Effect of Mixed Cultures on Microbiological Development in Berliner Weisse Beer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Count
2.2. Analytical Techniques
2.3. Quantification of Esters and Higher Alcohols
2.4. Wort Preparation
2.5. Bottle Conditioning
2.6. Microorganisms
2.7. Conditions of Storage, Detection, and Enumeration
2.8. Isolating Microorganisms Using Selective Media
2.9. Propagation of the Yeast and Bacterial Strains
2.10. Bright Field Microscopy
2.11. Cell Viability Determination
3. Results
3.1. Microorganismsand Isolating Using Selective Media
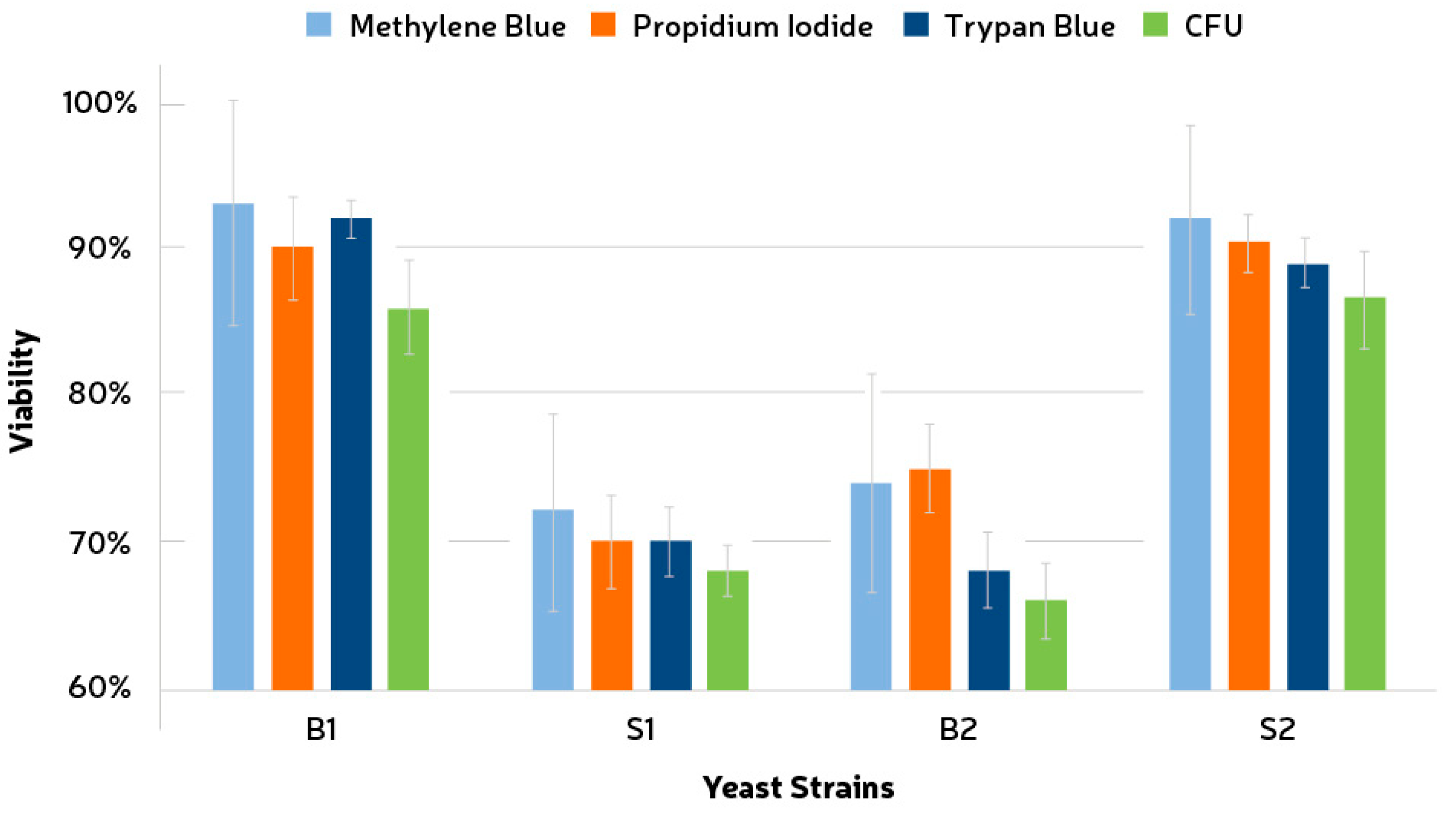
3.2. Viability Tests
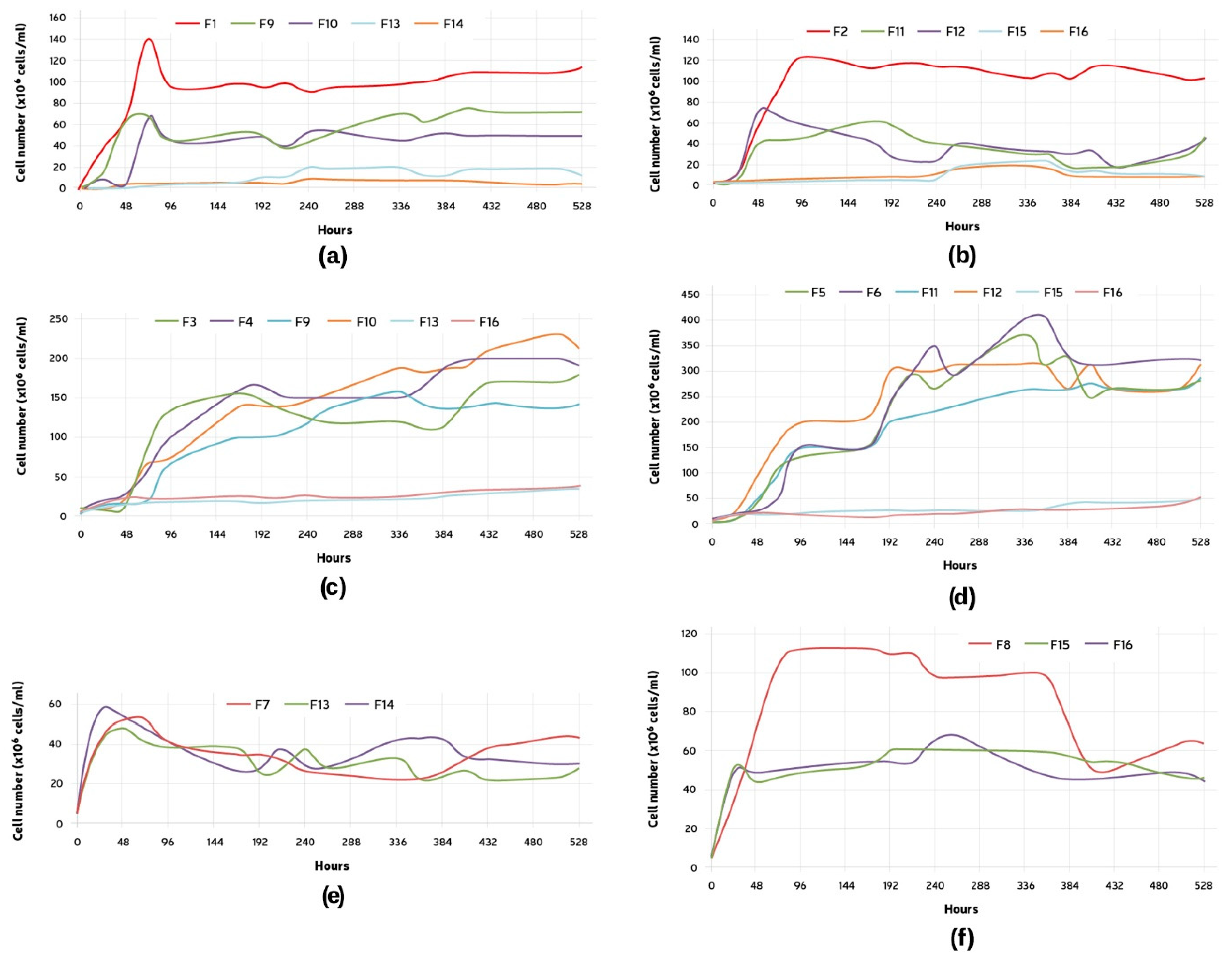
3.3. Comparison between Single and Mixed Fermentations: Cell Count
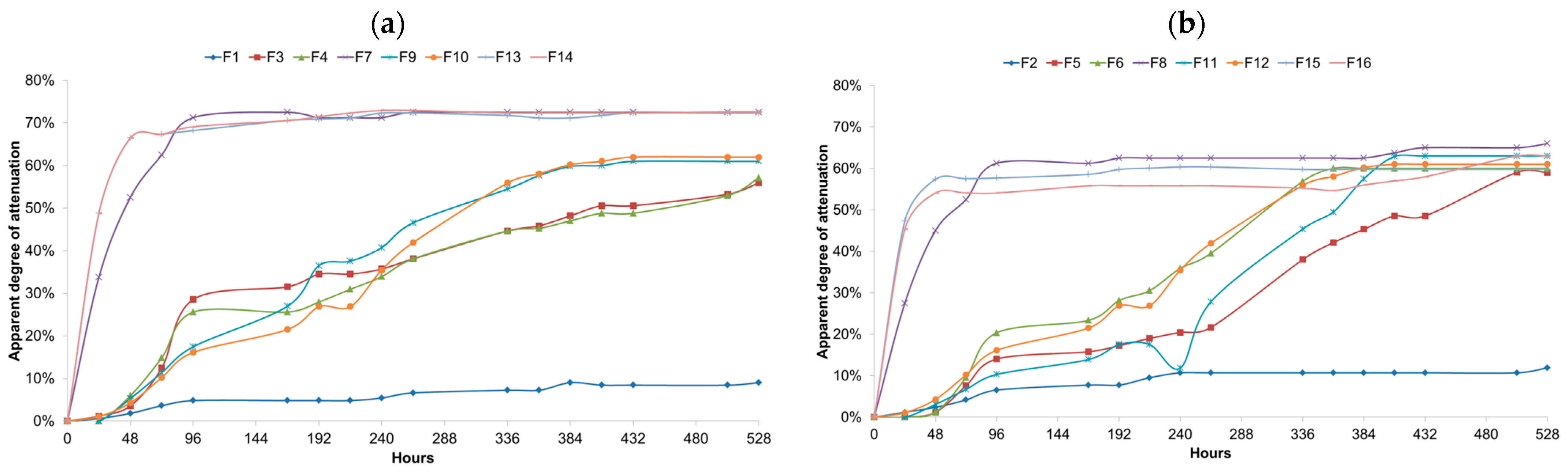
3.4. Comparison between Single and Mixed Fermentations: Titratable Acidity (TA) and pH
3.5. Comparison between Single and Mixed Fermentations: Yeast Cell Viability
3.6. Comparison of the Fermentation Parameters during the Mixed Fermentations
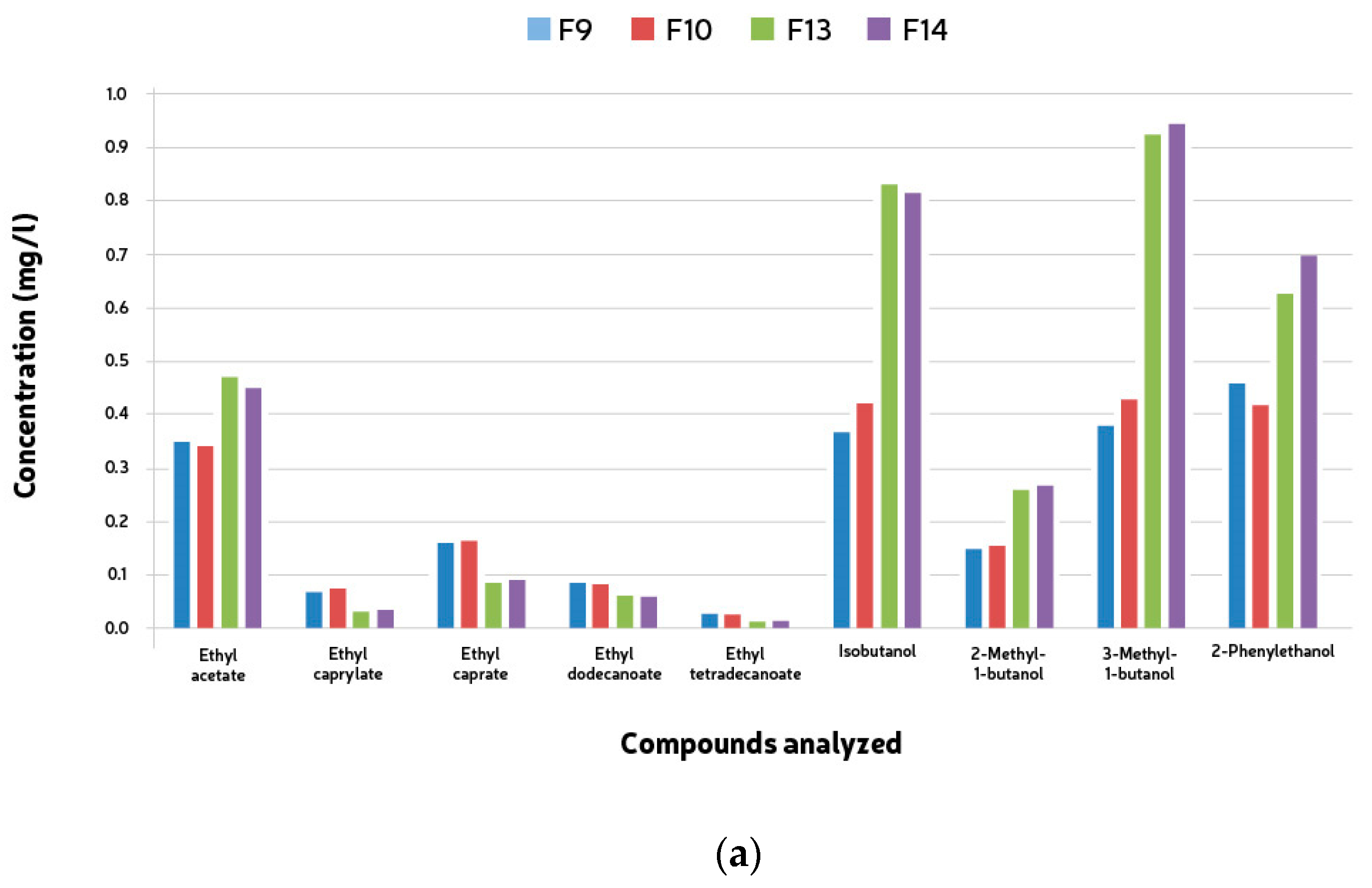
3.7. Quantification of Esters and Higher Alcohols
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dziedziński, M.; Stachowiak, B.; Kobus-Cisowska, J.; Kozłowski, R.; Stuper-Szablewska, K.; Szambelan, K.; Górna, B. Supplementation of beer with Pinus sylvestris L. shoots extracts and its effect on fermentation, phenolic content, antioxidant activity and sensory profiles. Electron. J. Biotechnol. 2023, 63, 10–17. [Google Scholar] [CrossRef]
