Exploring Phenotype, Genotype, and the Production of Promising GABA Postbiotics by Lactiplantibacillus plantarum: A Comprehensive Investigation
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Isolation of LAB and Optimum Growth Conditions
2.3. Phenotypic Characterization of LAB Isolates
2.3.1. Catalase Assay
2.3.2. Spore Staining and Motility Analysis
2.4. In Vitro Characterization of Candidate LAB Isolates
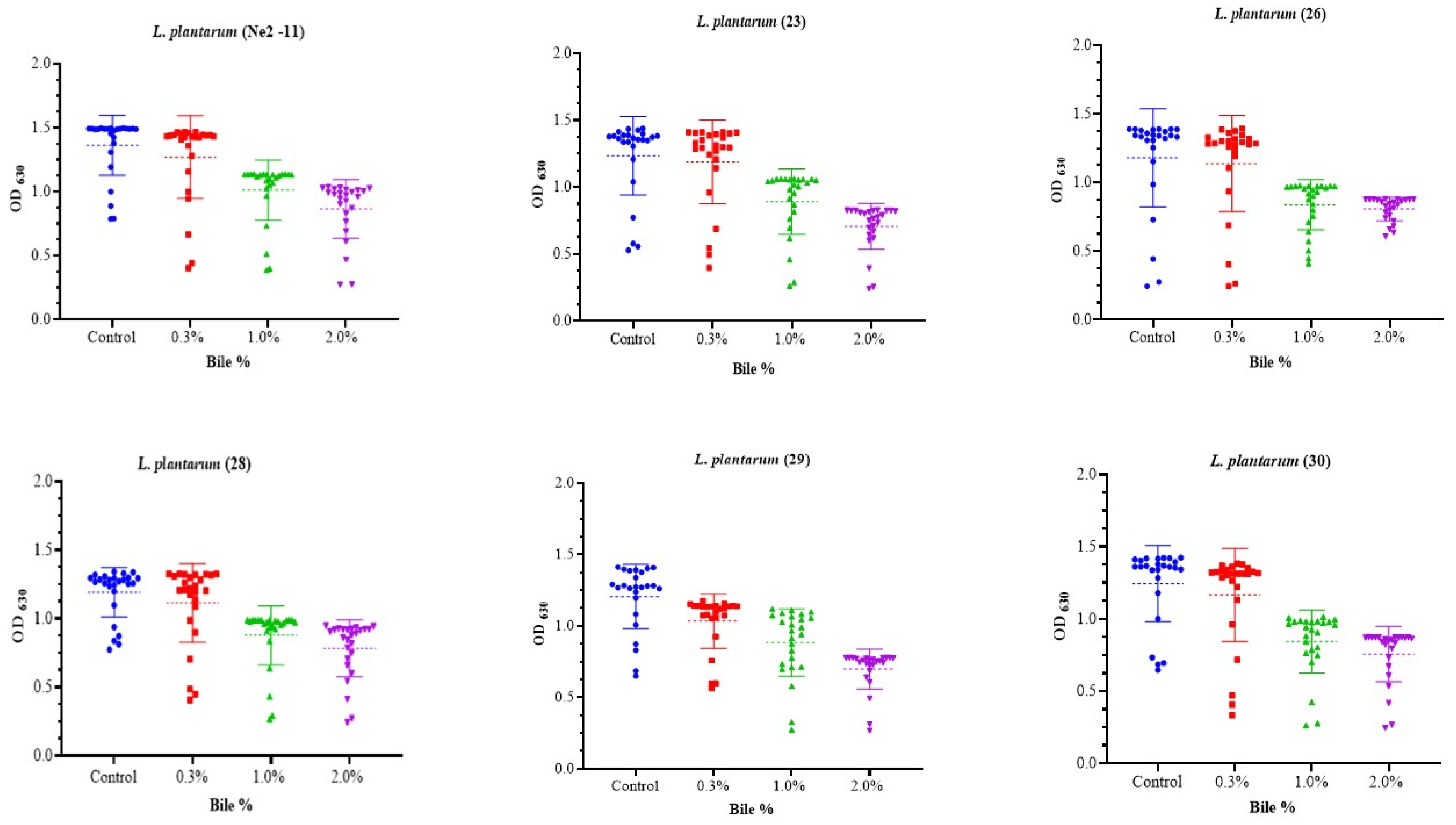
2.4.1. Bile Tolerance Assessment
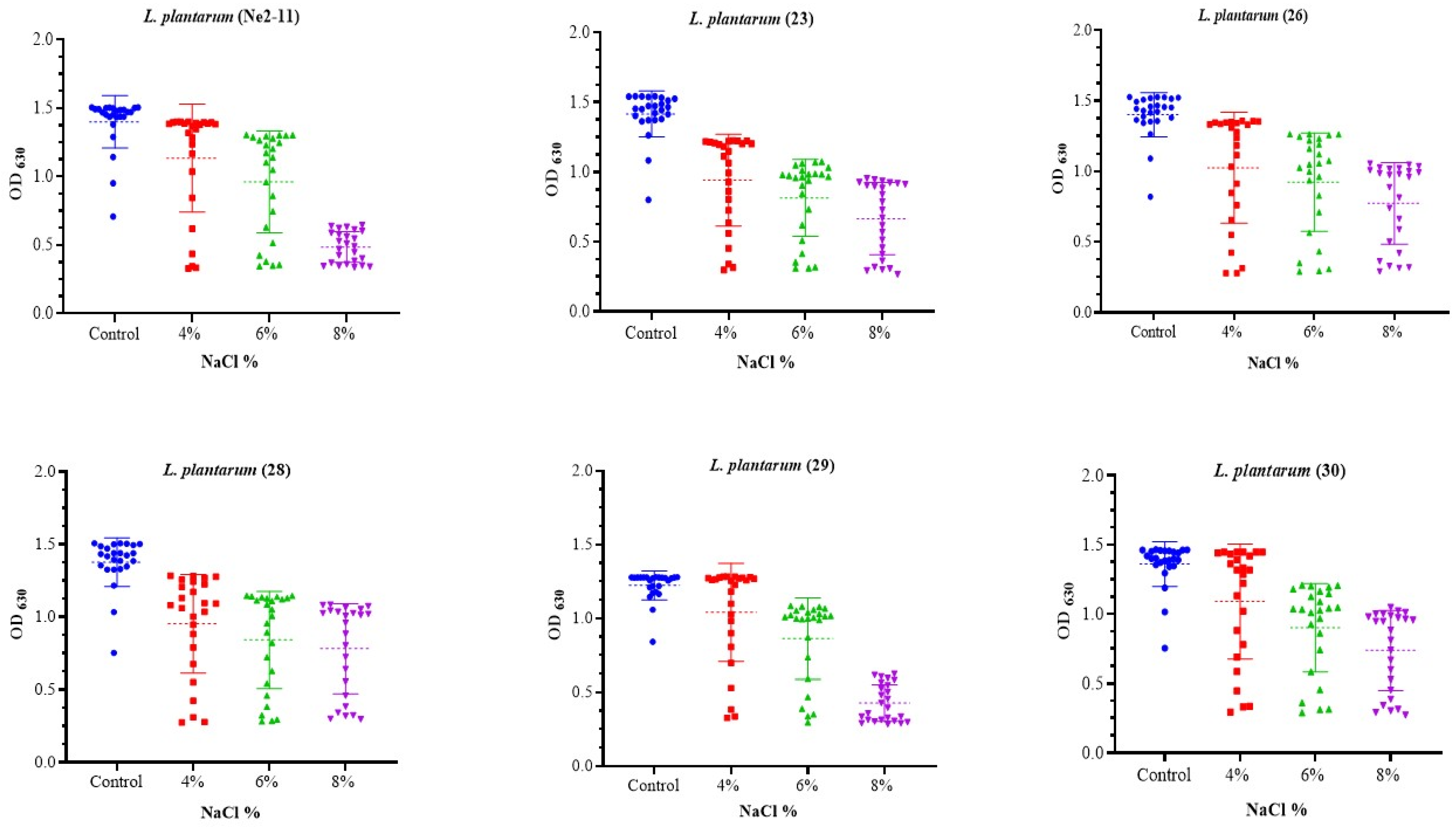
2.4.2. NaCl Tolerance Evaluation
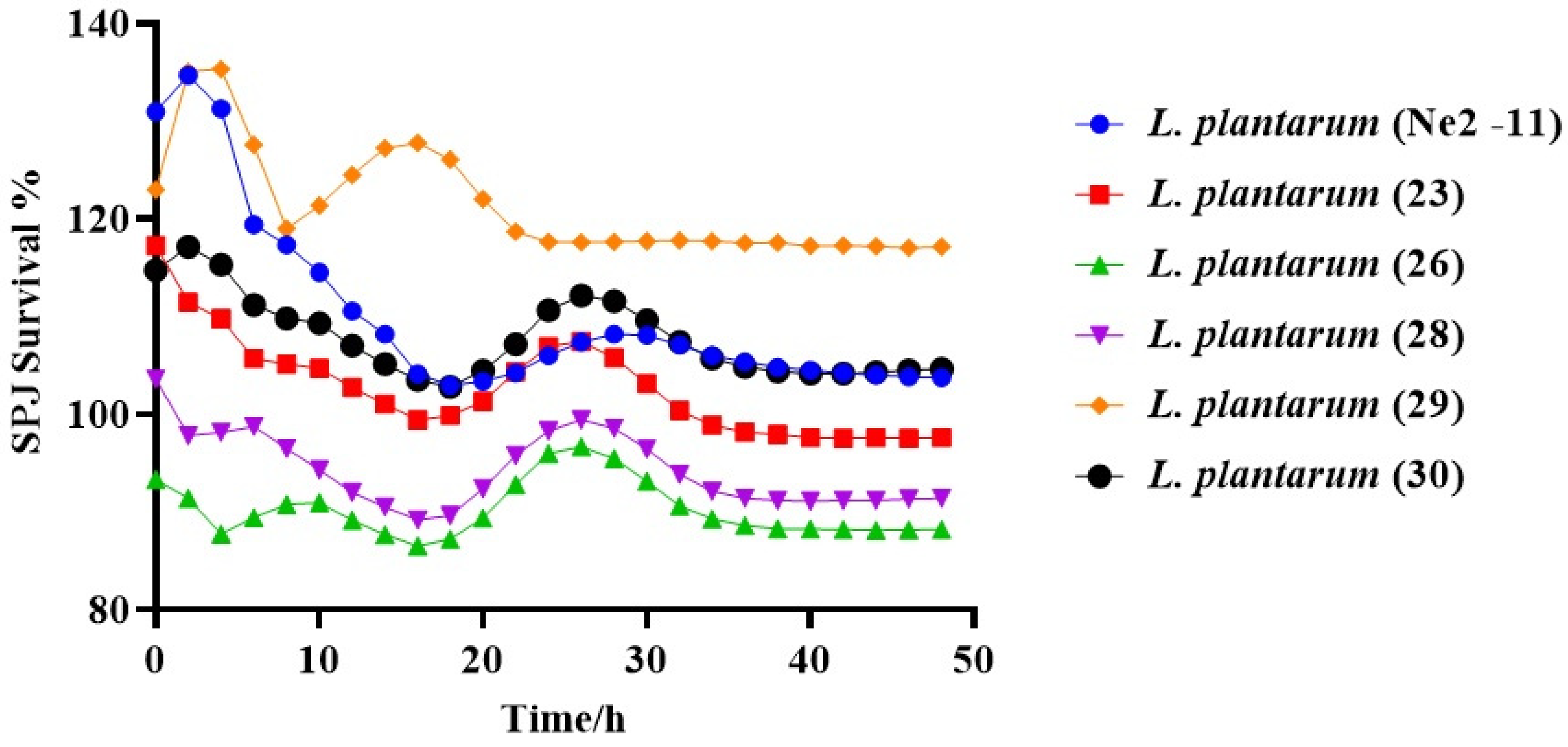
2.4.3. Tolerance of Simulated Pancreatic Juice (SPJ)
2.4.4. Susceptibility of Prospective Isolated Strains to Various Antibiotic Types
2.4.5. Determination of Antimicrobial Activity
2.5. Bioinformatics Analysis
2.5.1. Genetic Profiling of Isolated Strains
2.5.2. PCR Amplification of the 16S rRNA Sequence and Phylogenetic Analysis of Isolates
2.5.3. Detection of Glutamate Decarboxylase (gad) Gene
Colorimetric GAD Assay
Molecular Identification of gad Gene
2.6. Determination of Biochemical Analysis
2.6.1. Screening of β-D-Galactosidase
2.6.2. Sugar Fermentation Pattern
2.6.3. Determination of Biochemical Composition and Postbiotic Metabolites of L. plantarum Strains
2.6.4. Data Preprocessing and Annotation
2.7. Statistical Analysis
3. Results
3.1. Bioinformatics Analysis
3.2. Phenotypic Characteristics of Isolated Strains
3.3. In Vitro Experiments
3.3.1. Bile Tolerance
3.3.2. NaCl Resistance Assessment
3.3.3. Survivability of Lactiplantibacillus plantarum under SPJ Conditions
3.3.4. Antibiotic Susceptibility
3.3.5. Antimicrobial Activity
3.4. Biochemical Analysis of Isolated Strains
3.4.1. Sugar Fermentation Pattern Evaluation
3.4.2. β-D-Galactosidase Detection
3.4.3. Determination of Biochemical Postbiotics of Lactiplantibacillus plantarum Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, P.G.; Patel, A.C.; Chakraborty, P.; Gosai, H.B. Impact of Dietary Habits, Ethnicity, and Geographical Provenance in Shaping Human Gut Microbiome Diversity. In Probiotics, Prebiotics, Synbiotics, and Postbiotics: Human Microbiome and Human Health; Springer: Berlin/Heidelberg, Germany, 2023; pp. 3–27. [Google Scholar]
- Yuan, H.; Wang, Z.; Wang, Z.; Zhang, F.; Guan, D.; Zhao, R. Trends in Forensic Microbiology: From Classical Methods to Deep Learning. Front. Microbiol. 2023, 14, 1163741. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.S.; Mak, W.Q.; Tan, L.K.S.; Ng, C.X.; Chan, H.H.; Yeow, S.H.; Foo, J.B.; Ong, Y.S.; How, C.W.; Khaw, K.Y. Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases. Signal Transduct. Target. Ther. 2024, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Hols, P.; Bernard, E.; Rolain, T.; Zhou, M.; Siezen, R.J.; Bron, P.A. The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 2010, 34, 199–230. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.-F.; Zhang, L.-J.; Pang, X.-H.; Gu, X.-X.; Abdelazez, A.; Liang, Y.; Sun, S.-R.; Meng, X.-C. Complete genome sequence of bacteriocin-producing Lactobacillus plantarum KLDS1.0391, a probiotic strain with gastrointestinal tract resistance and adhesion to the intestinal epithelial cells. Genomics 2017, 109, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Abdelazez, A.; Mohamed, D.M.; Refaey, M.M.M.; Niu, J. Intervention effect of freeze-dried probiotic and unripe banana pulp combination on Set-Type bio-yogurt production during storage. J. Food Meas. Charact. 2023, 1, 18. [Google Scholar] [CrossRef]
- Abdelazez, A.; Abdelmotaal, H.; Evivie, S.E.; Bikheet, M.; Sami, R.; Mohamed, H.; Meng, X. Verification of Lactobacillus brevis Tolerance to Simulated Gastric Juice and the Potential Effects of Postbiotic Gamma-Aminobutyric Acid in Streptozotocin-Induced Diabetic Mice. Food Sci. Hum. Wellness 2022, 11, 165–176. [Google Scholar] [CrossRef]
- Pannerchelvan, S.; Rios-Solis, L.; Faizal Wong, F.W.; Zaidan, U.H.; Wasoh, H.; Mohamed, M.S.; Tan, J.S.; Mohamad, R.; Halim, M. Strategies For Improvement of Gamma-Aminobutyric Acid (GABA) Biosynthesis via Lactic Acid Bacteria (LAB) Fermentation. Food Funct. 2023, 14, 3929–3948. [Google Scholar] [CrossRef]
