Inhibitor Tolerance Capacity of Pichia kudriavzevii NBRC1279 and NBRC1664
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Media
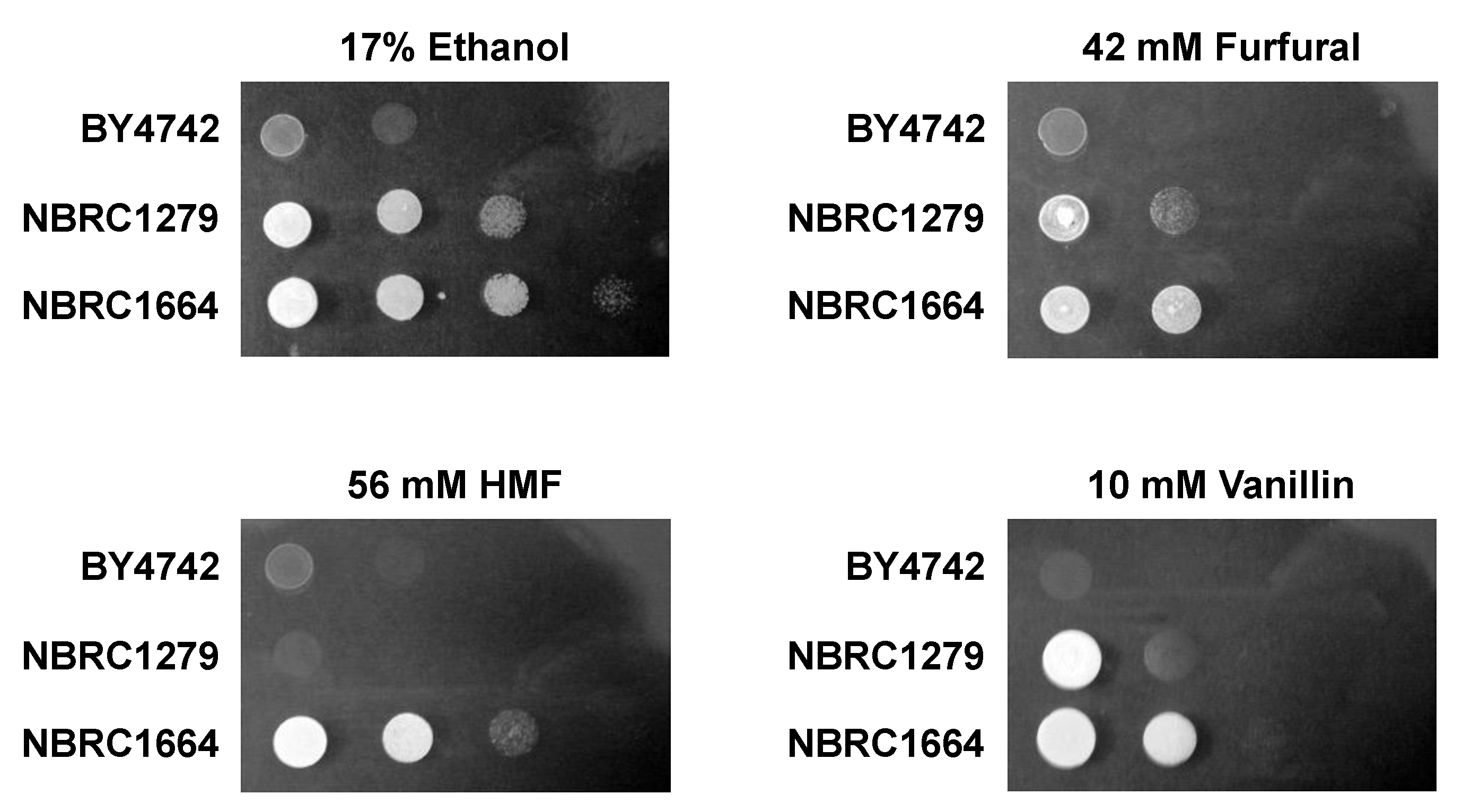
2.2. Spot Assay
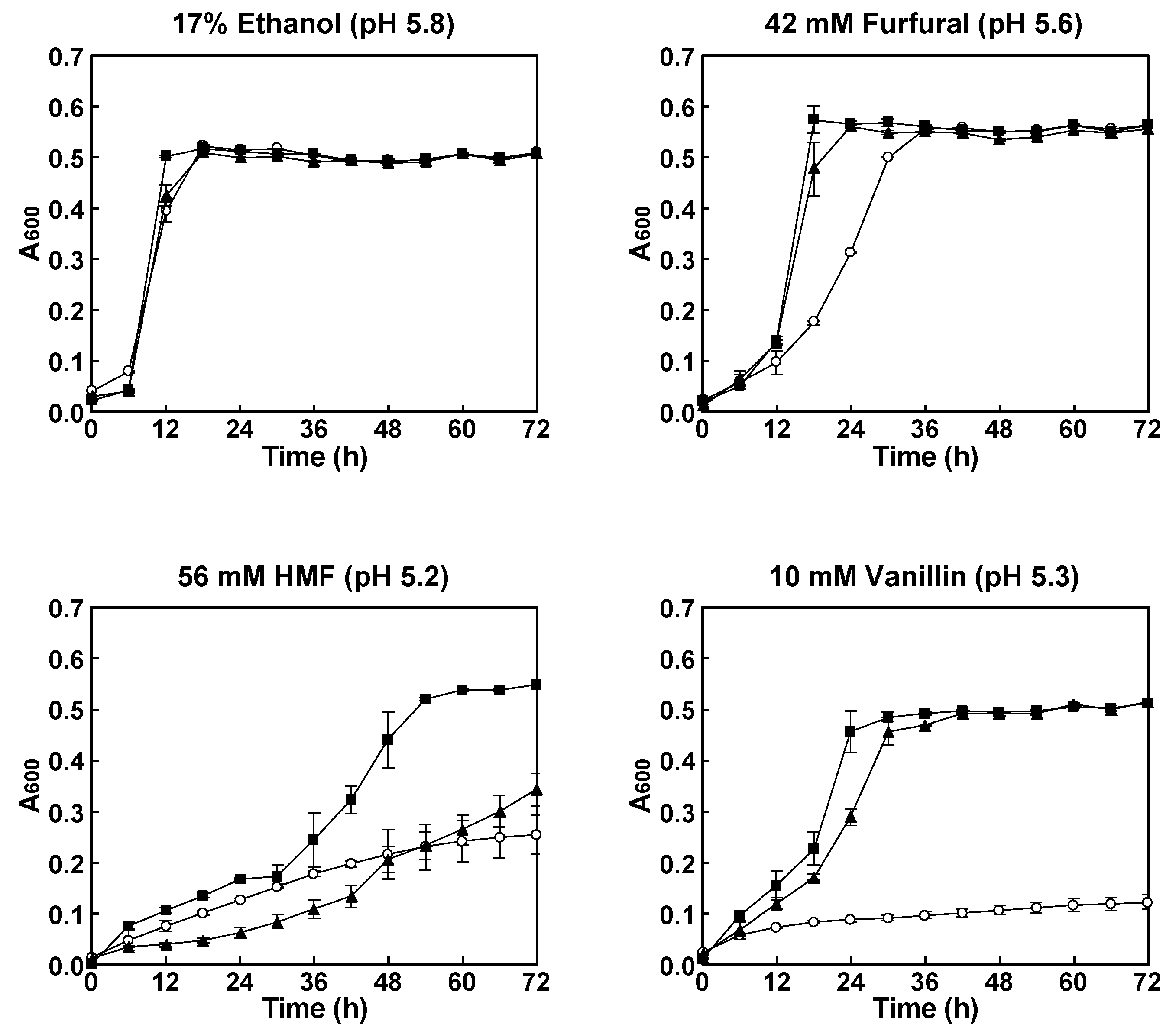
2.3. Aerobic Growth Experiment
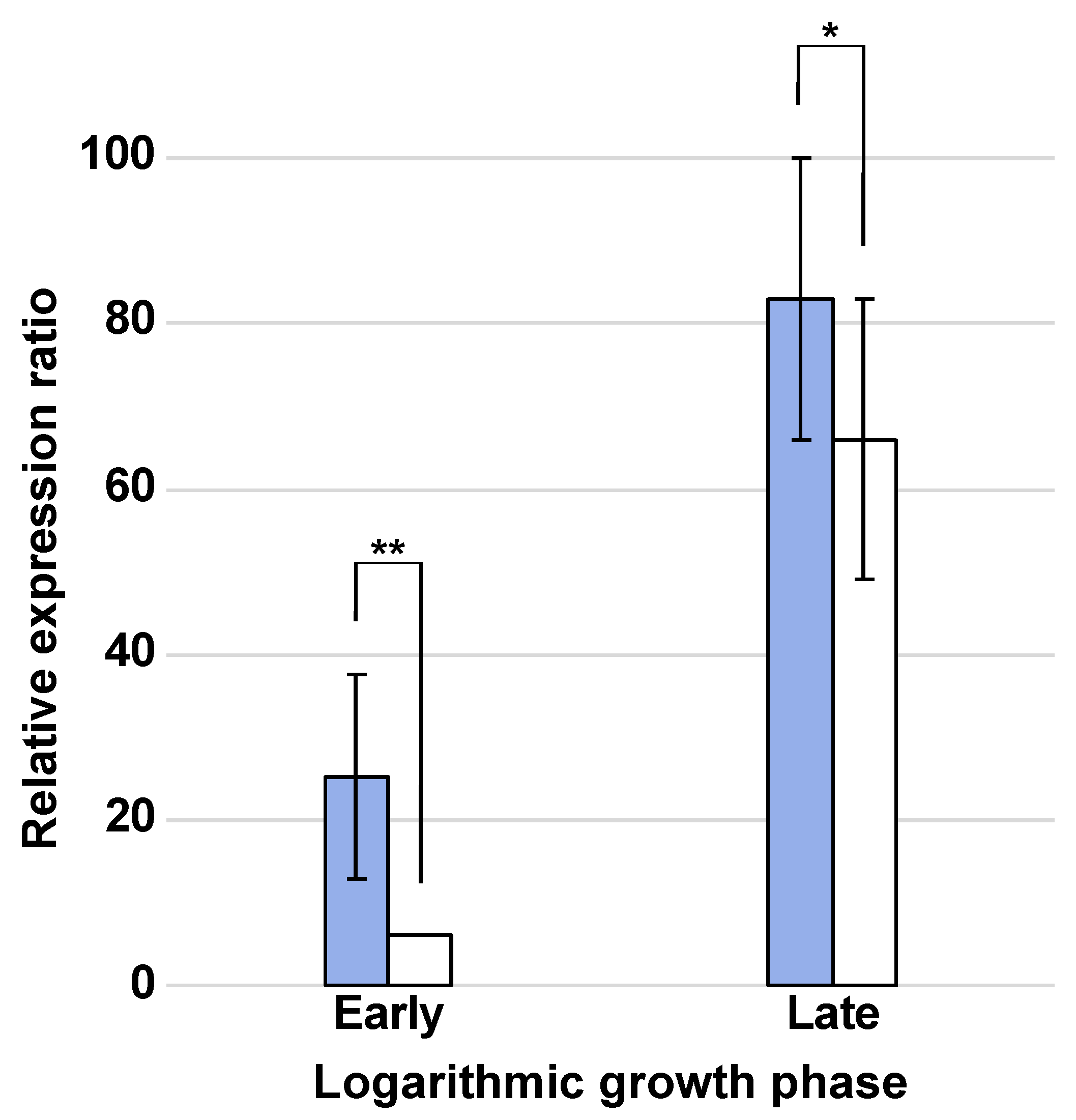
2.4. RT-qPCR Analysis
2.5. Comparative Analysis of Primary Structures
3. Results and Discussion
3.1. Effect of Inhibitors on Growth
3.2. RT-qPCR Analysis of ADHs under HMF Stress
3.3. Elucidation of the HMF Tolerance Capacity of P. kudriavzevii NBRC1664
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Cruz-Morales, P.; Zargar, A.; Belcher, M.S.; Pang, B.; Englund, E.; Dan, Q.; Yin, K.; Keasling, J.D. Biofuels for a sustainable future. Cell 2021, 184, 1636–1647. [Google Scholar] [CrossRef] [PubMed]
- Rego-Costa, A.; Huang, I.-T.; Desai, M.M.; Gombert, A.K. Yeast population dynamics in Brazilian bioethanol production. G3 2023, 13, jkad104. [Google Scholar] [CrossRef] [PubMed]
- Broda, M.; Yelle, D.J.; Serwańska, K. Bioethanol production from lignocellulosic biomass—Challenges and solutions. Molecules 2022, 27, 8717. [Google Scholar] [CrossRef]
- Demeke, M.M.; Echemendia, D.; Belo, E.; Foulquié-Moreno, M.R.; Thevelein, J.M. Enhancing xylose-fermentation capacity of engineered Saccharomyces cerevisiae by multistep evolutionary engineering in inhibitor-rich lignocellulose hydrolysate. FEMS Yeast Res. 2024, 24, foae013. