Abstract
The biodiversity of several beekeeping environments and honey samples was studied. The bacterial and yeast counts ranged from 0.00 to 5.19 Log CFU/g and from 0.00 to 3.33 Log CFU/g, respectively, presenting significant differences between the values. Of the honey samples, two of them exceeded the legislative limit established for total aerobic bacteria (5.19 Log CFU/g and 5.03 Log CFU/g). A total of ninety-eight yeast strains were isolated, with eight different species: Candida albicans, Dekkera anomala, Zygosaccharomyces rouxii, Z. mellis, Kazachstania unispora, Meyerozyma guillermondii, Saccharomyces cerevisiae, and S. unisporus. This implies a low microbial biodiversity and a low genetic variability index (D = 0.116 and 6–19%, respectively) due to the large number of genetically identical individuals found in each species. To select the most adequate strains for mead elaboration (with pure and mixed cultures), the fermentation capacity and organoleptic characteristics were studied. The best yeasts were chosen for the “pure culture fermentation” of honey (Saccharomyces spp. M11A2) and for mixed sequential inoculation (K. unispora M17A2). Both of the yeasts were isolated from honey and selected for their high fermentative capacity and resistance to ethanol. The results of the sensory analysis of the meads that were produced, in addition to one commercial product, were compared and indicated that the one inoculated with the pure culture had the best overall impression.
1. Introduction
At present, beekeeping occupies a very important place in the agricultural economy. The main products found in a beehive are honey, pollen, royal jelly, and propolis, which enter the international market after being produced with “clean technology”, leading to an increase in their demand by consumers [1]. Since ancient times, these products have been used for their medicinal and nutritional properties. From an environmental perspective, they are widely recognized for their benefits in pollination, which result in a significant increase in crop yields. The products (nectar and pollen) are also known for conducting indirect pest control by competing with plant food for the attention of insects.
However, of all the aforementioned products, honey is currently the most important. A major product of the hive, honey, is defined, according to the Codex Alimentarius [2], as “the natural sweet substance produced by Apis mellifera bees from the nectar of plants or from secretions of living parts of plants or excretions of plant-sucking insects left on living parts of plants, which the bees collect, transform and combine with specific substances of their own, and deposit, dehydrate, store and leave in the comb to mature and age”. Honey is essentially composed of different sugars, predominantly fructose and glucose, in addition to other substances such as organic acids, enzymes, and solid particles derived from the collection. The color of honey varies from almost colorless to dark brown. Its consistency may be fluid, viscous, or fully or partially crystallized. Flavor and aroma vary but derive from the plant of origin. The composition of honey is very difficult to determine, since it depends mainly on the origin of the flowers. It is a food that provides energy and is easily assimilated [3].
The Codex Alimentarius [2] and Royal Decree 1049/2003 [4] outline a series of characteristics and parameters with which a honey must comply to be considered a quality product. During the whole process of harvesting, packaging, and marketing honey and pollen, compliance must be maintained either with Regulation (EC) No. 852/2004 of the European Parliament of April 29, 2004 [5] on the hygiene of foodstuffs, or with regulations that replace it. In addition, the national or regional provisions that develop the aforementioned regulation must be followed.
Conversely, Codex Alimentarius Standard 12-1981 [2] indicates that apiculture products must comply with the microbiological criteria established in Commission Regulation (EC) No. 2073 of 15 November 2005 [6], as amended by Regulation (EC) No. 1441/2007 [7].
Honey is a medium with high osmotic pressure that hinders the growth of microorganisms. Even so, different genera of bacteria, yeasts, and molds with normally low microbial loads may be present. They can come from the same bacterial and fungal biota of the bees, as well as from flower nectar, pollen, or honeydew [8,9]. Generally, the biota will be composed of bacteria of the genus Bacillus and Clostridium and by yeasts of the genus Saccharomyces, with S. bisporus, S. rouxii, and S. bailii [8,10] being the most frequent. Other yeasts such as Zygosaccharomyces spp., Candida spp., and Rhodotorula spp. may also be present [9,11]. The presence of molds is associated with improper handling and storage of honey. Recognized among the molds are genera such as Aspergillus, Chaetomium, Penicillium, Peyronelia, and Mucor. In addition, it must be considered that poor handling during honey collection, processing, and storage can lead to contamination with pathogenic microorganisms such as Staphylococcus aureus, St. epidermidis, Pseudomonas aeruginosa, Escherichia coli, Salmonella enteritidis, Listeria monocytogenes, and A. niger [12].
Mead, also known as honey wine or honey water, is a traditional alcoholic beverage obtained from the fermentation of a mixture of water and honey. Its alcohol content ranges from 8 to 18%v/v [13,14]. Mead is one of the oldest fermented beverages, having originated in Africa from spontaneous fermentation performed by yeasts present in a jar of honey diluted by rain. In Europe, mead was heavily consumed by the Greeks, Celts, Saxons, and northern barbarians [15]. From the eleventh century onward, viticulture began to expand, and mead was displaced as a popular alcoholic beverage [16]. At present, mead is in great demand and acceptance in certain countries of the world, especially Venezuela. Also, due to the growing social demand in Europe, a rising number of beekeepers are interested in the elaboration of the product in an artisanal/traditional way.
In the present day, this ancestral fermented beverage has regained increasing interest, not only for being a sensory delight but also for its nutritional richness, which includes vitamins and minerals [17,18]. Other bioactive compounds are present and contribute beneficial properties, such as antimicrobial, anti-inflammatory, energetic, and antibiotic activity, to the beverage [18,19].