- Cha, X.; Ding, J.; Ba, W.; You, S.; Qi, W.; Su, R. High Production of γ-Aminobutyric Acid by Activating the xyl Operon of Lactobacillus brevis. ACS Omega 2023, 8, 8101–8109. [Google Scholar] [CrossRef]
- Żółkiewicz, J.; Marzec, A.; Ruszczyński, M.; Feleszko, W. Postbiotics—A Step Beyond Pre- and Probiotics. Nutrients 2020, 12, 2189. [Google Scholar] [CrossRef]
- Spiering, M.J. The discovery of GABA in the brain. J. Biol. Chem. 2018, 293, 19159–19160. [Google Scholar] [CrossRef]
- Hepsomali, P.; Groeger, J.A.; Nishihira, J.; Scholey, A. Effects of oral gamma-aminobutyric acid (GABA) administration on stress and sleep in humans: A systematic review. Front. Neuro. Sci. 2020, 14, 923. [Google Scholar] [CrossRef]
- AOKI, H.; Furuya, Y.; Endo, Y.; Fujimoto, K. Effect of γ-Aminobutyric Acid-enriched Tempeh-like Fermented Soybean (GABA-Tempeh) on the Blood Pressure of Spontaneously Hypertensive Rats. Biosci. Biotechnol. Biochem. 2003, 67, 1806–1808. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Liu, C.; Wang, C.; Hu, Y.; Qiu, L.; Xu, P. Neurotransmitter γ-aminobutyric acid-mediated inhibition of the invasive ability of cholangiocarcinoma cells. Oncol. Lett. 2011, 2, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Liwinski, T.; Lang, U.E.; Brühl, A.B.; Schneider, E. Exploring the therapeutic potential of gamma-aminobutyric acid in stress and depressive disorders through the gut-brain axis. Biomedicines 2023, 11, 3128. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Wang, J.-Y.; Chen, H.-L.; Wang, Y.; Cui, L.-Y.; Duan, J.-Y.; Guo, W.-Z.; Zhao, Y.-Q.; Li, Y.-F. Activation of σ-1 receptor mitigates estrogen withdrawal-induced anxiety/depressive-like behavior in mice via restoration of GABA/glutamate signaling and neuroplasticity in the hippocampus. J. Pharmacol. Sci. 2024, 154, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Braga, J.D.; Thongngam, M.; Kumrungsee, T. Gamma-aminobutyric acid as a potential postbiotic mediator in the gut–brain axis. NPJ Sci. Food 2024, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-H.; Kim, H.; Kong, M.-J.; Hong, Y.-T.; Oh, S.-J.; Choi, K.-C. Gamma-aminobutyric acid in rice bran extract exerts antistress effects in mouse models with depressive-like behaviors. J. Med. Food 2024, 27, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Khushboo; Karnwal, A.; Malik, T. Characterization and selection of probiotic lactic acid bacteria from different dietary sources for development of functional foods. Front. Microbiol. 2023, 14, 1170725. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.T. Anaerobic Bacteriology: Clinical and Laboratory Practice; Butterworth-Heinemann: Oxford, UK, 2014. [Google Scholar]
- Mulaw, G.; Sisay Tessema, T.; Muleta, D.; Tesfaye, A. In Vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int. J. Microbiol. 2019, 2019, 7179514. [Google Scholar] [CrossRef]
- Rashmi, B.S.; Gayathri, D. Molecular Characterization of Gluten Hydrolysing Bacillus sp. and Their Efficacy and Biotherapeutic Potential as Probiotics Using Caco-2 Cell Line. J. Appl. Microbiol. 2017, 123, 759–772. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Z.-Y.; Dong, K.; Yuan, J.-P.; Guo, X.-K. Antibiotic Resistance of Probiotic Strains of Lactic Acid Bacteria Isolated from Marketed Foods and Drugs. Biomed. Environ. Sci. 2009, 22, 401–412. [Google Scholar] [CrossRef]
- Mohammadi, R.; Moradi, M.; Tajik, H.; Molaei, R. Potential Application of Postbiotics Metabolites from Bioprotective Culture To Fabricate Bacterial Nanocellulose Based Antimicrobial Packaging Material. Int. J. Biol. Macromol. 2022, 220, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Keresztény, T.; Libisch, B.; Orbe, S.C.; Nagy, T.; Kerényi, Z.; Kocsis, R.; Posta, K.; Papp, P.P.; Olasz, F. Isolation and Characterization of Lactic Acid Bacteria with Probiotic Attributes from Different Parts of the Gastrointestinal Tract of Free-Living Wild Boars in Hungary. Probiotics Antimicrob. Proteins 2023. [Google Scholar] [CrossRef] [PubMed]
