Simultaneous Saccharification and Fermentation for Isobutanol Production from Banana Peel
Abstract
1. Introduction
2. Materials and Methods
2.1. Enzymatic Hydrolysis
2.2. Determination of Isobutanol Productivity
2.3. Simultaneous Saccharification and Fermentation for Isobutanol Production
2.4. Quantification of Sugar and Isobutanol
3. Results and Discussion
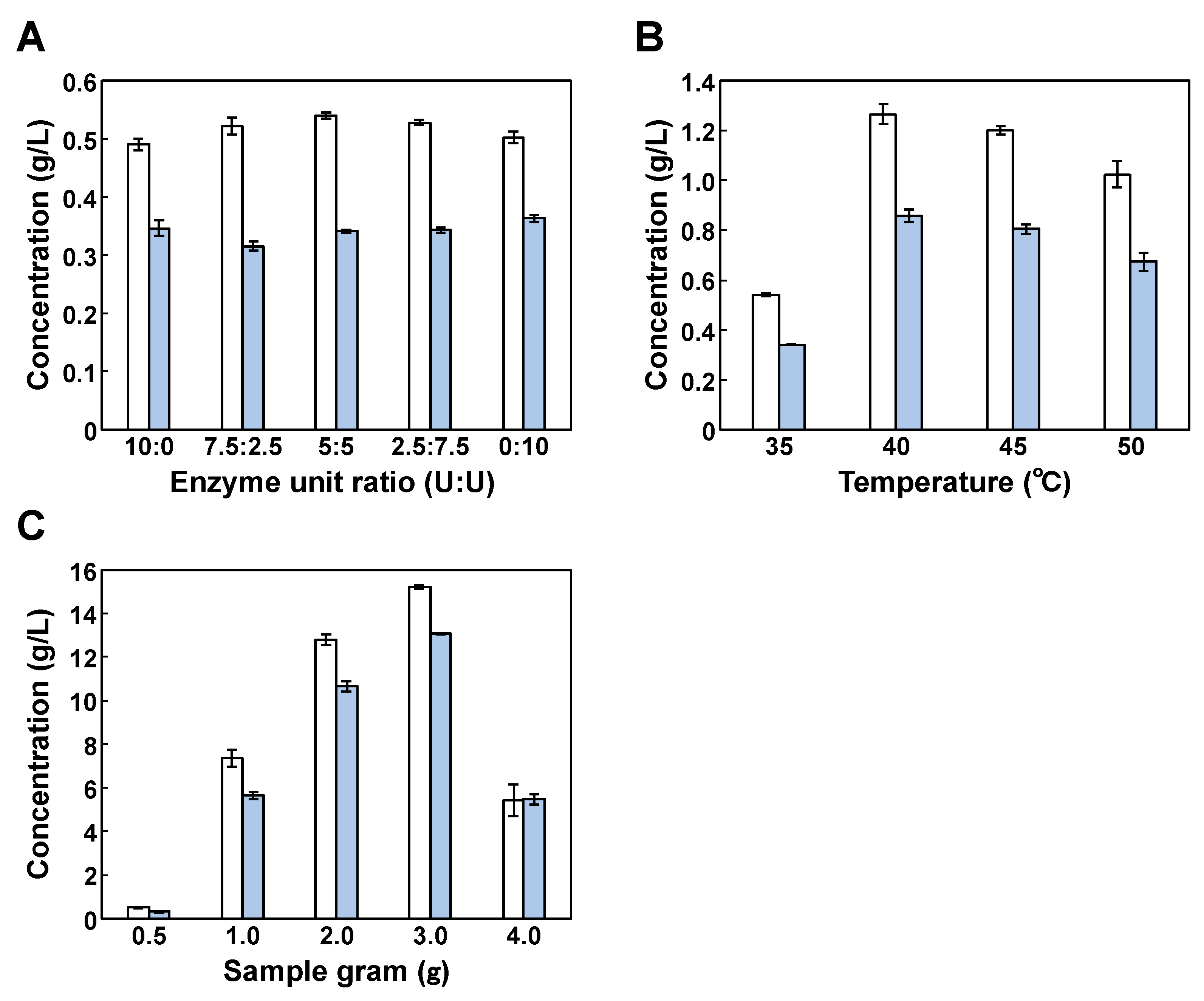
3.1. Use of Cellulase ONOZUKA R-10 and Macerozyme R-10 for Enzymatic Hydrolysis
3.2. Effect of Stirring Speed on Isobutanol Production
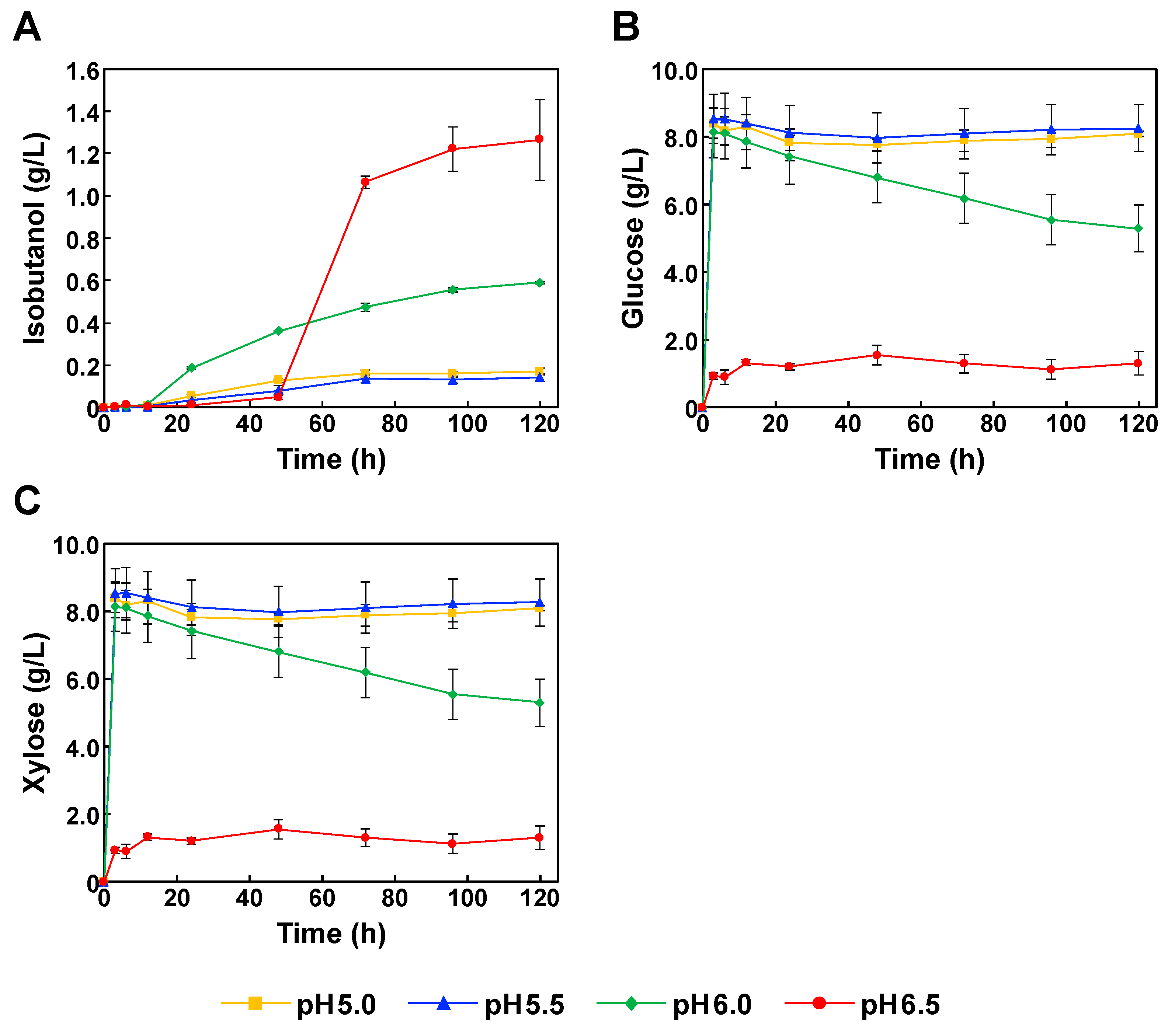
3.3. SSF for Isobutanol Production
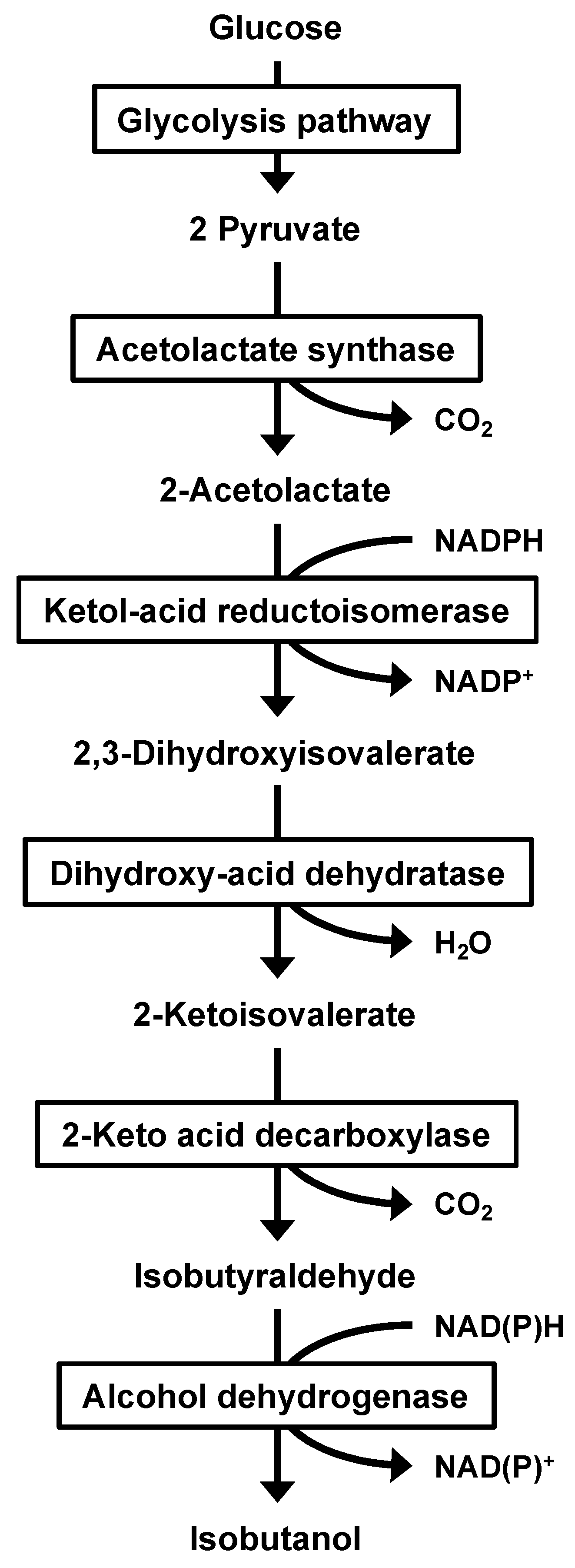