The objectives of the present study are focused on the microbiological analysis of bee products and environments as well as the identification of possible isolates. This will be used to select the yeast with the best fermentative properties for mead production, which are specific to the beekeeping ecosystem and perfectly adapted to the characteristics of the product.
2. Materials and Methods
2.1. Collection of Samples
Samples were provided directly by beekeepers in four different municipalities of Sierra de Cuenca, Spain. Twenty-three honey samples were taken from harvests between 2017 and 2021, including monofloral honeys (rosemary, thyme, safflower, saffron, lavender, morquera), milflores, and holm oak honeydew.
A sampling of the beekeeping environment was conducted in seven different apiaries located in the honey collection points mentioned above. Notably, samples were taken of the atmosphere, the surface of the hives, honeycombs, pollen, bees, and flowers located in the apiaries themselves. Honey extracted from wax and honey samples without elimination of impurities was also taken.
For atmosphere sampling, a microbiological control air sampler (Spin Air; IUL Instruments: Barcelona, Spain) was used that operated according to the impaction method. Microorganisms from a defined volume of air (100 L) at a flow rate of 100 L/min were retained on a Petri dish. The culture media used for yeast growth was Yeast extract Peptone Dextrose agar (YPD agar) (Pronadisa, Madrid, Spain) supplemented with biphenyl (400 μg/L), chloramphenicol (200 μg/L), and ampicillin (200 μg/L) to inhibit the growth of molds and bacteria. For bacterial growth, Tryptic Soy Agar (TSA) was used in addition to biphenyl (400 μg/L) and cycloheximide (200 μg/L) for the inhibition of molds and yeasts.
Surface sampling was performed on wooden hives populated with bees and on the comb. A sterile swab was used, and lawn seeding was performed on YPD and TSA with the corresponding antibiotics and antifungals, as mentioned above. Pollen and flowers were collected with sterile tweezers and placed in sterile stomacher bags (Figure 1). To extract possible yeasts present on the bees, the insects were retained on a plate for a few seconds, with their legs (the part with the highest microbial load) placed on the agar.

Figure 1.
Pollen sample collection.
All samples were kept at 4 °C until analysis was completed within a maximum of 24 h.
2.2. Microbial Counts
To obtain bacterial and yeast counts in the honey samples, 5 g of previously homogenized honey was taken and added to 45 mL of 0.9% (w/v) saline solution in a sterile flask. The corresponding dilutions were then prepared and seeded onto supplemented TSA and YPD agar plates, in duplicate, using the Eddy Jet 2 spiral slide (IUL Instruments, Barcelona, Spain). The bacterial and yeast plates were incubated at 37 °C and 30 °C, respectively, for 2–10 days each. Thereafter, the plates were counted with the Flash & Go automatic colony counter (IUL Instruments, Barcelona, Spain). The counts were expressed as CFU/g of honey. The results were compared with Commission Regulation (EC) No. 2073 of 15 November 2005 [6], as amended by Regulation (EC) No. 1441/2007 [7].
2.3. Yeast Isolation and Purification
For the isolation and purification of yeasts in the honey samples, the plates that were counted in the previous section were used. For the identification of yeasts obtained from the rest of the beekeeping environments (surface, pollen, flowers, and bees), yeasts were grown by sample incubation in YPD broth. In the specific case of pollens and flowers, the samples were homogenized with a stomacher (Masticator Homogeniser, IUL Instruments, Barcelona, Spain) in peptone water.
In all cases, samples were incubated in flasks at 30 °C for 48 h and with agitation (120 rpm). Then, the samples were seeded on YPD agar plates and incubated at 30 °C for 2–10 days. The colonies obtained with different morphologies were selected and reseeded with the 3-spin technique in order to obtain pure cultures.
2.4. Genotyping, Identification, and Sequencing of Yeasts
A RAPD-PCR (random amplification of polymorphic DNA) analysis was performed to genotype the samples, following the methodology of Padilla et al. [20]. The M13 (5′-GAGGGTGGCGGTTCT-3′) primer was used, having been supplied by Macrogen (Madrid, Spain). The reactions were performed in a total volume of 20 μL, and the amplification was conducted using a Life Touch thermal cycler (Bioer, Barcelona, Spain). The conditions included an initial denaturation step at 94 °C for 120 s, followed by 40 cycles of 94 °C for 60 s, 45 °C for 20 s, and 72 °C for 120 s. The final elongation step was performed at 72 °C for 10 min. The amplified products were resolved by electrophoresis (70 V for 1.5 h) on 2% (w/v) agarose stained with Gel Green (6x). A DNA molecular weight marker (100 bp ladder (Biotools) was used as a positive control.
Next, following the methodology of Padilla et al., the PCR-RFLP (Restriction Fragment Length Polymorphism) technique was applied to the different isolates that were found in order to identify them at the species level [20]. The rRNA gene region was amplified in the Life Touch thermal cycler. The primer pairs used to amplify the ITS region were ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Macrogen, Madrid, Spain). The thermal cycling parameters consisted of an initial denaturation at 95 °C for 10 min, followed by 36 cycles of denaturation at 95 °C for 60 s, annealing at 55.5 °C for 60 s, and extension at 72 °C for 90 s, with a final extension at 72 °C for 10 min. The PCR products were digested with the restriction endonucleases Hae III, Hinf I, and Hha I (Thermo Scientific, Vilnius, Lithuania). The samples were incubated, along with the corresponding enzymes, at 37 °C for 8 h, with slight agitation. Then, the results were visualized by electrophoresis (in the same conditions mentioned above).