- Angmo, K.; Kumari, A.; Savitri; Bhalla, T.C. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT-Food Sci. Technol. 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Olier, M.w.; Rousseaux, S.; Piveteau, P.; Lemaître, J.-P.; Rousset, A.; Guzzo, J. Screening of Glutamate Decarboxylase Activity and Bile Salt Resistance of Human Asymptomatic Carriage, Clinical, Food, and Environmental Isolates of Listeria monocytogenes. Int. J. Food Microbiol. 2004, 93, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Abdelazez, A.; Abdelmotaal, H.; Evivie, S.E.; Melak, S.; Jia, F.-F.; Khoso, M.H.; Zhu, Z.-T.; Zhang, L.-J.; Sami, R.; Meng, X.-C. Screening Potential Probiotic Characteristics of Lactobacillus brevis Strains In Vitro and Intervention Effect on Type I Diabetes In Vivo. BioMed Res. Int. 2018, 2018, 7356173. [Google Scholar] [CrossRef] [PubMed]
- Gheytanchi, E.; Heshmati, F.; Shargh, B.K.; Nowroozi, J.; Movahedzadeh, F. Study on Β-Galactosidase Enzyme Produced by Isolated Lactobacilli from Milk and Cheese. Afr. J. Microbiol. Res. 2010, 4, 454–458. [Google Scholar]
- Nataraj, B.H.; Ali, S.A.; Behare, P.V.; Yadav, H. Postbiotics-Parabiotics: The New Horizons in Microbial Biotherapy and Functional Foods. Microb. Cell Fact. 2020, 19, 168. [Google Scholar] [CrossRef] [PubMed]
- Abdelazez, A.; Alshehry, G.; Algarni, E.; Al Jumayi, H.; Abdel-Motaal, H.; Meng, X.-C. Postbiotic Gamma-Aminobutyric Acid and Camel Milk Intervention as Innovative Trends against Hyperglycemia and Hyperlipidemia in Streptozotocin-Induced C57BL/6J Diabetic Mice. Front. Microbiol. 2022, 13, 943930. [Google Scholar] [CrossRef]
- Petersen, J.; McLaughlin, S. Laboratory Exercises in Microbiology: Discovering the Unseen World through Hands-On Investigation; CUNY Academic Works; City University of New York (CUNY): New York, NY, USA, 2016. [Google Scholar]
- Gutarowska, B.; Szulc, J.; Jastrząbek, K.; Kręgiel, D.; Śmigielski, K.; Cieciura-Włoch, W.; Mroczyńska-Florczak, M.; Liszkowska, W.; Rygała, A.; Berłowska, J. Effectiveness of Ozonation for Improving the Microbiological Safety of Fresh-Cut Parsley (Petroselinum crispum) Leaves. Appl. Sci. 2023, 13, 8946. [Google Scholar] [CrossRef]
- Kouadri Boudjelthia, N.; Belabbas, M.; Bekenniche, N.; Monnoye, M.; Gérard, P.; Riazi, A. Probiotic Properties of Lactic Acid Bacteria Newly Isolated from Algerian Raw Cow’s Milk. Microorganisms 2023, 11, 2091. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, F.D.; Pino, A.; Maciel, G.L.R.; Sanfilippo, R.R.; Caggia, C.; de Carvalho, A.F.; Randazzo, C.L. Technological Characterization of Lactic Acid Bacteria Strains for Potential Use in Cheese Manufacture. Foods 2023, 12, 1154. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhang, R.; Tian, X.; Zhou, X.; Pan, X.; Wong, A. Assessing the Risk of Probiotic Dietary Supplements in the Context of Antibiotic Resistance. Front. Microbiol. 2017, 8, 264345. [Google Scholar] [CrossRef] [PubMed]
- de Almeida Júnior, W.L.G.; de Souza Ferrari ÍdS, J.V.; da Silva, C.D.A.; da Costa, M.M.; Dias, F.S. Characterization and Evaluation of Lactic Acid Bacteria Isolated from Goat Milk. Food Control 2015, 53, 96–103. [Google Scholar] [CrossRef]
- Tulumoglu, S.; Yuksekdag, Z.N.; Beyatli, Y.; Simsek, O.; Cinar, B.; Yaşar, E. Probiotic Properties of Lactobacilli Species Isolated from Children’s Feces. Anaerobe 2013, 24, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Karapetkov, N.; Georgieva, R.; Rumyan, N.; Karaivanova, E. Antibiotic Susceptibility of Different Lactic Acid Bacteria Strains. Benef. Microbes 2011, 2, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, Z.; Ramzan, R.; Abdelazez, A.; Amjad, A.; Afzaal, M.; Zhang, S.; Pan, S. Assessment of the Antimicrobial Potentiality and Functionality of Lactobacillus plantarum Strains Isolated from the Conventional Inner Mongolian Fermented Cheese against Foodborne Pathogens. Pathogens 2019, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.N.; Fernandes, N.; Carvalho, L.; Faria, A.S.; Teixeira, J.A.; Rodrigues, C.; Gonzales-Barron, U.; Cadavez, V. Lactic Acid Bacteria from Artisanal Raw Goat Milk Cheeses: Technological Properties and Antimicrobial Potential. Ital. J. Food Saf. 2023, 12, 11559. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-H.; Ren, L.-Q.; Zhou, Y.; Ye, B.-C. Characterization of Antimicrobial Activity of Three Lactobacillus plantarum Strains Isolated from Chinese Traditional Dairy Food. Food Sci. Nutr. 2019, 7, 1997–2005. [Google Scholar] [CrossRef]