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cao, L.; Li, Q.; Wijayawardene, N.N.; Zhao, J.; Cheng, M.; Li, Q.R.; Li, X.; Promputtha, I.; Kang, Y.Q. Overexpressing GRE3 in Saccharomyces cerevisiae enables high ethanol production from different lignocellulose hydrolysates. Front. Microbiol. 2022, 13, 1085114. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, M.; Mierke, F.; Nygård, Y.; Olsson, L. Nutrient-supplemented propagation of Saccharomyces cerevisiae improves its lignocellulose fermentation ability. AMB Express 2020, 10, 157. [Google Scholar] [CrossRef]
- Fujii, T.; Murakami, K.; Endo, T.; Fujimoto, S.; Minowa, T.; Matsushika, A.; Yano, S.; Sawayama, S. Bench-scale bioethanol production from eucalyptus by high solid saccharification and glucose/xylose fermentation method. Bioprocess Biosyst. Eng. 2014, 37, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Akita, H.; Goshima, T.; Suzuki, T.; Itoiri, Y.; Kimura, Z.-I.; Matsushika, A. Application of Pichia kudriavzevii NBRC1279 and NBRC1664 to simultaneous saccharification and fermentation for bioethanol production. Fermentation 2021, 7, 83. [Google Scholar] [CrossRef]
- Liu, Z.L.; Blaschek, H.P. Biomass conversion inhibitors and in situ detoxification. Biomass Biofuels Strateg. Glob. Ind. 2010, 233–259. [Google Scholar] [CrossRef]
- Iwaki, A.; Ohnuki, S.; Suga, Y.; Izawa, S.; Ohya, Y. Vanillin inhibits translation and induces messenger ribonucleoprotein (mRNP) granule formation in saccharomyces cerevisiae: Application and validation of high-content, image-based profiling. PLoS ONE 2013, 8, e61748. [Google Scholar] [CrossRef]