The obtained results were entered into the Yeast-ID database to determine which microorganism was involved. Yeast-ID allows the rapid identification of yeasts based on a restriction analysis of the 5,8S rRNA coding gene region and the 5,8S-ITS transcribed intergenic regions.
A representative strain of each species was sent to the company Macrogen SPAIN (Madrid, Spain). The purpose of doing so was to obtain the sequence of each of the identified species and corroborate the identification that had been made. The results that were received were introduced into the bioinformatics tool BLAST (Basic Local Alignment Search Tool), which has the objective of finding regions of local similarity between proteins, namely DNA or RNA sequences. The findings are based on sequences from different databases, and the percentage of coincidence with each of the nomenclatures (species) offered is returned.
2.5. Biodiversity of Beekeeping Environments
To determine the biodiversity of beekeeping environments, the Simpson Index (D) was used. This is defined as the coefficient of biodiversity of species in an environment or community, D, which varies between 0 (no biodiversity) and 1 (infinite biodiversity). To calculate D, the following formula is applied, where “n” is the number of isolates per species and where “N” is the total number of isolates per sample.
The genetic variability that exists within the same species was calculated. A greater relationship with biodiversity equates to greater genetic variability and greater biodiversity.
2.6. Development of Mead
2.6.1. Selection of the Saccharomyces spp. Strain
Due to its high fermentative power, its production of and tolerance to ethanol, and its ability to avoid causing defects in the final product, the genus Saccharomyces was selected as the yeast that would perform the alcoholic fermentation of the substrate [21]. These starting conditions for mead production were chosen based on the works of Rodríguez Ardila and Rubiano Cepeda [22] and Hernández et al. [23].
All strains identified as Saccharomyces spp. were characterized in order to select the most suitable one based on its fermentative capacity, microbial viability during fermentation, and sensory analysis. A control strain, S. cerevisiae UCLMS 1, isolated from wine (Yeast Collection of the Yeast Biotechnology Laboratory, UCLM) and chosen for its good capacities, was used in parallel. To evaluate the fermentative capacity, fermentations were performed in a 100 mL sterile flask, where 25 g of homogenized honey and 90 mL of sterile distilled water were added.
According to Acosta Romero [8], the Brix value to start mead fermentation should be approximately 17° Brix to obtain 10%v/v ethanol. Conversely, the initial pH value established by Campos Chavez and Lapa Carrera [24] ranges from 3.1 to 3.8, and the cell population required for starting fermentation is a minimum of 6.3 Log CFU/mL [8].
The mixture of honey and water was pasteurized at 60 °C for 30 min in a water bath, and the Brix degrees and pH were adjusted to 15–20° Brix (Pocket Digital Refractometer|ATAGO CO., LTD, Bellevue, WA, USA) and 3.1–3.8 (Micro pH 2001, Crison, Barcelona, Spain), respectively.
Each flask was inoculated with the corresponding yeast strain at a concentration of 6.70 Log CFU/mL and incubated at 25 °C. This assay was performed in duplicate. The fermentation was monitored by analyzing the Brix degrees, sugar concentration, weight loss, pH, microbiological counts, and ethanol production. Fermentations were considered finished when the flask weight remained constant for at least 48 h.
To determine the content of reducing sugars (D-glucose/D-fructose), an enzymatic colorimetric method (Chemelex S.A., Barcelona, Spain) was used. Weight loss was monitored using a calibrated scale, which determines the release of carbon dioxide produced by the conversion of simple sugars (fructose and glucose). Ethanol was determined enzymatically (R-Biopharm AG, Darmstadt, Germany) by measuring absorbance in the ultraviolet region.
Yeast counts were performed every 48 h using the spiral plater and YPD agar plates. They were incubated for 48 h at 30 °C, and the colonies were counted with the automatic colony counter.
For the sensory analysis after the fermentation was finished, a consensus sensory profile was conducted in accordance with UNE-EN ISO 13299:2017 [25]. The panel was composed of seven highly trained experts in sensory analysis, all of whom belong to the staff of the Food Technology Area at the University of Castilla-La Mancha. First, in a previous session, the panelists selected attributes to describe the off-flavors present in the samples, as well as their intensities. Thereafter, the three samples were tested, and each expert evaluated the samples individually using the symbol “+” (3 as the maximum), according to the intensity perception. Once the attribute scores were collected by the panel leader, a debate was performed to assign the intensity of each attribute by consensus.
2.6.2. Elaboration of the Final Product
Once the best Saccharomyces spp. strain was chosen for the elaboration of the final mead, two fermentations were proposed: one using a pure culture (the Saccharomyces one was chosen) and another with a mixed culture by sequential inoculation (non-Saccharomyces plus Saccharomyces strains).
To select the non-Saccharomyces strain from all yeasts obtained, the indications of Jood et al. [26] were followed regarding the GRAS status and resistance to ethanol.
To perform the trial, 333 g of honey was homogenized in 1200 mL of sterile distilled water. The Brix degrees and the pH were adjusted to 15–20° Brix and 3.1–3.8, respectively. One of the flasks was inoculated with the S. cerevisiae chosen in pure culture (6.70 Log CFU/mL). The other flask was inoculated by sequential inoculation. A non-Saccharomyces strain was inoculated first, followed by the Saccharomyces strain after 48 h (both strains were inoculated with the same population, achieving a final population of 6.70 Log CFU/mL).
Fermentations were maintained at 25 °C, and the process was controlled by measuring the same variables and method as before: Brix degrees, sugar concentration, weight loss, pH, microbiological counts, and ethanol production. Again, fermentation was considered finished when the weight of the flask remained constant for at least 48 h.