- Fossi, B.T.; Ekue, N.B.; Nchanji, G.T.; Ngah, B.; Anyangwe, I.A.; Wanji, S. Probiotic Properties of Lactic Acid Bacteria Isolated from Fermented Sap of Palm Tree (Elaeis guineensis). J. Microbiol. AntiMicrob. 2015, 7, 42–52. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, T.; Zhang, D.; Wang, X.; Zhang, Z.; Lim, C.; Lee, S. Probiotic Characterization of Lactiplantibacillus plantarum HOM3204 and Its Restoration Effect on Antibiotic-Induced Dysbiosis in Mice. Lett. Appl. Microbiol. 2022, 74, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- Hedberg, M.; Hasslöf, P.; Sjöström, I.; Twetman, S.; Stecksén-Blicks, C. Sugar Fermentation in Probiotic Bacteria—An In Vitro Study. Oral Microbiol. Immunol. 2008, 23, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Fassio, F.; Facioni, M.S.; Guagnini, F. Lactose Maldigestion, Malabsorption, and Intolerance: A Comprehensive Review with a Focus on Current Management and Future Perspectives. Nutrients 2018, 10, 1599. [Google Scholar] [CrossRef] [PubMed]
- Saburi, W.; Ota, T.; Kato, K.; Tagami, T.; Yamashita, K.; Yao, M.; Mori, H. Function and Structure of Lacticaseibacillus casei GH35 β-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lactobiose I and Galactobiose. J. Appl. Glycosci. 2023, 70, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Romero-Velarde, E.; Delgado-Franco, D.; García-Gutiérrez, M.; Gurrola-Díaz, C.; Larrosa-Haro, A.; Montijo-Barrios, E.; Muskiet, F.A.J.; Vargas-Guerrero, B.; Geurts, J. The importance of lactose in the human diet: Outcomes of a Mexican consensus meeting. Nutrients 2019, 11, 2737. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Vinuesa, L.; Sánchez-Arroyo, A.; Moreno, F.J.; de las Rivas, B.; Muñoz, R. Dual 6Pβ-Galactosidase/6Pβ-Glucosidase GH1 Family for Lactose Metabolism in the Probiotic Bacterium Lactiplantibacillus plantarum WCFS1. J. Agric. Food Chem. 2023, 71, 10693–10700. [Google Scholar] [CrossRef] [PubMed]
- Rozhkova, I.V.; Yurova, E.A.; Leonova, V.A. Evaluation of the Amino Acid Composition and Content of Organic Acids of Complex Postbiotic Substances Obtained on the Basis of Metabolites of Probiotic Bacteria Lacticaseibacillus paracasei ABK and Lactobacillus helveticus H9. Fermentation 2023, 9, 460. [Google Scholar] [CrossRef]
- Yue, T.; Zhang, X.; Gong, P.; Li, J.; Wang, X.; Li, X.; Ma, Y.; Chen, X.; Zhang, X.; Cheng, S.; et al. Antitumor Effect of Invasive Lactobacillus plantarum Delivering Associated Antigen Gene Shsp Between Trichinella Spiralis and Lewis Lung Cancer Cells. Int. Immunopharmacol. 2023, 115, 109708. [Google Scholar] [CrossRef]
- Cai, H.; Li, X.; Li, D.; Liu, W.; Han, Y.; Xu, X.; Yang, P.; Meng, K. Optimization of Gamma-Aminobutyric Acid Production by Lactiplantibacillus plantarum FRT7 from Chinese Paocai. Foods 2023, 12, 3034. [Google Scholar] [CrossRef]
- Sun, Y.; Mehmood, A.; Giampieri, F.; Battino, M.A.; Chen, X. Insights into the Cellular, Molecular, and Epigenetic Targets of Gamma-Aminobutyric Acid against Diabetes: A Comprehensive Review on Its Mechanisms. Crit. Rev. Food Sci. Nutr. 2023, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, K.; Motomura, E.; Okada, M. A Novel Gliotransmitter, L-β-Aminoisobutyric Acid, Contributes to Pathophysiology of Clinical Efficacies and Adverse Reactions of Clozapine. Biomolecules 2023, 13, 1288. [Google Scholar] [CrossRef]
- Chaudhry, T.S.; Senapati, S.G.; Gadam, S.; Mannam, H.P.S.S.; Voruganti, H.V.; Abbasi, Z.; Abhinav, T.; Challa, A.B.; Pallipamu, N.; Bheemisetty, N.; et al. The Impact of Microbiota on the Gut–Brain Axis: Examining the Complex Interplay and Implications. J. Clin. Med. 2023, 12, 5231. [Google Scholar] [CrossRef]
- Hafza, N.; Li, N.; Luqman, A.; Götz, F. Identification of a Serotonin N-Acetyltransferase from Staphylococcus pseudintermedius ED99. Front. Microbiol. 2023, 14, 1073539. [Google Scholar] [CrossRef]
- Nimgampalle, M.; Chakravarthy, H.; Sharma, S.; Shree, S.; Bhat, A.R.; Pradeepkiran, J.A.; Devanathan, V. Neurotransmitter Systems in the Etiology of Major Neurological Disorders: Emerging Insights and Therapeutic Implications. Ageing Res. Rev. 2023, 89, 101994. [Google Scholar] [CrossRef]
- Gharahdaghi, N.; Rudrappa, S.; Brook, M.S.; Idris, I.; Crossland, H.; Hamrock, C.; Aziz, M.H.A.; Kadi, F.; Tarum, J.; Greenhaff, P.L.; et al. Testosterone Therapy Induces Molecular Programming Augmenting Physiological Adaptations to Resistance Exercise in Older Men. J. Cachexia Sarcopenia Muscle 2019, 10, 1276–1294. [Google Scholar] [CrossRef]