3.4. Comparison of Isobutanol Productivity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Cruz-Morales, P.; Zargar, A.; Belcher, M.S.; Pang, B.; Englund, E.; Dan, Q.; Yin, K.; Keasling, J.D. Biofuels for a sustainable future. Cell 2021, 184, 1636–1647. [Google Scholar] [CrossRef]
- Francis, J. Arctic meltdown and unruly tropical storms: Are they connected? Arct. Antarct. Alp. Res. 2021, 53, 223–224. [Google Scholar]
- Tebaldi, C.; Ranasinghe, R.; Vousdoukas, M.; Rasmussen, D.J.; Vega-Westhoff, B.; Kirezci, E.; Kopp, R.E.; Sriver, R.; Mentaschi, L. Extreme sea levels at different global warming levels. Nat. Clim. Chang. 2021, 11, 746–751. [Google Scholar] [CrossRef]
- Xu, R.B.; Yu, P.; Abramson, M.J.; Johnston, F.H.; Samet, J.M.; Bell, M.L.; Haines, A.; Ebi, K.L.; Li, S.; Guo, Y. Wildfires, global climate change, and human health. N. Engl. J. Med. 2020, 383, 2173–2181. [Google Scholar] [CrossRef]
- Turco, M.; Rosa-Cánovas, J.J.; Bedia, J.; Jerez, S.; Montávez, J.P.; Llasat, M.C.; Provenzale, A. Exacerbated fires in Mediterranean Europe due to anthropogenic warming projected with non-stationary climate-fire models. Nat. Commun. 2018, 9, 3821. [Google Scholar] [CrossRef]
- Chen, X.M.; Sharma, A.; Liu, H. The Impact of climate change on environmental sustainability and human mortality. Environments 2023, 10, 165. [Google Scholar] [CrossRef]
- United Nations Report. Nature’s Dangerous Decline ‘Unprecedented’; Species Extinction Rates ‘Accelerating’. Available online: https://www.un.org/sustainabledevelopment/blog/2019/05/nature-decline-unprecedented-report/ (accessed on 30 January 2024).