All fermentations were carried out in triplicate.
2.6.3. Sensory Analysis
To characterize the products and determine whether there were differences between the mead made with the pure culture and that made with the mixed culture, a sensory analysis was performed. Moreover, other objectives of this work were to determine the organoleptic characteristics of the product and to observe whether the parameters were adequate for this fermented beverage. The analysis included a commercial product already available on the market.
A sensory profile test (ISO 13299:2016) [25] was conducted. Specifically, the test involved a quantitative descriptive profile, where each taster (ten in total) evaluated the samples using a list of attributes and scoring their intensity. Finally, an ordering test (UNE-ISO 8587:2010) [27] was performed for the “overall impression”, rated from lowest to highest. In each trial session, three different samples were evaluated.
- Sample M11A2: pure culture (Saccharomyces strain);
- Sample M11A2 + M17A1: mixed culture (non-Saccharomyces + Saccharomyces);
- Commercial sample: commercial product.
The experimental design was performed with a complete block design (BCE) that completed two sessions. A literature search was conducted to obtain the most appropriate attributes of the product description associated with mead [8], with the following conclusions.
- Olfactory attributes: intensity, frankness, fruity, fermentation, and overall quality;
- Taste attributes: sweetness, bitterness, acidity, smoothness, effervescence, balance, astringency, persistence, and overall quality.
2.7. Statistical Analysis
To perform the statistical analysis, the IBM SPSS Statistics 21 statistical program was used. An analysis of variance of one factor (ANOVA) was used together with Duncan’s test (p < 0.05) on the yeast isolation stage to evaluate if there were significant differences between the bacterial and yeast counts of samples.
These same analyses were used to evaluate whether there were significant differences between the final mead fermentation parameters (sugars (g/L), weight loss (g/L), pH, population (UFC/mL), and alcoholic strength (%v/v)) in both the preliminary test and the final elaborations.
Regarding the sensory analysis results, a one-factor analysis of variance (ANOVA) and a post hoc test (Duncan’s test) was applied to evaluate statistically if there were significant differences between samples. For the ordination test, the Friedman test was applied (at a significance level of 5%). To determine between which samples there were significant differences, the Minimum Significant Differences (MDSs) test was applied for a balanced complete block design, with a risk of α = 0.05 and Z = 1.96 (α = 0.05, bilateral normal probability).
3. Results and Discussion
3.1. Microbial Counts
The bacterial and yeast counts of the honey samples are shown in Table 1. The mean bacterial count was 2.72 Log CFU/g, and the yeast count was 1.22 Log CFU/g.

Table 1.
Bacteria and yeast counts in the different honey samples analyzed on the TSA and YPD media, respectively.
All the honey samples that were analyzed complied with the microbiological criteria established in Commission Regulation (EC) No. 2073 of 15 November 2005, [6] and its amendment, Regulation (EC) No. 1441/2007 [7]. The limits for total aerobic bacteria and yeasts were <5.00 Log CFU/g and <4.70 Log CFU/g, respectively, except for honey samples 3 and 4 (5.19 Log CFU/g and 5.03 Log CFU/g, respectively), which exceeded the limit established for total aerobic bacteria, and honey samples 12 and 13 (6.08 Log CFU/g and 6.42 Log CFU/g, respectively) for the yeast counts.
The results of the microorganism counts revealed significant differences (p = 0.00) between both bacterial and yeast counts, generating four groups for bacteria (F = 41.46) and six groups for yeast counts (F = 660.70).
Finola et al. [28] analyzed twenty-three honeys and obtained yeast and mold counts below 1 Log CFU/g. According to Reyes Rojas Zamorano [29], the ranges of these microorganisms are from 1.82 to 2.92 Log CFU/g and 0.87 to 1.34 Log CFU/g in unpasteurized and pasteurized honey, respectively.
Total aerobic bacteria were also quantified as ranging from 2.52 to 2.99 Log CFU/g and 1.43 to 2.39 Log CFU/g in unpasteurized and pasteurized honey, respectively. In all the data collected, the values were lower than those found in the present work. Commercial honeys are usually pasteurized and, therefore, have lower microorganism counts [30]. However, the honey samples in this study had not been subjected to any heat treatment, and most of them were not packaged. Rather, they were stored in previously sterilized glass jars after being cut at the sampling point. Nevertheless, as indicated above, the honey samples complied with current regulations.
3.2. Yeast Isolation and Purification
Yeasts from honey samples were isolates from the count plates. The rest of the samples (honeycomb, pollen, flowers, bees, hive surface) required a pre-conditioning phase in enriched broth using the methodology explained previously in Section 2.3.
After incubation, a total of ninety-eight yeasts were isolated from the honey samples and beekeeping environments. To obtain pure cultures of each yeast, the colonies were streak-seeded on a plate with the three-spin technique.
3.3. Genotyping, Identification, and Sequencing of Yeasts
To differentiate the genetic profiles of the isolates, the RAPDs-PCR technique was applied. An example is shown in Figure 2.
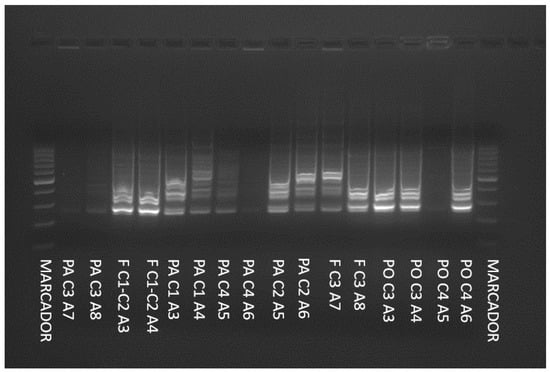
Figure 2.