- Hani, U.; Ali Bu Sinnah, Z.; Obaidullah, A.J.; Huwaimel, B.; Alanazi, M.; Nafea Alharby, T.; Lahiq, A.A.; Alshehri, A.A. Computational Intelligence Modeling of Nanomedicine Preparation Using Advanced Processing: Solubility of Fludrocortisone Acetate in Supercritical Carbon Dioxide. Case Stud. Therm. Eng. 2023, 45, 102968. [Google Scholar] [CrossRef]
- Bercea, M.; Plugariu, I.-A.; Gradinaru, L.M.; Avadanei, M.; Doroftei, F.; Gradinaru, V.R. Hybrid Hydrogels for Neomycin Delivery: Synergistic Effects of Natural/Synthetic Polymers and Proteins. Polymers 2023, 15, 630. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Dou, X.; Song, X.; Chang, J.; Zeng, X.; Zhu, L.; Yi, H.; Xu, C. Replacing Dietary Sodium Selenite with Biogenic Selenium Nanoparticles Improves the Growth Performance and Gut Health of Early-Weaned Piglets. Anim. Nutr. 2023, 15, 99–113. [Google Scholar] [CrossRef]
- Coimbra, J.T.S.; Fernandes, P.A.; Ramos, M.J. Deciphering the Catalytic Mechanism of Virginiamycin B Lyase with Multiscale Methods and Molecular Dynamics Simulations. J. Chem. Inf. Model. 2023, 63, 6354–6365. [Google Scholar] [CrossRef]
- Rafique, A.; Bulbul, Y.E.; Usman, A.; Raza, Z.A.; Oksuz, A.U. Effect of Cold Plasma Treatment on Polylactic Acid and Polylactic Acid/Poly (Ethylene glycol) Films Developed as A Drug Delivery System for Streptomycin Sulfate. Int. J. Biol. Macromol. 2023, 235, 123857. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Rainville, P. Liquid Chromatography–Tandem Mass Spectrometry Analysis of Aminoglycosides in Foods Using an Ethylene-Bridged Hybrid Zwitterionic Stationary Phase and Hydrophilic–Lipophilic-Balanced Solid-Phase Extraction Cartridges. J. Agric. Food Chem. 2023, 71, 7593–7603. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Tang, Q.; An, Z.; Qi, J. Minocycline Relieves Neuropathic Pain in Rats with Spinal Cord Injury via Activation of Autophagy and Suppression of PI3K/Akt/mTOR Pathway. J. Pharmacol. Sci. 2023, 153, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yu, Y.; Hua, X. Resistance Mechanisms of Tigecycline in Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 2023, 13, 1141490. [Google Scholar] [CrossRef] [PubMed]
- Leporati, M.; Pullara, F.; Canevaro, A.; Valesella, P.; Vitale, C.; Cosseddu, D.; Marangella, M.; Petrarulo, M. Identification of Sulfamethoxazole’s Residues in Sulfamethoxazole Induced Kidney Stones by Mass Spectrometry. Clin. Chem. Lab. Med. 2024, 62, e94–e98. [Google Scholar] [CrossRef] [PubMed]
- Trubitsyn, V.E.; Suzina, N.E.; Rivkina, E.M.; Shcherbakova, V.A. A New Methanogenic, Hydrogenotrophic Archaeon from Spitsbergen Permafrost. Microbiology 2023, 92, 119–128. [Google Scholar] [CrossRef]
- Vairo, C.; Villar Vidal, M.; Maria Hernandez, R.; Igartua, M.; Villullas, S. Colistin- and Amikacin-Loaded Lipid-Based Drug Delivery Systems For Resistant Gram-Negative Lung and Wound Bacterial Infections. Int. J. Pharm. 2023, 635, 122739. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Sun, W.; Murchie, A.I.H.; Chen, D. Genome-Wide Identification of Kanamycin B Binding RNA in Escherichia coli. BMC Genom. 2023, 24, 120. [Google Scholar] [CrossRef]
- Ailincai, D.; Cibotaru, S.; Anisiei, A.; Coman, C.G.; Pasca, A.S.; Rosca, I.; Sandu, A.-I.; Mititelu-Tartau, L.; Marin, L. Mesoporous Chitosan Nanofibers Loaded with Norfloxacin and Coated with Phenylboronic Acid Perform as Bioabsorbable Active Dressings to Accelerate the Healing of Burn Wounds. Carbohydr. Polym. 2023, 318, 121135. [Google Scholar] [CrossRef]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, 10. [Google Scholar] [CrossRef]
- Ozdogan, M.S.; Gungormus, M. Salivary Opiorphin Levels in Denture-Related Traumatic Ulcers. Oral Dis. 2023, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Matos, B.; Howl, J.; Jerónimo, C.; Fardilha, M. CAVPENET Decreases Prostate Cancer Cells Proliferation and Invasion through Modulation of Protein Phosphatase Activity. Biol. Life Sci. Forum. 2023, 21, 17. [Google Scholar] [CrossRef]