- Bongaerts, D.; De Roos, J.; De Vuyst, L. Technological and environmental features determine the uniqueness of the lambic beer microbiota and production process. Appl. Environ. Microbiol. 2021, 87, e00612-21. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Fancello, F.; Balmas, V.; Dettori, M.; Motroni, A.; Zara, G.; Budroni, M. Microbial communities and malt quality of durum wheat used in brewing. J. Inst. Brew. 2019, 125, 222–229. [Google Scholar] [CrossRef]
- Walker, G.M. Yest: Physiology and Biotechnology; Wiley: Hobokrn, NJ, USA, 1998. [Google Scholar]
- De Roos, J.; De Vuyst, L. Microbial acidification, alcoholization, and aroma production during spontaneous lambic beer production. J. Sci. Food Agric. 2019, 99, 25–38. [Google Scholar] [CrossRef]
- Vermote, L.; De Roos, J.; Cnockaert, M.; Vandamme, P.; Weckx, S.; De Vuyst, L. New insights into the role of key microorganisms and wooden barrels during lambic beer fermentation and maturation. Int. J. Food Microbiol. 2023, 394, 110163. [Google Scholar] [CrossRef]
- Dysvik, A.; La Rosa, S.L.; De Rouck, G.; Rukke, E.O.; Westereng, B.; Wicklund, T. Microbial dynamics in traditional and modern sour beer production. Appl. Environ. Microbiol. 2020, 86, e00566-20. [Google Scholar] [CrossRef]
- Annemüller, G.; Manger, H.J.; Lietz, P. Die Berliner Weiße; VLB Berlin: Berlin, Germany, 2008. [Google Scholar]
- Tonsmeire, M. American Sour Beer: Innovative Techniques for Mixed Fermentations; Brewers Publications: Boulder, CO, USA, 2014. [Google Scholar]