- Akita, H.; Matsushika, A. Transcription analysis of the acid tolerance mechanism of Pichia kudriavzevii NBRC1279 and NBRC1664. Fermentation 2023, 9, 559. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interface. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Métoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef]
- Elahi, A.; Rehman, A. Bioconversion of hemicellulosic materials into ethanol by yeast, Pichia kudriavzevii 2-KLP1, isolated from industrial waste. Rev. Argent. Microbiol. 2018, 50, 417–425. [Google Scholar] [CrossRef]
- Dandi, N.D.; Dandi, B.N.; Chaudhari, A.B. Bioprospecting of thermo- and osmo-tolerant fungi from mango pulp-peel compost for bioethanol production. Antonie Van Leeuwenhoek 2013, 103, 723–736. [Google Scholar] [CrossRef]
- Yuan, S.F.; Guo, G.L.; Hwang, W.S. Ethanol production from dilute-acid steam exploded lignocellulosic feedstocks using an isolated multistress-tolerant Pichia kudriavzevii strain. Microb. Biotechnol. 2017, 10, 1581–1590. [Google Scholar] [CrossRef]
- Ma, M.; Liu, Z.L. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genom. 2010, 11, 660. [Google Scholar] [CrossRef]
- Paes, B.G.; Steindorff, A.S.; Formighieri, E.F.; Pereira, I.S.; Almeida, J.R.M. Physiological characterization and transcriptome analysis of Pichia pastoris reveals its response to lignocellulose-derived inhibitors. AMB Express 2021, 11, 2. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Z.; Li, R.; Miao, Y.; Weng, P.; Wang, L. Integrated transcriptomic and proteomic analysis of the acetic acid stress in Issatchenkia orientalis. J. Food Biochem. 2020, 44, e13203. [Google Scholar] [CrossRef]