- Putra, N.R.; Aziz, A.H.A.; Faizal, A.N.M.; Che Yunus, M.A. Methods and potential in valorization of banana peels waste by various extraction processes: In review. Sustainability 2022, 14, 10571. [Google Scholar] [CrossRef]
- Hikal, W.M.; Said-Al, A.H.A.H.; Bratovcic, A.; Tkachenko, K.G.; Sharifi-Rad, J.; Kačániová, M.; Elhourri, M.; Atanassova, M. Banana peels: A waste treasure for human being. Evid. Based Complement. Alternat. Med. 2022, 2022, 7616452. [Google Scholar] [CrossRef]
- Ragab, M.; Osman, M.F.; Khalil, M.E.; Gouda, M.S. Banana (Musa sp.) peels as a source of pectin and some food nutrients. J. Agric. Res. Kafr El-Sheikh Univ. 2016, 42, 88–102. [Google Scholar]
- Martín, C.; Dixit, P.; Momayez, F.; Jönsson, L.J. Hydrothermal pretreatment of lignocellulosic feedstocks to facilitate biochemical conversion. Front. Bioeng. Biotechnol. 2022, 10, 846592. [Google Scholar] [CrossRef]
- Nakashima, N.; Akita, H.; Hoshino, T. Establishment of a novel gene expression method, BICES (biomass-inducible chromosome-based expression system), and its application to the production of 2,3-butanediol and acetoin. Metab. Eng. 2014, 25, 204–214. [Google Scholar] [CrossRef]
- Matsushika, A.; Hoshino, T. Increased ethanol production by deletion of HAP4 in recombinant xylose-assimilating Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2015, 42, 1623–1631. [Google Scholar] [CrossRef]
- Akita, H.; Nakashima, N.; Hoshino, T. Bacterial production of isobutanol without expensive reagents. Appl. Microbiol. Biotechnol. 2015, 99, 991–999. [Google Scholar] [CrossRef]
- Akita, H.; Goshima, T.; Suzuki, T.; Itoiri, Y.; Kimura, Z.-i.; Matsushika, A. Application of Pichia kudriavzevii NBRC1279 and NBRC1664 to simultaneous saccharification and fermentation for bioethanol production. Fermentation 2021, 7, 83. [Google Scholar] [CrossRef]
- Akita, H.; Matsushika, A. Transcription analysis of the acid tolerance mechanism of Pichia kudriavzevii NBRC1279 and NBRC1664. Fermentation 2023, 9, 559. [Google Scholar] [CrossRef]
- Mattedi, A.; Sabbi, E.; Farda, B.; Djebaili, R.; Mitra, D.; Ercole, C.; Cacchio, P.; Del Gallo, M.; Pellegrini, M. Solid-state fermentation: Applications and future perspectives for biostimulant and biopesticides production. Microorganisms 2023, 11, 1408. [Google Scholar] [CrossRef]
- Beldman, G.; Searle-Van, L.M.F.; Rombouts, F.M.; Voragen, F.G. The cellulase of Trichoderma viride. Purification, characterization and comparison of all detectable endoglucanases, exoglucanases and β-glucosidases. Eur. J. Biochem. 1985, 146, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Kiso, K.; Sekiya, J.; Yasuda, T. Examination of the conditions for protoplast isolation from tobacco cells cultured in vitro. Agric. Biol. Chem. 1972, 36, 1055–1059. [Google Scholar] [CrossRef]
- Schirmer, C.; Nussbaumer, T.; Schöb, R.; Pörtner, R.; Eibl, R.; Eibl, D. Development, engineering and biological characterization of stirred tank bioreactors. In Biopharmaceuticals; Yeh, M., Chen, Y., Eds.; IntechOpen: London, UK, 2018; pp. 87–107. [Google Scholar]
- Akita, H.; Yusoff, M.Z.M.; Fujimoto, S. Preparation of oil palm empty fruit bunch hydrolysate. Fermentation 2021, 7, 81. [Google Scholar] [CrossRef]
- Mansur, M.C.; O’Donnell, M.K.; Rehmann, M.S.; Zohaib, M. ABE fermentation of sugar in Brazil. Snr. Des. Rep. 2010, 2010 17, 1–139. [Google Scholar]