RAPDs-PCR amplification patterns for some yeasts isolated from honey and beekeeping environments.
According to the results, which are outlined in Table 2, of the ninety-eight strains of yeast that originated from honey and beekeeping environments, only fourteen isolates were genetically differentiated.

Table 2.
Isolates of each species, isolate representative of each strain, and the confidence level of each identification by RAPDs-PCR.
After one representative yeast of each species was selected, the PRC-RFLP technique was used for identification (amplification of the ITS region and enzymatic restriction with Hae III, Hinf I, and Hha I). At the same time, all of the yeast species were sequenced. The YEAST ID was used for assigning an adequate name, and several bibliographic sources were used to contrast the results [31,32].
Of the fourteen yeasts, only ten were identified. The results are shown in Table 3, where the amplification and cut-off sizes of each of the restriction enzymes used are indicated, with 80% confidence.

Table 3.
Identification of the strains by PCR-RFLP and DNA sequencing by ID database and BLAST. The indicated species has 80% confidence in its identification.
The yeasts present in honey are generally of the Saccharomyces spp. genus [29]. However, other authors indicate that there is a probability of growth of other yeast genera such as Candida spp., specifically, the species C. magnoliae and C. sorbosivorans; Pichia spp. (P. membranifaciens); or Debaryomyces spp. Species of the genus Zygosaccharomyces spp., such as Z. rouxii and Z. mellis, have also been identified [33].
Carvalho et al. [34] isolated twenty-four yeast strains in honey from Trás-os-Montes (Portugal). The strains belonged to nine different species: S. cerevisiae, C. magnoliae, C. parapsilosis, C. sorbosivorans, Rhodotorula mucilaginosa, Z. mellis, Z. rouxii, and P. membranifaciens. The genus Candida spp. accounted for more than 45% of the honey isolates.
It is increasingly recognized that the gut microbial community regulates a wide variety of important functions in their animal hosts, including health. However, the complex interactions between bee gut microorganisms and the environment remain unclear [35], so an obvious comparison is not possible.
The results found in the present work can be summarized as follows. Regarding the yeasts isolated from honey, the genera Saccharomyces and Zygosaccharomyces were mainly found, with the species S. cerevisiae, S. unisporus, Z. mellis, and Z. rouxii being notable. In the pollen samples, a total of six strains of D. anomala, which was the most common yeast in this type of sample, were isolated. Among the honeycomb samples analyzed, a large variety of yeasts were found, including C. albicans, D. anomala, S. cerevisiae, Z. rouxii, K. unispora, and M. guillermondii. Few yeasts were identified in the hive surface samples due to the presence of molds. However, one strain of K. unispora and two strains of M. guillermondii were differentiated. From the bee leg samples, five genera were differentiated: Candida, Dekkera, Saccharomyces, Zygosaccharomyces, and Kazachstania.
3.4. Biodiversity in Beekeeping Environments
Of the ninety-eight total isolates from honey samples and beekeeping environments, fourteen strains corresponding to six genera and eight different species were differentiated.
To calculate the biodiversity in the beekeeping environments that were studied, the Simpson Index (D) was used. The result of the D value was 0.116, which indicated a low microbial biodiversity compared to that of other environments [36]. These data reveal the adaptation to the media that the microorganisms require. As noted previously, the substrate conditions are very demanding in that very few strains are able to survive in beekeeping environments.
The genetic variability that exists within a single species indicates the biodiversity of strains in that sample (Table 3). The results present a low Simpson Index despite the high number of isolates of the same species, since many of them were genetically identical.
3.5. Development of Mead
3.5.1. Selection of Saccharomyces spp. Strain
Of the ninety-eight isolates, twenty belonged to the Saccharomyces genus. However, only three strains were different: two S. cerevisiae (M11A2, M15A2) and one S. unisporus (M13A2). To select the best one for the final product, the fermentations of a mead were performed. S. cerevisiae UCLM1 was also used as a positive control.
The initial parameters of the fermentations were adjusted (as described in Section 2.6.1) in the different tanks; the Brix degrees ranged from 16.9 to 18.5° Brix, the pH values were between 3.72 and 3.77, and the initial inoculum was 6.7 Log (CFU/mL).
Fermentations were monitored by measuring sugar, weight loss, pH, microbial growth, and ethanol production. The fermentations finished after 14–18 days, reaching an ethanol concentration around 8%v/v, as also reported by Rodríguez Ardila and Ruibiano Cepeda [22].
The results are shown in Table 4, where one can observe that there were no significant differences (p = 0.01) between the parameters measured for the mead made with each of the strains (at 18 days). The only exception was the sugar content (p = 0.90) in the UCLM1 sample with respect to the M15A2, which was higher in the latter.

Table 4.
Parameters calculated at the initial and final times of mead fermentation with each of the Saccharomyces strains.
The average final pH value of the meads was 3.08. In Romero’s study [8], the starting pH was 3.6, which decreased to 3.2 with the course of fermentation. A similar final pH was found in the study of Buschmann [37], who bottled meads with pHs in the range of 3.40 to 3.80.