- Van Oevelen, D.; Spaepen, M.; Timmermans, P.; Verachtert, H. Microbiological aspects of spontaneous wort fermentation in the production of lambic and gueuze. J. Inst. Brew. 1977, 83, 356–360. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Bamforth, C.W. The microbiology of malting and brewing. Microbiol. Mol. Biol. Rev. 2013, 77, 157–172. [Google Scholar] [CrossRef]
- Keersmaecker, J. The mystery of lambic beer. Sci. Am. 1996, 275, 56–62. [Google Scholar] [CrossRef]
- BJCP—Beer Judge Certification Program. Beer Style Guide. Brazil, 2021. Available online: https://www.bjcp.org/bjcp-style-guidelines/ (accessed on 20 June 2024).
- Yakobson, C. Pure culture fermentation characteristics of Brettanomyces yeast species and their use in the brewing industry. In The Brettanomyces Project; Heriot-Watt University, Institute of Brewing and Distilling: Edinburgh, Scotland, 2010. [Google Scholar]
- Morneau, A.D.; Zuehlke, J.M.; Edwards, C.G. Comparison of media formulations used to selectively cultivate Dekkera/Brettanomyces. Lett. Appl. Microbiol. 2011, 53, 460–465. [Google Scholar] [CrossRef]
- Colomer, M.S.; Chailyan, A.; Fennessy, R.T.; Olsson, K.F.; Johnsen, L.; Solodovnikova, N.; Forster, J. Assessing population diversity of Brettanomyces yeast species and identification of strains for brewing applications. Front. Microbiol. 2020, 11, 637. [Google Scholar] [CrossRef] [PubMed]
- Freek Spitaels, F.; Wieme, A.D.; Janssens, M.; Aerts, M.; Landschoot, A.V.; Vuyst, L.; Vandamme, P. The microbial diversity of an industrially produced lambic beer shares members of a traditionally produced one and reveals a core microbiota for lambic beer fermentation. Food Microbiol. 2015, 49, 23–32. [Google Scholar] [CrossRef] [PubMed]