- Fujimoto, S.; Inoue, S.; Yoshida, M. High solid concentrations during the hydrothermal pretreatment of eucalyptus accelerate hemicellulose decomposition and subsequent enzymatic glucose production. Bioresour. Technol. Rep. 2018, 4, 16–20. [Google Scholar] [CrossRef]
- Zakaria, M.R.; Norrrahim, M.N.; Hirata, S.; Hassan, M.A. Hydrothermal and wet disk milling pretreatment for high conversion of biosugars from oil palm mesocarp fiber. Bioresour. Technol. 2015, 181, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Yusoff, M.Z.M.; Akita, H.; Hassan, M.A.; Fujimoto, S.; Yoshida, M.; Nakashima, N.; Hoshino, T. Production of acetoin from hydrothermally pretreated oil mesocarp fiber using metabolically engineered Escherichia coli in a bioreactor system. Bioresour. Technol. 2017, 245, 1040–1048. [Google Scholar] [CrossRef]
- Zakaria, M.R.; Hirata, S.; Hassan, M.A. Combined pretreatment using alkaline hydrothermal and ball milling to enhance enzymatic hydrolysis of oil palm mesocarp fiber. Bioresour. Technol. 2014, 169, 236–243. [Google Scholar] [CrossRef]
- Aulitto, M.; Fusco, S.; Bartolucci, S.; Franzén, C.J.; Contursi, P. Bacillus coagulans MA-13: A promising thermophilic and cellulolytic strain for the production of lactic acid from lignocellulosic hydrolysate. Biotechnol. Biofuels 2017, 10, 210. [Google Scholar] [CrossRef]
- Unden, G.; Bongaerts, J. Alternative respiratory pathways of Escherichia coli: Energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1997, 1320, 217–234. [Google Scholar] [CrossRef]
- Coutard, B.; Danchin, E.G.; Oubelaid, R.; Canard, B.; Bignon, C. Single pH buffer refolding screen for protein from inclusion bodies. Protein Expr. Purif. 2012, 82, 352–359. [Google Scholar] [CrossRef]
- Holtzclaw, W.D.; Chapman, L.F. Degradative acetolactate synthase of Bacillus subtilis: Purification and properties. J. Bacteriol. 1975, 121, 917–922. [Google Scholar] [CrossRef]
- Tadrowski, S.; Pedroso, M.M.; Sieber, V.; Larrabee, J.A.; Guddat, L.W.; Schenk, G. Metal ions play an essential catalytic role in the mechanism of ketol-acid reductoisomerase. Chemistry 2016, 22, 7427–7436. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.W. Dihydroxy acid dehydrase: An enzyme involved in the biosynthesis of isoleucine and valine. J. Biol. Chem. 1961, 236, 1414–1418. [Google Scholar] [CrossRef] [PubMed]
- Smit, B.A.; van Hylckama, V.J.E.; Engels, W.J.; Meijer, L.; Wouters, J.T.; Smit, G. Identification, cloning, and characterization of a Lactococcus lactis branched-chain alpha-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 2005, 71, 303–311. [Google Scholar] [CrossRef]
- Arnau, J.; Jørgensen, F.; Madsen, S.M.; Vrang, A.; Israelsen, H. Cloning of the Lactococcus lactis adhE gene, encoding a multifunctional alcohol dehydrogenase, by complementation of a fermentative mutant of Escherichia coli. J. Bacteriol. 1998, 180, 3049–3055. [Google Scholar] [CrossRef]
- Novak, K.; Baar, J.; Freitag, P.; Pflügl, S. Metabolic engineering of Escherichia coli W for isobutanol production on chemically defined medium and cheese whey as alternative raw material. J. Ind. Microbiol. Biotechnol. 2020, 47, 1117–1132. [Google Scholar] [CrossRef]