- Luo, M.; Zhang, Y.; Xu, Z.; Wu, C.; Ye, Y.; Liu, R.; Lv, S.; Wei, Q. Tautomycin and Enzalutamide Combination Yields Synergistic Effects on Castration-Resistant Prostate Cancer. Cell Death Discov. 2022, 8, 471. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Li, Z.; Wang, Z.; Wang, Q.; Qin, F.; Pan, H.; Lin, W.; Mu, X.; Wang, Y.; Jiang, Y.; et al. Immunosuppression by Opioids: Mechanisms of Action on Innate and Adaptive Immunity. Biochem. Pharmacol. 2023, 209, 115417. [Google Scholar] [CrossRef] [PubMed]
- Bhoir, S.; Uhelski, M.; Guerra-Londono, J.J.; Cata, J.P. The Role of Opioid Receptors in Cancer. Adv. Biol. 2023, 7, 2300102. [Google Scholar] [CrossRef] [PubMed]
- Castillo-González, J.; Ruiz, J.L.; Serrano-Martínez, I.; Forte-Lago, I.; Ubago-Rodriguez, A.; Caro, M.; Pérez-Gómez, J.M.; Benítez-Troncoso, A.; Andrés-León, E.; Sánchez-Navarro, M.; et al. Cortistatin Deficiency Reveals a Dysfunctional Brain Endothelium with Impaired Gene Pathways, Exacerbated Immune Activation, and Disrupted Barrier Integrity. J. Neuroinflamm. 2023, 20, 226. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Qiao, N.; Ning, F.; Huang, X.; Luo, L. Identification and Characterization of Plant-Derived Biomarkers and Physicochemical Variations in the Maturation Process of Triadica Cochinchinensis Honey Based on UPLC-QTOF-MS Metabolomics Analysis. Food Chem. 2023, 408, 135197. [Google Scholar] [CrossRef]
- Leite, A.A.; da Silva, L.C.; Borgato, G.B.; Santos-Silva, A.R.; Martins, M.D.; Lopes, M.A.; Vargas, P.A. Cephaeline Sensitizes Cisplatin and Reduces Oral Squamous Cell Carcinoma Cancer Stem Cells. Oral Surg. Oral Med. Oral Radiol. 2023, 136, e53. [Google Scholar] [CrossRef]
- Bhardwaj, G.; Sharma, A.; Thakur, S.; Sareen, S.; Sohal, H.S.; Mutreja, V.; Sharma, A. Gentiana Kurroo Royle: Himalayan Gentian. In Immunity Boosting Medicinal Plants of the Western Himalayas; Sharma, A., Nayik, G.A., Eds.; Springer Nature: Singapore, 2023; pp. 187–204. [Google Scholar]
- O’Brien, M.; Mochel, J.P.; Kersh, K.; Wang, C.; Troy, J. Phenylbutazone Concentrations in Synovial Fluid Following Administration via Intravenous Regional Limb Perfusion in the Forelimbs of Six Adult Horses. Front. Vet. Sci. 2023, 10, 1236976. [Google Scholar] [CrossRef]
- Sharma, B.K. Synthetic and natural compounds as anti-cancer agents—A review. Asian J. Chem. 2017, 10, 699–707. [Google Scholar] [CrossRef]
- Meng, X.; Yu, G.; Luo, T.; Zhang, R.; Zhang, J.; Liu, Y. Transcriptomics Integrated with Metabolomics Reveals Perfluorobutane Sulfonate (PFBS) Exposure Effect During Pregnancy and Lactation on Lipid Metabolism in Rat Offspring. Chemosphere 2023, 341, 140120. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lorca, A.; Novillo, A.; Gaibar, M.; Bandrés, F.; Fernández-Santander, A. Impacts of the Glucuronidase Genotypes UGT1A4, UGT2B7, UGT2B15 and UGT2B17 on Tamoxifen Metabolism in Breast Cancer Patients. PLoS ONE 2015, 10, e0132269. [Google Scholar] [CrossRef] [PubMed]
- Elks, J. The Dictionary of Drugs: Chemical Data: Chemical Data, Structures and Bibliographies; Springer: Easton, PA, USA, 2014. [Google Scholar]
- Blokdijk, G.J. Difluprednate: A Complete Guide; CreateSpace Independent Publishing Platform: Scotts Valley, CA, USA, 2018. [Google Scholar]
- Zhang, H.; Li, K.; Zhao, X.; Zou, H.; Zhao, L.; Li, X. Occurrence, Consumption Level, Fate and Ecotoxicology Risk of Beta-Agonist Pharmaceuticals in a Wastewater Treatment Plant in Eastern China. Environ. Monit. Assess. 2023, 195, 481. [Google Scholar] [CrossRef] [PubMed]
- Muhamad, R.; Akrivaki, A.; Papagiannopoulou, G.; Zavridis, P.; Zis, P. The Role of Vitamin B6 in Peripheral Neuropathy: A Systematic Review. Nutrients 2023, 15, 2823. [Google Scholar] [CrossRef] [PubMed]
- Holick, M.F. The One-Hundred-Year Anniversary of the Discovery of the Sunshine Vitamin D3: Historical, Personal Experience and Evidence-Based Perspectives. Nutrients 2023, 15, 593. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Fan, J.; Lin, L.; Zhao, M. Combination of Lycium barbarum L. and Laminaria japonica Polysaccharides as a Highly Efficient Prebiotic: Optimal Screening and Complementary Regulation of Gut Probiotics and Their Metabolites. Int. J. Biol. Macromol. 2023, 246, 125534. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wang, S.; Shi, Y.; Fei, X.; Cheng, B.; Lu, Y.; Chen, J. Discovery of Novel d-(+)-Biotin-Conjugated Resorcinol Dibenzyl Ether-Based PD-L1 Inhibitors for Targeted Cancer Immunotherapy. J. Med. Chem. 2023, 66, 10364–10380. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Ogawa, T.; Aono, T. Dethiobiotin Uptake and Utilization by Bacteria Possessing BioYB Operon. Res. Microbiol. 2023, 174, 104131. [Google Scholar] [CrossRef]