- Du, H.; Fu, Y.; Deng, N.; Xu, Y. Transcriptional profiling reveals adaptive response and tolerance to lactic acid stress in Pichia kudriavzevii. Foods 2022, 11, 2725. [Google Scholar] [CrossRef]
- Zwirzitz, A.; Alteio, L.; Sulzenbacher, D.; Atanasoff, M.; Selg, M. Ethanol production from wheat straw hydrolysate by Issatchenkia orientalis isolated from waste cooking oil. J. Fungi 2021, 7, 121. [Google Scholar] [CrossRef]
- Chen, R.E.; Thorner, J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2007, 1773, 1311–1340. [Google Scholar] [CrossRef]
- Khamwachirapithak, P.; Guillaume-Schoepfer, D.; Chansongkrow, P.; Teichmann, S.A.; Wigge, P.A.; Charoensawan, V. Characterizing different modes of interplay between Rap1 and H3 using inducible H3-depletion yeast. J. Mol. Biol. 2023, 435, 168355. [Google Scholar] [CrossRef]
- Guo, W.; Chen, Y.; Wei, N.; Feng, X. Investigate the metabolic reprogramming of Saccharomyces cerevisiae for enhanced resistance to mixed fermentation inhibitors via 13C metabolic flux analysis. PLoS ONE 2016, 11, e0161448. [Google Scholar] [CrossRef]
- Wasylenko, T.M.; Stephanopoulos, G. Metabolomic and 13C-metabolic flux analysis of a xylose-consuming Saccharomyces cerevisiae strain expressing xylose isomerase. Biotechnol. Bioeng. 2015, 112, 470–483. [Google Scholar] [CrossRef]
- Joshi, A.; Vermam, K.K.; Rajput, V.D.; Minkina, T.; Arora, J. Recent advances in metabolic engineering of microorganisms for advancing lignocellulose-derived biofuels. Bioengineered 2022, 13, 8135–8163. [Google Scholar] [CrossRef]