- de Jong, S.; Hoefnagels, R.; Wetterlund, E.; Pettersson, K.; Faaij, A.; Junginger, M. Cost optimization of biofuel production—The impact of scale, integration, transport and supply chain configurations. Appl. Energy 2017, 195, 1055–1070. [Google Scholar] [CrossRef]
- Su, H.; Lin, J.; Wang, G. Metabolic engineering of Corynebacterium crenatium for enhancing production of higher alcohols. Sci. Rep. 2016, 6, 39543. [Google Scholar] [CrossRef] [PubMed]
- Bumrungtham, P.; Promdonkoy, P.; Prabmark, K.; Bunterngsook, B.; Boonyapakron, K.; Tanapongpipat, S.; Champreda, V.; Runguphan, W. Engineered production of isobutanol from sugarcane trash hydrolysates in Pichia pastoris. J. Fungi 2022, 8, 767. [Google Scholar] [CrossRef]
- Baez, A.; Cho, K.M.; Liao, J.C. High-flux isobutanol production using engineered Escherichia coli: A bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 2011, 90, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.M.; Lee, J.Y.; Lee, J.H.; Oh, M.K. Improved production of isobutanol in pervaporation-coupled bioreactor using sugarcane bagasse hydrolysate in engineered Enterobacter aerogenes. Bioresour. Technol. 2018, 259, 373–380. [Google Scholar] [CrossRef]
- Sommer, B.; von Moeller, H.; Haack, M.; Qoura, F.; Langner, C.; Bourenkov, G.; Garbe, D.; Loll, B.; Brück, T. Detailed structure-function correlations of Bacillus subtilis acetolactate synthase. Chembiochem 2015, 16, 110–118. [Google Scholar] [CrossRef]
- Atsumi, S.; Li, Z.; Liao, J.C. Acetolactate synthase from Bacillus subtilis serves as a 2-ketoisovalerate decarboxylase for isobutanol biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 2009, 75, 6306–6311. [Google Scholar] [CrossRef]
- Singh, R.N.; Akimenko, V.K. Isolation of a cellobiohydrolase of Clostridium thermocellum capable of degrading natural crystalline substrates. Biochem. Biophys. Res. Commun. 1993, 192, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Romero-Borbon, E.; Grajales-Hernandez, D.; Armendariz-Ruiz, M.; Ramirez-Velasco, L.; Rodriguez-Gonzalez, J.; Cira-Chavez, L.; Estrada-Alvarado, M.; Mateos-Diaz, J. Type C feruloyl esterase from Aspergillus ochraceus A butanol specific biocatalyst for the synthesis of hydroxycinnamates in a ternary solvent system. Electron. J. Biotechnol. 2018, 35, 1–9. [Google Scholar] [CrossRef]
- Nishitani, K.; Nevins, D.J. Enzymic analysis of feruloylated arabinoxylans (Feraxan) derived from Zea mays cell walls. I. Purification of novel enzymes capable of dissociating Feraxan fragments from Zea mays coleoptile cell wall. Plant Physiol. 1988, 87, 883–890. [Google Scholar] [CrossRef] [PubMed]
| Saccharification Enzyme | Activity | Source | Supplier |
|---|---|---|---|
| Accellerase 1500 | Cellulase, xylanase | Trichoderma reesei | Genencor |
| Acremonium cellulase | β-Glucosidase, cellulase, xylanase | Acremonium cellulolyticus | Meiji Seika |
| Biocellulase A | β-Glucosidase | Aspergillus niger | Quest Intl |
| Bio-feed Beta L | β-glucosidase, cellulase, xylanase | Trichoderma longibrachiatum, T. reesei | Novozymes |
| Cellubrix | β-Glucosidase, cellulase, xylanase | T. longibrachiatum, A. niger | Novozymes |
| Cellulase 2000 L | Cellulase, xylanase | T. longibrachiatum, T. reesei | Rhodia-Danisco |
| Cellulase AP | β-Glucosidase, cellulase | A. niger | Amano Enzyme |
| Cellulase ONOZUKA R-10 | α-Amylase, cellulase, hemicellulase, pectinase | Trichoderma virid | Yakult |