- Chen, H.; Qin, L.; Gao, R.; Jin, X.; Cheng, K.; Zhang, S.; Hu, X.; Xu, W.; Wang, H. Neurodevelopmental Effects of Maternal Folic Acid Supplementation: A Systematic Review and Meta-Analysis. Crit. Rev. Food Sci. Nutr. 2023, 63, 3771–3787. [Google Scholar] [CrossRef]
- Tucaliuc, A.; Cîșlaru, A.; Kloetzer, L.; Blaga, A.C. Strain Development, Substrate Utilization, and Downstream Purification of Vitamin C. Processes 2022, 10, 1595. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, F.; Xu, Y.; Qiu, J.; Qian, Y. Gut Flora-Mediated Metabolic Health, the Risk Produced by Dietary Exposure to Acetamiprid and Tebuconazole. Foods 2021, 10, 835. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhao, Y.; Sun, N.; Li, C.; Jiang, Q.; Zhang, Y.; Wei, H.; Li, Y.; Hu, Q.; Li, X. Comparison of the Gut Microbiota and Untargeted Gut Tissue Metabolome of Chinese Mitten Crabs (Eriocheir sinensis) with Different Shell Colors. Front. Microbiol. 2023, 14, 1218152. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Wang, S.; Tian, X.; Liu, W.; Gao, P. Fecal Microbiota Transplantation Confirmed That 919 Syrup Reduced the Ratio of Erucamide to 5-Avab in Hippocampus to Alleviate Postpartum Depression By Regulating Gut Microbes. Front. Immunol. 2023, 14, 1203015. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, C.; Guo, D.; Chen, W.; Xie, Q.; Zhai, Q. Dose-Related Effects of Sn-2 Palmitate, A Specific Positional Distributed Human Milk Fatty Acid, Intake in Early Life on the Composition and Metabolism of the Intestinal Microbiota. J. Dairy Sci. 2023, 106, 8272–8286. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Pak, S.; Li, D.; Dong, L.; Chen, F.; Hu, X.; Ma, L. Bamboo Shoots Modulate Gut Microbiota, Eliminate Obesity in High-Fat-Diet-Fed Mice and Improve Lipid Metabolism. Foods 2023, 12, 1380. [Google Scholar] [CrossRef] [PubMed]
- Morizumi, H.; Okamoto, K.; Akahane, S.; Takemae, H.; Oba, M.; Okada, Y.; Kitano, Y.; Mizutani, T.; Chiba, K. Stereoselective Production of Imino-d-ribitol and C-Azanucleosides through Electrochemical C−H Functionalization. Eur. J. Org. Chem. 2023, 26, e202201046. [Google Scholar] [CrossRef]
- Manolios, N.; Pham, S.; Hou, G.; Du, J.; Quek, C.; Hibbs, D. Non-Antigenic modulation of antigen receptor (TCR) Cβ-FG loop modulates signalling: Implications of external factors influencing T-Cell responses. Int. J. Mol. Sci. 2023, 24, 9334. [Google Scholar] [CrossRef]
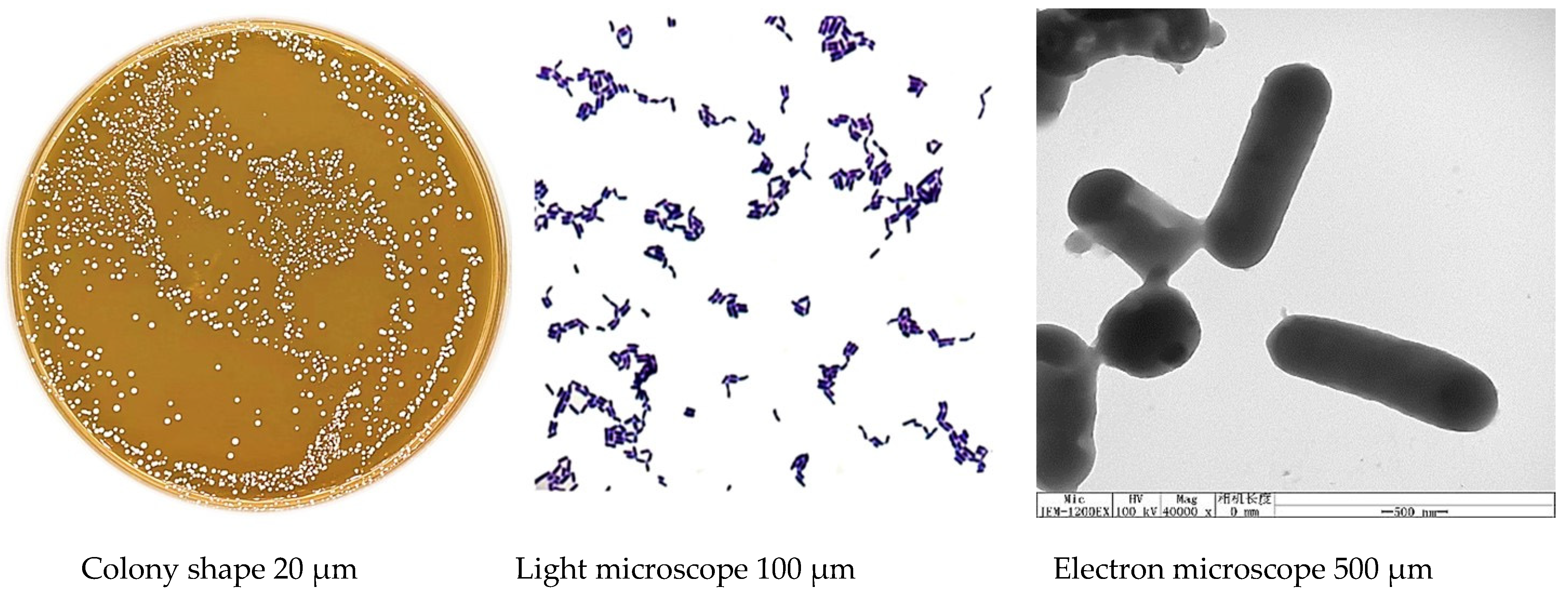
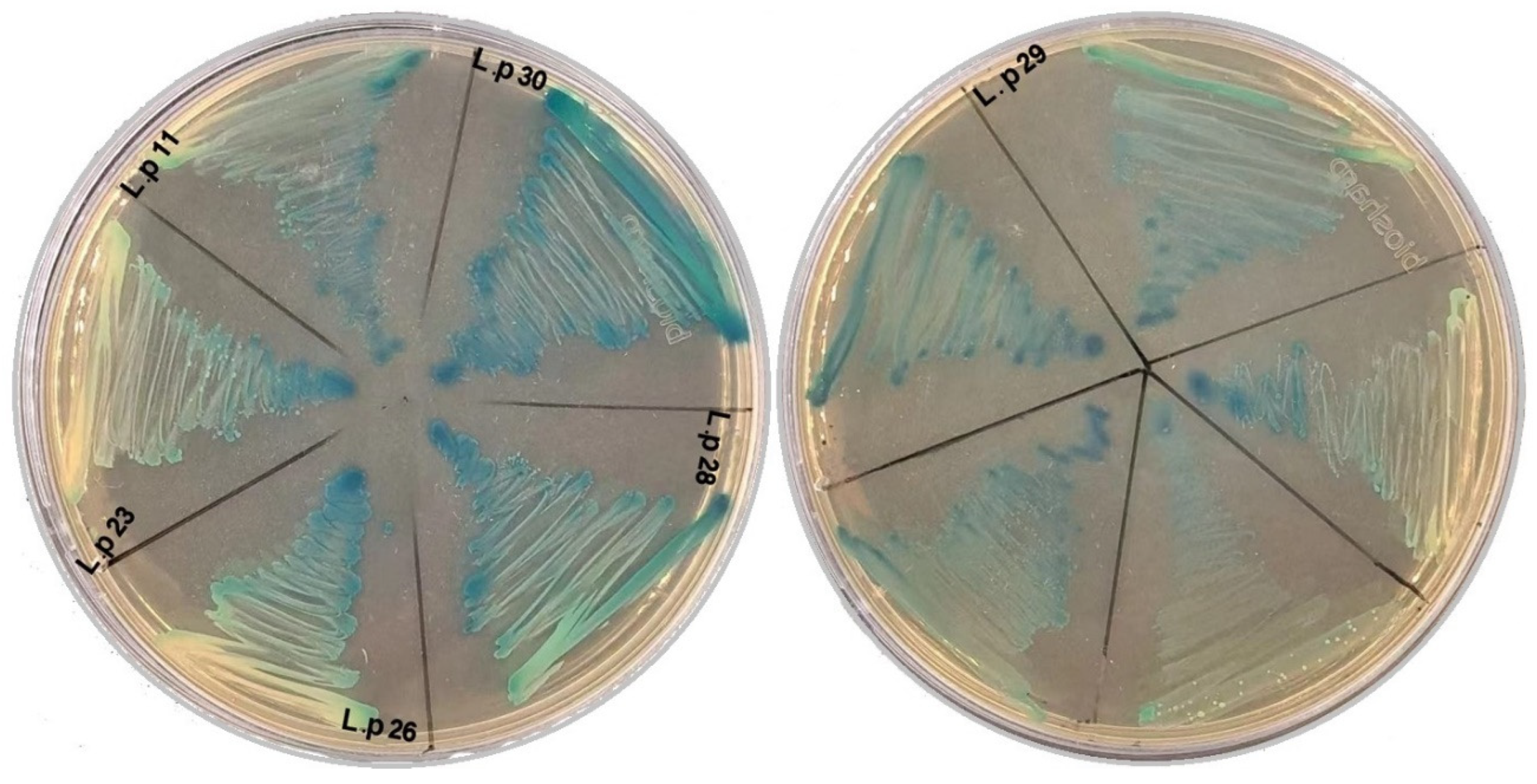



| Names of LAB-Isolated Strains | Sources of Isolated Strains | Growth Media | Temperature |
|---|---|---|---|
| Lactiplantibacillus plantarum (Ne2-11) | Processed cheese, Egypt | MRS | 37, 45 °C |
| Lactiplantibacillus plantarum (23) | Camel manure, China | ||
| Lactiplantibacillus plantarum (26) | |||
| Lactiplantibacillus plantarum (28) | Sand Lake Water, China | ||
| Lactiplantibacillus plantarum (30) | |||
| Lactiplantibacillus plantarum (29) | Baby stool, China |
| Isolated Strains | Accession Version | Reference Strains | Accession Version | Similarity % (Per. Identity) |
|---|---|---|---|---|
| Lactiplantibacillus plantarum (Ne2-11) | PP388282.1 | Lactiplantibacillus plantarum 3761 | MT538615.1 | 99.93% |
| Lactiplantibacillus argentoratensis M22 | OQ405821.1 | 100% | ||
| Lactiplantibacillus plantarum HBUR51045 | OR502116.1 | 99.93% | ||
| Lactiplantibacillus plantarum (23) | PP388283.1 | Lactiplantibacillus plantarum 3358 | MT613641.1 | 99.93% |
| Lactiplantibacillus plantarum 3549 | MT538442.1 | 99.93% | ||
| Lactiplantibacillus plantarum 6405 | MT515846.1 | 99.93% | ||
| Lactiplantibacillus plantarum (26) | PP388284.1 | Lactiplantibacillus plantarum 1968 | MT597774.1 | 99.93% |
| Lactiplantibacillus plantarum 1863 | MT597690.1 | 99.93% | ||
| Lactiplantibacillus plantarum 3733 | MT538590.1 | 99.93% | ||
| Lactiplantibacillus plantarum (28) | PP388285.1 | Lactiplantibacillus plantarum 8194 | MT538940.1 | 100% |
| Lactiplantibacillus plantarum 1985 | MT597789.1 | 100% | ||