- Akita, H.; Watanabe, M.; Suzuki, T.; Nakashima, N.; Hoshino, T. Characterization of the Kluyveromyces marxianus strain DMB1 YGL157w gene product as a broad specificity NADPH-dependent aldehyde reductase. AMB Express 2015, 5, 17. [Google Scholar] [CrossRef]
- Castellví, A.; Pequerul, R.; Barracco, V.; Juanhuix, J.; Parés, X.; Farrés, J. Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3. Commun. Biol. 2022, 5, 354. [Google Scholar] [CrossRef]
- Moretti, A.; Li, J.; Donini, S.; Sobol, R.W.; Rizzi, M.; Garavaglia, S. Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid. Sci. Rep. 2016, 6, 35710. [Google Scholar] [CrossRef]
- Sobreira, T.J.P.; Marlétaz, F.; Simões-Costa, M.; Schechtman, D.; Pereira, A.C.; Brunet, F.; Sweeney, S.; Pani, A.; Aronowicz, J.; Lowe, C.J.; et al. Structural shifts of aldehyde dehydrogenase enzymes were instrumental for the early evolution of retinoid-dependent axial patterning in metazoans. Proc. Natl. Acad. Sci. USA 2011, 108, 226–231. [Google Scholar] [CrossRef]
- Wang, X.; Gao, Q.; Bao, J. Transcriptional analysis of Amorphotheca resinae ZN1 on biological degradation of furfural and 5-hydroxymethylfurfural derived from lignocellulose pretreatment. Biotechnol. Biofuels 2015, 8, 136. [Google Scholar] [CrossRef]

| Primer Sequence (5′–3′) | |||
|---|---|---|---|
| Locus | Protein | Forward | Reverse |
| 3340 | Actin | ATCCTTCAGAGTGCCACACC | ACCGCTTTCTCTGTTGACGA |
| 21,230 | Aldehyde dehydrogenase (ADH1) | TTGCCCAACTTTTCTCGTGC | CAAGTTCCAAAACCACCGGC |