| Cellulase TRL | β-Glucosidase | T. longibrachiatum, T. reesei | Solvay Enzymes |
| Cellulyve 50 L | Cellulase, xylanase | T. longibrachiatum, T. reesei | Lyven |
| Energex L | β-Glucosidase, cellulase, xylanase | T. longibrachiatum, T. reesei | Novozymes |
| GC220 | β-Glucosidase, cellulase, xylanase | T. longibrachiatum, T. reesei | Genencor |
| GC440 | β-Glucosidase, cellulase, xylanase | T. longibrachiatum, T. reesei | Genencor |
| GC880 | β-Glucosidase, cellulase, xylanase | T. longibrachiatum, T. reesei | Genencor |
| Macerozyme R-10 | Hemicellulase, pectinase | Rhizopus sp. | Yakult |
| Novozyme 188 | α-Amylase, β-glucosidase, cellulase, glucoamylase, xylanase | A. niger | Novozymes |
| Optimash BG | β-Glucosidase, cellulase, xylanase | Not defined | Genencor International |
| Rohament CL | β-Glucosidase, cellulase, xylanase | T. longibrachiatum, T. reesei | AB Enzymes |
| Spezyme CP | Cellulase, xylanase | T. longibrachiatum, T. reesei | Genencor |
| Ultraflo L | β-Glucosidase, cellulase, xylanase | T. longibrachiatum, T. reesei | Novozymes |
| Ultraflo XL | α-Amylase | Bacillus amyloliquefaciens, Humicola insolens | Novozymes |
| Viscoferm | β-Glucosidase, cellulase, xylanase | Not defined | Novozymes |
| Viscostar 150 L | β-Glucosidase, cellulase, xylanase | T. longibrachiatum, T. reesei | Dyadic |
| Viscozyme L | β-Glucosidase, cellulase, xylanase | Aspergillus sp. | Novozymes |
| Stirring Speed (rpm) | Concentration (g/L) | Productivity [(g/(L·h)] |
|---|---|---|
| 500 | 4.62 ± 0.122 | 0.193 ± 0.00512 |
| 750 | 4.26 ± 0.184 | 0.178 ± 0.00768 |
| 1000 | 3.79 ± 0.369 | 0.158 ± 0.0154 |
| Strain | Use of Expression Plasmid | Source Material | Production Process | Concentration (g/L) | Productivity [(g/(L·h)] | Reference |
|---|---|---|---|---|---|---|
| Corynebacterium crenatium MA11C | Yes | Duckweed | 2 steps | 5.61 | 0.0688 | [37] |
| E. coli mlcXT7-LAFC-AAKCD | No | Banana peel | 2 steps | 1.27 ± 0.191 | 0.0148 ± 0.00200 * | This study |
| E. coli mlcXT7-LAFC-AAKCD | No | Japanese cedar | 3 steps | 3.70 ± 0.0356 | 0.0386 ± 0.000376 | [14] |
| Pichia pastoris PPY0311 | Yes | Sugarcane trash | 3 steps | 0.0482 ± 0.00170 | 0.000669 ± 0.0000236 | [38] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akita, H.; Shibata, S.; Komoriya, T.; Kamei, S.; Asamoto, H.; Matsumoto, M. Simultaneous Saccharification and Fermentation for Isobutanol Production from Banana Peel. Fermentation 2024, 10, 161. https://doi.org/10.3390/fermentation10030161
Akita H, Shibata S, Komoriya T, Kamei S, Asamoto H, Matsumoto M. Simultaneous Saccharification and Fermentation for Isobutanol Production from Banana Peel. Fermentation. 2024; 10(3):161. https://doi.org/10.3390/fermentation10030161
Chicago/Turabian StyleAkita, Hironaga, Shodai Shibata, Tomoe Komoriya, Shinnosuke Kamei, Hiromichi Asamoto, and Masakazu Matsumoto. 2024. "Simultaneous Saccharification and Fermentation for Isobutanol Production from Banana Peel" Fermentation 10, no. 3: 161. https://doi.org/10.3390/fermentation10030161
APA StyleAkita, H., Shibata, S., Komoriya, T., Kamei, S., Asamoto, H., & Matsumoto, M. (2024). Simultaneous Saccharification and Fermentation for Isobutanol Production from Banana Peel. Fermentation, 10(3), 161. https://doi.org/10.3390/fermentation10030161