| Lactiplantibacillus plantarum 8614 | MT464373.1 | 100% | ||
| Lactiplantibacillus plantarum (29) | PP388286.1 | Lactiplantibacillus plantarum 2207 | MT604705.1 | 100% |
| Lactiplantibacillus plantarum 4294 | MT544837.1 | 100% | ||
| Lactiplantibacillus plantarum VP-2.1 | MF191695.1 | 100% | ||
| Lactiplantibacillus plantarum (30) | PP388295.1 | Lactiplantibacillus plantarum Z3 | OR259088.1 | 99.93% |
| Lactiplantibacillus plantarum LP1 | OR354385.1 | 99.93% | ||
| Lactiplantibacillus plantarum 3482 | MT538386.1 | 99.93% |
| L. plantarum (Ne2-11) | L. plantarum (23) | L. plantarum (26) | L. plantarum (28) | L. plantarum (29) | L. plantarum (30) | |
|---|---|---|---|---|---|---|
| Ami | I | R | R | R | R | R (-) |
| Poly | R (-) | R | R | R (-) | R (-) | R (-) |
| Cef | R (-) | S | I | I | S | S |
| Sul | I | S | S | S | S | S |
| Cm | S | S | S | S | S | I |
| Sepc | I | R (-) | R | R (-) | R (-) | R (-) |
| Gn | I | R (-) | R (-) | R (-) | R (-) | R (-) |
| St | I | R (-) | R (-) | R (-) | R (-) | R (-) |
| Amp | S | S | S | S | R (-) | S |
| Kan | I | R (-) | R (-) | R (-) | R (-) | R (-) |
| Van | R (-) | R (-) | R (-) | R (-) | R (-) | R (-) |
| Nor | R (-) | ND | ND | ND | ND | ND |
| Oxa | R (-) | I | S | R (-) | I | R (-) |
| Tet | I | S | S | I | S | I |
| Cli | S | S | S | I | I | S |
| Pen | R (-) | ND | ND | ND | ND | ND |
| Ery | ND | S | S | S | S | S |
| Pathogens | L. plantarum Strains | |||||
|---|---|---|---|---|---|---|
| (Ne2-11) | (23) | (26) | (28) | (29) | (30) | |
| E. coli | +++ | +++ | +++ | +++ | +++ | +++ |
| S. Typhimurium | +++ | +++ | +++ | +++ | +++ | +++ |
| E. faecium | +++ | +++ | +++ | +++ | +++ | +++ |
| P. aeruginosa | +++ | +++ | +++ | +++ | +++ | +++ |
| Kleb. spp. | +++ | +++ | +++ | +++ | +++ | +++ |
| S. saureus | +++ | +++ | +++ | +++ | +++ | +++ |
| B. cereus | +++ | +++ | +++ | +++ | +++ | +++ |
| C. freundii | +++ | +++ | +++ | +++ | +++ | +++ |
| A. veronii | +++ | +++ | +++ | +++ | +++ | +++ |
| C. youngae | +++ | +++ | +++ | +++ | +++ | +++ |
| C. sakazakii | +++ | +++ | +++ | +++ | +++ | +++ |
| W. chitiniclastica | +++ | +++ | +++ | +++ | +++ | +++ |
| Leclercia sp. | +++ | +++ | +++ | +++ | +++ | +++ |
| P. agglomerans | +++ | +++ | +++ | +++ | +++ | +++ |
| F. nucleatum | +++ | +++ | +++ | +++ | +++ | +++ |
| Isolated Strain | Sugar Names | |||||||
|---|---|---|---|---|---|---|---|---|
| Glucose | Sucrose | Galactose | Mannose | Maltose | Lactose | Arabinose | Xylose | |
| L. plantarum (Ne2-11) | + | + | + | + | + | + | + | + |
| L. plantarum (23) | + | + | + | + | + | + | + | + |
| L. plantarum (26) | + | + | + | + | + | + | + | + |
| L. plantarum (28) | + | + | + | + | + | + | + | + |
| L. plantarum (29) | + | + | + | + | + | + | + | + |
| L. plantarum (30) | + | + | + | + | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdel-motaal, H.; Abdelazez, A.; Wang, P.; Abady, G.; Abozaed, S.; Ye, B.; Xu, L.; Zhao, Y.; Niu, J.; Alshehry, G.; et al. Exploring Phenotype, Genotype, and the Production of Promising GABA Postbiotics by Lactiplantibacillus plantarum: A Comprehensive Investigation. Fermentation 2024, 10, 309. https://doi.org/10.3390/fermentation10060309
Abdel-motaal H, Abdelazez A, Wang P, Abady G, Abozaed S, Ye B, Xu L, Zhao Y, Niu J, Alshehry G, et al. Exploring Phenotype, Genotype, and the Production of Promising GABA Postbiotics by Lactiplantibacillus plantarum: A Comprehensive Investigation. Fermentation. 2024; 10(6):309. https://doi.org/10.3390/fermentation10060309
Chicago/Turabian StyleAbdel-motaal, Heba, Amro Abdelazez, Peikun Wang, Ghada Abady, Shaima Abozaed, Bin Ye, Linan Xu, Yuzhong Zhao, Jianrui Niu, Garsa Alshehry, and et al. 2024. "Exploring Phenotype, Genotype, and the Production of Promising GABA Postbiotics by Lactiplantibacillus plantarum: A Comprehensive Investigation" Fermentation 10, no. 6: 309. https://doi.org/10.3390/fermentation10060309
APA StyleAbdel-motaal, H., Abdelazez, A., Wang, P., Abady, G., Abozaed, S., Ye, B., Xu, L., Zhao, Y., Niu, J., Alshehry, G., Algarni, E., Aljumayi, H., & Zhang, X. (2024). Exploring Phenotype, Genotype, and the Production of Promising GABA Postbiotics by Lactiplantibacillus plantarum: A Comprehensive Investigation. Fermentation, 10(6), 309. https://doi.org/10.3390/fermentation10060309