Regarding microbial growth during mead fermentation, it was observed that strains M11A2 and M15A2 followed the same kinetics trend, with populations of 7.67 Log CFU/mL and 7.30 Log CFU/mL, respectively. Both strains reached their maximum population on the sixth day, finishing the fermentation after 15 days. Conversely, strain M13A2 reached its maximum population (6.94 Log CFU/mL) after 8 days, reaching a constant weight on the eighteenth day. Nevertheless, the control strain, UCLM1, was faster, reaching its maximum population (7.11 Log CFU/mL) on the fourth day.
For the results of the consensus sensory profile, the panelists selected, after a previous debate, the four odor attributes that best described the off-flavors of the mead samples: sweat, solvent, medicinal odor, and sulfur. Three scores were used to describe the aroma intensity of each sample: low (+), medium (++), and high (+++). Mead fermented with the strain M11A2 did not demonstrate any off-flavor. A low intensity of sulfur was evident in the mead fermented with M15A2. Lastly, the mead fermented with strain M13A2 had a high intensity of sweat odor and a low intensity of solvent and medicinal odors.
Considering all the results and the physicochemical, microbiological, and organoleptic analyses, it was decided that the most suitable strain for making the final mead was M11A2.
3.5.2. Elaboration of the Final Product
Among the strains isolated from honey samples and bee environments, the non-Saccharomyces M17A1 was identified as Kazachstania unispora, which is found with low frequency in nature on the surfaces of grapes in different countries, and several strains have shown good fermentation capacity similar to S. cerevisiae [25]. This strain was selected for sequential inoculation with a mixed culture, based on literature references and its wide use in the food industry shows that it is suitable for the development of this type of products [25].
Two fermentations were conducted: one with a pure culture of S. cerevisiae (M11A2) and the other with a sequential inoculation (M17A1 + M11A2), as described in the Materials and Methods section (Section 2.6.2).
The process was controlled with measurements of weight loss, sugar concentration, and yeast counts every 48 h, until the flasks achieved a constant weight for 48 h after 17 days.
As shown in Table 5, among the results of the final parameters, no significant differences were observed between the two products in terms of sugar content (p = 0.344) and alcohol content (p = 0.430). However, there were significant differences in weight loss (p = 0.009), which was lower in the mead inoculated with the mixed culture (M11A2 + M17A1). Regarding the final yeast counts, there were also significant differences (p = 0.002).

Table 5.
Initial and final parameters of the elaborated mead.
The results of the sensory analysis, specifically, the scalar test of the elaborated meads and the commercial product, are shown visually in Figure 3.
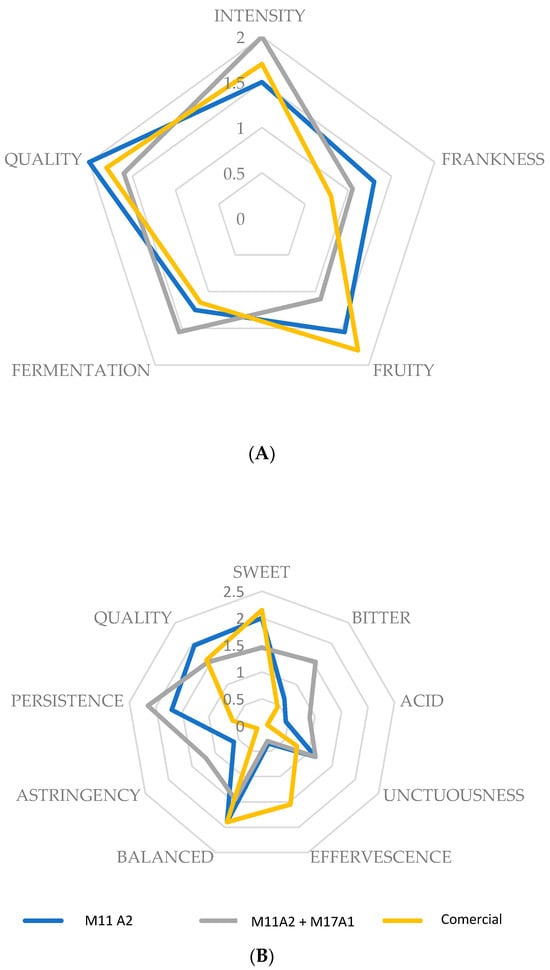
Figure 3.
Sensory spider plot of the olfactive (A) and taste (B) analysis scores of the mead fermented with strains M11A2 and M11A2 + M17A1, as well as a commercial mead. The intensity of each attribute was rated by the assessors using the scale (0) null, (1) light, (2) medium, and (3) strong.
In the olfactory attributes, no significant differences were observed (p = 0.180), except for fruitiness (p = 0.045), between the sample inoculated with mixed culture and the commercial sample, with the latter presenting higher values.
Regarding taste attributes, there were no significant differences in smoothness (p = 0376), balance (p = 0.182), and overall quality (p = 0.184). However, for the sweet attribute, there were differences (p = 0.008) between the same meads, as in the previous case (mixed and commercial cultivation), with the commercial one being sweeter. With respect to acid (p = 0.016), bitterness (p = 0.004), and astringency (p = 0.002), the M11 + M17A2 sample presented significantly higher values. Finally, the commercial sample showed higher effervescence (p = 0.000) but lower persistence (p = 0.00) than the other two samples. This could be due to external gassing, although the labelling did not include this information.
The experimental F value is 13.40, taking into account that j = 10 judges, p = three samples, Rn = the sum of product ordinations, and the tabulated F = 6.20 (95% confidence level). Since the F exp (13.40) was greater than the tabulated F (6.20), the null hypothesis was rejected with a risk of error of 5%; that is, there were significant differences between the three samples with respect to the overall impression attribute.