| 39,770 | Aldehyde dehydrogenase (ADH2) | TGATCCAAGAATCCGCAGCA | CCAACGTTAACACCAGTGGC |
| 42,380 | Aldehyde dehydrogenase (ADH3) | AAGGTGCACAAACCACTCCA | TCAAGCCTTTCGGAAACCCA |
| 44,250 | Aldehyde dehydrogenase family protein (ADHF1) | TGTCGGCTGCATCATTCCAT | GAAAGCGCTGAGCCAACATC |
| 51,420 | Aldehyde dehydrogenase family protein (ADHF2) | ATATCAGTATTGAGCAAGCTTGTAG | ACATGACAGATCGCCTCACC |
| 60,390 | Aldehyde dehydrogenase (ADH4) | GTTTGTTTTCAGTCCTTGTATCCT | TGTCCTTGTGTGCCTCCATC |
| Yeast | 17% Ethanol | 42 mM Furfural | 56 mM HMF | 10 mM Vanillin |
|---|---|---|---|---|
| S. cerevisiae BY4742 | 0.267 | 0.0173 | 0.0920 | 0.0229 |
| P. kudriavzevii NBRC1279 | 0.396 | 0.0403 | 0.131 | 0.0783 |
| P. kudriavzevii NBRC1664 | 0.411 | 0.0464 | 0.169 | 0.0849 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akita, H.; Matsushika, A. Inhibitor Tolerance Capacity of Pichia kudriavzevii NBRC1279 and NBRC1664. Fermentation 2024, 10, 331. https://doi.org/10.3390/fermentation10070331
Akita H, Matsushika A. Inhibitor Tolerance Capacity of Pichia kudriavzevii NBRC1279 and NBRC1664. Fermentation. 2024; 10(7):331. https://doi.org/10.3390/fermentation10070331
Chicago/Turabian StyleAkita, Hironaga, and Akinori Matsushika. 2024. "Inhibitor Tolerance Capacity of Pichia kudriavzevii NBRC1279 and NBRC1664" Fermentation 10, no. 7: 331. https://doi.org/10.3390/fermentation10070331
APA StyleAkita, H., & Matsushika, A. (2024). Inhibitor Tolerance Capacity of Pichia kudriavzevii NBRC1279 and NBRC1664. Fermentation, 10(7), 331. https://doi.org/10.3390/fermentation10070331