- He, Z.W.; Li, A.H.; Tang, C.C.; Zhou, A.J.; Liu, W.; Ren, Y.X.; Li, Z.; Wang, A. Biochar Regulates Anaerobic Digestion: Insights to the Roles of Pore Size. Chem. Eng. J. 2024, 480, 148219. [Google Scholar] [CrossRef]
- Postigo, V.; Sánchez, A.; Cabellos, J.M.; Arroyo, T. New Approaches for the Fermentation of Beer: Non-Saccharomyces Yeasts from Wine. Fermentation 2022, 8, 280. [Google Scholar] [CrossRef]
- Dysvik, A.; Liland, K.H.; Myhrer, K.S.; Westereng, B.; Rukke, E.-O.; de Rouck, G.; Wicklund, T. Pre-Fermentation with Lactic Acid Bacteria in Sour Beer Production. J. Inst. Brew. 2019, 125, 342–356. [Google Scholar] [CrossRef]
- He, Y.; Zhao, J.; Yin, H.; Deng, Y. Transcriptome Analysis of Viable but Non-Culturable Brettanomyces Bruxellensis Induced by Hop Bitter Acids. Front. Microbiol. 2022, 13, 902110. [Google Scholar] [CrossRef] [PubMed]
- Nasuti, C.; Ruffini, J.; Sola, L.; Di Bacco, M.; Raimondi, S.; Candeliere, F.; Solieri, L. Sour Beer as Bioreservoir of Novel Craft Ale Yeast Cultures. Microorganisms 2023, 11, 2138. [Google Scholar] [CrossRef]
- Lopitz-Otsoa, F.; Rementeria, A.; Elguezabal, N.; Garaizar, J. Kefir: Una Comunidad Simbiótica de Bacterias y Levaduras Con Propiedades Saludables. Rev. Iberoam. Micol. 2006, 23, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Ghesti, G.F.; Carvalho, I.; Carmo, T.; Suarez, P.A.Z. A newer source of microorganism to produce Catharina Sour beers. Food Sci. Technol. 2023, 43, 10–22. [Google Scholar] [CrossRef]
