99 ± 0.025 | 64.13 ± 0.076 |
| 10 | 4.82 ± 0.010 | 22.9 ± 0.039 | 26.21 ± 0.040 | 48.19 ± 0.030 |
| 11 | 4.88 ± 0.010 | 23.6 ± 0.044 | 26.17 ± 0.025 | 44.50 ± 0.057 |
| 12 | 4.12 ± 0.010 | 25.7 ± 0.030 | 26.67 ± 0.030 | 25.93 ± 0.021 |
| 13 | 4.00 ± 0.015 | 26.0 ± 0.011 | 27.01 ± 0.020 | 15.50 ± 0.015 |
| 14 | 6.81 ± 0.020 | 22.82 ± 0.017 | 25.55 ± 0.072 | 82.30 ± 0.055 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Xu, Y.; Guo, K.; Cui, W.; Li, Y.; Hou, M. Improving the Nutritional Value and Safety of Cotton Stalk Feed via Response Surface Methodology and Co-Fermentation Techniques. Fermentation 2025, 11, 124. https://doi.org/10.3390/fermentation11030124
Li K, Xu Y, Guo K, Cui W, Li Y, Hou M. Improving the Nutritional Value and Safety of Cotton Stalk Feed via Response Surface Methodology and Co-Fermentation Techniques. Fermentation. 2025; 11(3):124. https://doi.org/10.3390/fermentation11030124
Chicago/Turabian StyleLi, Kunyi, Yuansheng Xu, Kai Guo, Weidong Cui, Yang Li, and Min Hou. 2025. "Improving the Nutritional Value and Safety of Cotton Stalk Feed via Response Surface Methodology and Co-Fermentation Techniques" Fermentation 11, no. 3: 124. https://doi.org/10.3390/fermentation11030124
APA StyleLi, K., Xu, Y., Guo, K., Cui, W., Li, Y., & Hou, M. (2025). Improving the Nutritional Value and Safety of Cotton Stalk Feed via Response Surface Methodology and Co-Fermentation Techniques. Fermentation, 11(3), 124. https://doi.org/10.3390/fermentation11030124