In determining between which products these differences occurred with a risk of α = 0.05 and Z = 1.96 (α = 0.05, bilateral normal probability), a value of 7.16 was obtained. Therefore, the M11A2 sample presents significant differences from the sample inoculated with the mixed culture (M11A2 + M17A1) and the commercial sample, which are statistically equal to each other. The best overall impression obtained was from the sample inoculated only with S. cerevisiae (M11A2).
4. Conclusions
The bacterial and yeast counts in the beekeeping environments and honey samples that were studied did not exceed the established legislative limits, except for four honey samples (3, 4, 12, and 13) that showed counts of bacteria higher than stabilized. Among the samples used, a total of ninety-eight yeast strains were isolated, corresponding to C. albicans, D. anomala, Z. rouxii, S. cerevisiae, S. unisporus, Z. mellis, K. unispora, and M. guillermondii, implying a low microbial biodiversity (Simpson index D = 0.116) and a low genetic variability index (between 6 and 19%).
The strain of the genus Saccharomyces spp. that presented the best fermentation capacity based on the data obtained for reducing sugars, weight loss, yeast counts, and ethanol production, as well as organoleptic characteristics, was M11A2. The selected candidate for mixed sequential inoculation was the non-Saccharomyces yeast (M17A2), belonging to the species Kazachstania uniparaelected, for its suitable characteristics demonstrated in food industries as referenced.
For the future final production of mead, the best overall process would be the one carried out in pure fermentation, using the strain S. cerevisiae M11A2 based on the results obtained in the present study.
Author Contributions
Conceptualization, M.A.-V.; methodology, M.A.-V. and P.F.-P.; validation, M.A.-V. and P.F.-P.; formal analysis, N.V.-L.; investigation, N.V.-L.; resources, M.A.-V. and P.F.-P.; data curation, N.V.-L. and P.F.-P.; writing—original draft preparation, N.V.-L. and P.F.-P.; writing—review and editing, M.A.-V. and P.F.-P.; visualization, M.A.-V.; supervision, M.A.-V. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ballesteros, H.H.; Vásquez, R.E. Determination of royal jelly production in rearing hives of different dimensions. Cienc. Tecnol. Agropecuaria 2007, 8, 75–81. [Google Scholar] [CrossRef]
- Codex Alimentarius. International Honey Standards CXS 12-1981. 2019. Available online: https://www.fao.org/fao-who-codexalimentarius/sh-proxy/en/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FStandards%252FCXS%2B12-1981%252FCXS_012s.pdf (accessed on 1 April 2024).
- Insuasty-Santacruz, E.; Martínez-Benavides, J.; Jurado-Gámez, H. Flora identification and nutritional analysis of bee honey for beekeeping production. Biotechnol. Agric. Agroind. Sect. 2016, 14, 37–44. [Google Scholar]
- BOE-A-2003-15598; Royal Decree 1049/2003, of 1 August 2003, Approving the Quality Standard for Honey. BOE: Madrid, Spain, 2003.
- BOE. Regulation (EC) 852/2004 of the European Parliament and of the Council of 29 April 2004 on the Hygiene of Foodstuffs; BOE: Madrid, Spain, 2004. [Google Scholar]
- BOE. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs; BOE: Madrid, Spain, 2005. [Google Scholar]
- BOE. Commission Regulation (EC) No 1441/2007 of 5 December 2007 Amending Regulation (EC) No 2073/2005 on Microbiological Criteria for Foodstuffs; BOE: Madrid, Spain, 2007. [Google Scholar]
- Acosta Romero, C. Evaluation of Alcoholic Fermentation for Mead Production. Master’s Thesis, Universidad Nacional de Colombia, Bogotá, Colombia, 2012. [Google Scholar]
- Narváez Arauz, M.E.; Ortiz Arana, C.O. Presence of Microorganisms in Bee Honey from Apiaries in the Municipalities of León and El Sauce as well as in Honeys Offered in Local Commerce, June–November 2010. National Autonomous University of Nicaragua 2010. Available online: http://riul.unanleon.edu.ni:8080/jspui/bitstream/123456789/3532/1/217935.pdf (accessed on 25 February 2024).
- Medina, A. Fermentation of Honey for Obtaining Hidromiel. Ferment. Honey Obtaining Hidromiel 2019, 1, 21–30. Available online: http://infometrica.org/index.php/syh/article/view/63/62 (accessed on 5 March 2024).
- García Flores, M. Physicochemical, Microbiological and Biotechnological Characterization of Bee Products. Master’s Thesis, Universidad de Castilla-La Mancha, Ciudad Real, Spain, 2019. [Google Scholar]
- Estrada, H.; del Mar Gamboa, M.; Chaves, C.; Arias, M.L. Evaluation of the antimicrobial activity of bee honey against Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa, Escherichia coli, Salmonella enteritidis, Listeria monocytogenes and Aspergillus niger. Arch. Latinoam. Nutr. 2005, 55, 167–171. [Google Scholar] [PubMed]
- Mendes-Ferreira, A.; Cosme, F.; Barbosa, C.; Falco, V.; Inês, A.; Mendes-Faia, A. Optimization of honey-must preparation and alcoholic fermentation by Saccharomyces cerevisiae for mead production. Int. J. Food Microbiol. 2010, 144, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Ramalhosa, E.; Gomes, T.; Pereira, A.P.; Dias, T.; Estevinho, L.M. Mead production: Tradition versus modernity. Adv. Food Nutr. Res. 2011, 63, 101–118. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Agriculture. FoodData Central. 2019. Available online: https://fdc.nal.usda.gov/fdc-app.html#/food-details/169640/nutrients (accessed on 10 March 2024).