- Iattici, F.; Catallo, M.; Solieri, L. Designing New Yeasts for Craft Brewing: When Natural Biodiversity Meets Biotechnology. Beverages 2020, 6, 3. [Google Scholar] [CrossRef]
- Ciosek, A.; Fulara, K.; Hrabia, O.; Satora, P.; Poreda, A. Chemical Composition of Sour Beer Resulting from Supplementation the Fermentation Medium with Magnesium and Zinc Ions. Biomolecules 2020, 10, 1599. [Google Scholar] [CrossRef] [PubMed]
- Peyer, L.C.; Zannini, E.; Jacob, F.; Arendt, E.K. Growth Study, Metabolite Development, and Organoleptic Profile of a Malt-Based Substrate Fermented by Lactic Acid Bacteria. J. Am. Soc. Brew. Chem. 2015, 73, 303–313. [Google Scholar] [CrossRef]
- Ramanan, M.; Fox, G.P.; Marco, M.L. Beer for Live Microbe Delivery. J. Funct. Foods 2024, 113, 105987. [Google Scholar] [CrossRef]
- Fu, X.; Guo, L.; Li, Y.; Chen, X.; Song, Y.; Li, S. Transcriptional Analysis of Mixed-Culture Fermentation of Lachancea Thermotolerans and Saccharomyces Cerevisiae for Natural Fruity Sour Beer. Fermentation 2024, 10, 180. [Google Scholar] [CrossRef]
- Reitenbach, A.F.; Iwassa, I.J.; Barros, B.C.B. Production of Functional Beer with the Addition of Probiotic: Saccharomyces Boulardii. Res. Soc. Dev. 2021, 10, e5010212211. [Google Scholar] [CrossRef]


| Strain | ID | Method of Identification |
|---|---|---|
| S1 | Saccaromyces cerevisiae (OK3) | RT-PCR |
| S2 | Saccaromyces cerevisiae (S2RA) | RT-PCR |
| B1 | Dekkera bruxellensis | RT-PCR |
| B2 | Dekkera anomala | RT-PCR |
| L1 | Lactobacillus brevis (Dsn7 623 s) | |
| L2 | Lactobacillus parabrevis (B201 TUM) |
| Selective Media | VLB-S7-S | NBB®-A | MYPG + Cycloheximide | ||||
|---|---|---|---|---|---|---|---|
| Growth Condition | Aerobic | Anaerobic | Aerobic | Anaerobic | Aerobic | Anaerobic | |
| Strains | S1 | − | − | − | − | − | − |
| S2 | − | − | − | − | − | − | |
| B1 | +/− | +/− | ++ | ++ | ++ | ++ | |
| B2 | +/− | − | ++ | + | ++ | ++ | |
| L1 | +/− | ++ | − | + | − | +/− | |
| L2 | +/− | ++ | − | ++ | − | +/− | |
| Fermentation ID | Final pH | Microorganisms |
|---|---|---|
| F6 | 3.29 | B2 b |
| F1 | 3.32 | L1 |
| F5 | 3.35 | B2 a |
| F2 | 3.37 | L2 |
| F3 | 3.53 | B1 a |
| F9 | 3.58 | L1 + B1 a |
| F10 | 3.60 | L1 + B1 b |
| F4 | 3.63 | B1 b |
| F12 | 3.64 | L2 + B2 b |
| F11 | 3.67 | L2 + B2 a |
| F14 | 3.85 | L1 + S1 + B1 b |
| F13 | 3.90 | L1 + S1 + B1 a |
| F15 | 3.94 | L2 + S2 + B2 a |
| F16 | 4.00 | L2 + S2 + B2 b |
| F7 | 4.12 | S1 |
| F8 | 4.20 | S2 |
| Fermentation ID | Strains | Days | ||||
|---|---|---|---|---|---|---|
| 0 | 1 | 8 | 15 | 22 | ||
| F9 | L1:B1 | 1:5 | 1:0.4 | 1:2 | 1:2.3 | 1:2 |
| F11 | L2:B2 | 1:5 | 1:8 | 1:3.5 | 1:9 | 1:7 |
| F13 | L1:B1:S1 | 1:5:5 | 1:9:43 | 1:1.5:2 | 1:1.8:1.8 | 1:2.8:2 |
| F15 | L2:B2:S2 | 1:5:5 | 1:8:21 | 1:7:16 | 1:1.5:2.8 | 1:5:4.8 |
| Ethyl Acetate (g/mL) | Ethyl Caprylate (g/mL) | Ethyl Caprate (g/mL) | Ethyl Dodecanoate (g/mL) | Ethyl Tetradecanoate (g/mL) | Isobutanol (g/mL) | 2-Methyl-1-Butanol (g/mL) | 3-Methyl-1-Butanol(g/mL) | 2-Phenylethanol (g/mL) | |
|---|---|---|---|---|---|---|---|---|---|