- Barrios, C.; Principal, J.; Sánchez, J.; Guédez, J.C. Physicochemical characterization and sensory analysis of a handcrafted mead. Zootec. Trop. 2010, 28. Available online: http://ve.scielo.org/scielo.php?script=sci_arttext&pid=S0798-72692010000300002 (accessed on 10 March 2024).
- Azeredo, L.C.; Azeredo, M.A.A.; Dutra, V.M.L. Protein contents and physicochemical properties in honey samples of Apis mellifera of different floral origins. Food Chem. 2003, 80, 249–2543. [Google Scholar] [CrossRef]
- Silva, P.M.; Gauche, C.; Gonzaga, L.V.; Costa, A.C.O.; Feit, R. Honey: Chemical composition, stability, and authenticity. Food Chem. 2016, 196, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Abadio Finco, F.D.B.; Moura, L.L.; Silva, I.G. Physical and chemical properties of Apis mellifera L. hone. Food Sci. Technol. 2010, 30, 706–712. [Google Scholar] [CrossRef]
- Padilla, B.; Manzanares, P.; Belloch, C. Yeast species and genetic heterogeneity within Debaryomyces hansenii along the ripening process of traditional ewes’ and goats’ cheeses. Food Microbiol. 2014, 38, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Oteruelo Morales, B. Study of the Tolerance to High Sugar Concentrations of wine Saccharomyces Cerevisiae. Bachelor’s Thesis, Universidad de Valladolid, Valladolid, Spain, 2018. [Google Scholar]
- Rodríguez Ardila, N.; Rubiano Cepeda, M.E. Proceso de elaboración de hidromiel. Mosquera, Cundinamarca: Servicio Nacional de Aprendizaje (SENA). Centro de Biotecnología Agropecuaria, 2019. ISBN 978-958-15-0674-3. Available online: https://repositorio.sena.edu.co/bitstream/handle/11404/7441/Proceso_elaboracion_hidromiel.pdf?sequence=1&isAllowed=y (accessed on 12 March 2024).
- Hernández, C.; Blanco, A.; Quicazán, M.C. Establishment of mead brewing conditions by design of experiments. 2014. Available online: https://www.researchgate.net/publication/295548282 (accessed on 20 March 2024).
- Campos Chavez, C.J.; Lapa Carrera, E.W. Determination of the Optimum Parameters in the Production of Mead, Using Two Types of Natural Binders as Clarifiers. Bachelor’s Thesis, National University of Central Peru, Huancayo, Peru, 2014. [Google Scholar]
- UNE-EN ISO 13299:2017; Análisis sensorial. Metodología. Guía general para establecer un perfil sensorial. Asociación Española de Normalización y Certificación: Madrid, Spain, 2017.
- Jood, I.; Hoff, J.W.; Mathabatha; Setati, E. Evaluating fermentation characteristics of Kazachstania spp. and their potential influence on wine quality. World J. Microbiol. Biotechnol. 2017, 33, 129. [Google Scholar] [CrossRef]
- UNE-EN ISO 8587; Análisis Sensorial. Metodología. Ordenación. Asociación Española de Normalización y Certificación: Madrid, Spain, 2006.
- Finola, M.S.; Lasagno, M.C.; Marioli, J.M. Microbiological and chemical characterization of honeys from central Argentina. Food Chem. 2007, 100, 1649–1653. [Google Scholar] [CrossRef]
- Reyes Rojas, H.D. Efecto de la Pasteurización y Proveedor Apícola en las Características Microbiológicas y Químicas de la Miel de Abeja. Bachelor’s Thesis, Universidad Zamorano, Tegucigalpa, Honduras, 2012. [Google Scholar]
- Pascual Anderson, M.; Calderon, Y.; Pascual, V. Microbiología Alimentaria. Metodología Analítica para Alimentos y Bebidas, 2nd ed.; Ediciones Diaz de Santos: Madrid, Spain, 2007; ISBN 978-84-7978-424-9. [Google Scholar]
- Esteve-Zarzoso, B.; Zorman, T.; Belloch, C.; Querol, A. Molecular Characterisation of the Species of the Genus Zygosaccharomyces. Syst. Appl. Microbiol. 2003, 26, 404–411. [Google Scholar] [CrossRef] [PubMed]
- García-Béjar, B.; Árevalo-Villena, M.; Briones, A. Characterization of yeast population from unstudied natural sources in La Mancha region. J. Appl. Microbiol. 2021, 130, 650–664. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Rocha, A.; Estevinho, M.L.F.; Choupina, A. Identification of honey yeast species based on RFLP analysis of the ITS region. CYTA-J. Food 2009, 5, 11–17. [Google Scholar] [CrossRef][Green Version]
- Carvalho, C.; Merinho, S.; Estevinho, M.; Choupina, A. Especies de levaduras asociadas a la miel: Métodos diversos de identificación. Zootecnia 2010, 59, 103–113. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Fernández-Pacheco, P.; Rosa, I.Z.; Arévalo-Villena, M.; Gomes, E.; Pérez, A.B. Study of potential probiotic and biotechnological properties of non-Saccharomyces yeasts from fruit Brazilian ecosystems. Braz. J. Microbiol. 2021, 52, 2129–2144. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, R.A.H. Producción y Caracterización de Mead (Hidromiel) Espumante. Ph.D. Thesis, Universidad Austral de Chile, Los Ríos, Chile, 2015. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).