| F9 | 0.32 | 0.10 | 0.18 | 0.10 | 0.02 | 0.40 | 0.20 | 0.42 | 0.50 |
| F10 | 0.31 | 0.12 | 0.20 | 0.11 | 0.02 | 0.42 | 0.17 | 0.45 | 0.43 |
| F11 | 0.33 | 0.05 | 0.14 | 0.10 | 0.02 | 0.28 | 0.12 | 0.48 | 0.33 |
| F12 | 0.35 | 0.05 | 0.13 | 0.10 | 0.02 | 0.3 | 0.15 | 0.49 | 0.30 |
| F13 | 0.48 | 0.02 | 0.07 | 0.06 | 0.01 | 0.83 | 0.30 | 0.90 | 0.63 |
| F14 | 0.46 | 0.02 | 0.07 | 0.06 | 0.02 | 0.80 | 0.3 | 0.92 | 0.70 |
| F15 | 0.58 | 0.01 | 0.13 | 0.07 | 0.01 | 0.63 | 0.20 | 0.78 | 0.70 |
| F16 | 0.56 | 0.01 | 0.13 | 0.06 | 0.01 | 0.69 | 0.21 | 0.80 | 0.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hübbe, T.; Reitenbach, A.F.; Burin, V.M.; Ghesti, G.F.; Jürgen, F. Effect of Mixed Cultures on Microbiological Development in Berliner Weisse Beer. Fermentation 2024, 10, 363. https://doi.org/10.3390/fermentation10070363
Hübbe T, Reitenbach AF, Burin VM, Ghesti GF, Jürgen F. Effect of Mixed Cultures on Microbiological Development in Berliner Weisse Beer. Fermentation. 2024; 10(7):363. https://doi.org/10.3390/fermentation10070363
Chicago/Turabian StyleHübbe, Thomas, Amanda Felipe Reitenbach, Vívian Maria Burin, Grace Ferreira Ghesti, and Frank Jürgen. 2024. "Effect of Mixed Cultures on Microbiological Development in Berliner Weisse Beer" Fermentation 10, no. 7: 363. https://doi.org/10.3390/fermentation10070363
APA StyleHübbe, T., Reitenbach, A. F., Burin, V. M., Ghesti, G. F., & Jürgen, F. (2024). Effect of Mixed Cultures on Microbiological Development in Berliner Weisse Beer. Fermentation, 10(7), 363. https://doi.org/10.3390/fermentation10070363





