Deciphering the Role of microRNA Mediated Regulation of Coronin 1C in Glioblastoma Development and Metastasis
(This article belongs to the Section Small Non-Coding RNA)
Abstract
1. Introduction
1.1. Epidemiology and Molecular Characteristics
1.2. MicroRNAs and Their Role in Glioblastoma Tumorigenesis and Diagnosis
1.3. Nanosensor-Based Diagnostic Approaches for GBM
1.4. Coronin 1C and Its Role in Glioblastoma Multiforme
2. Results
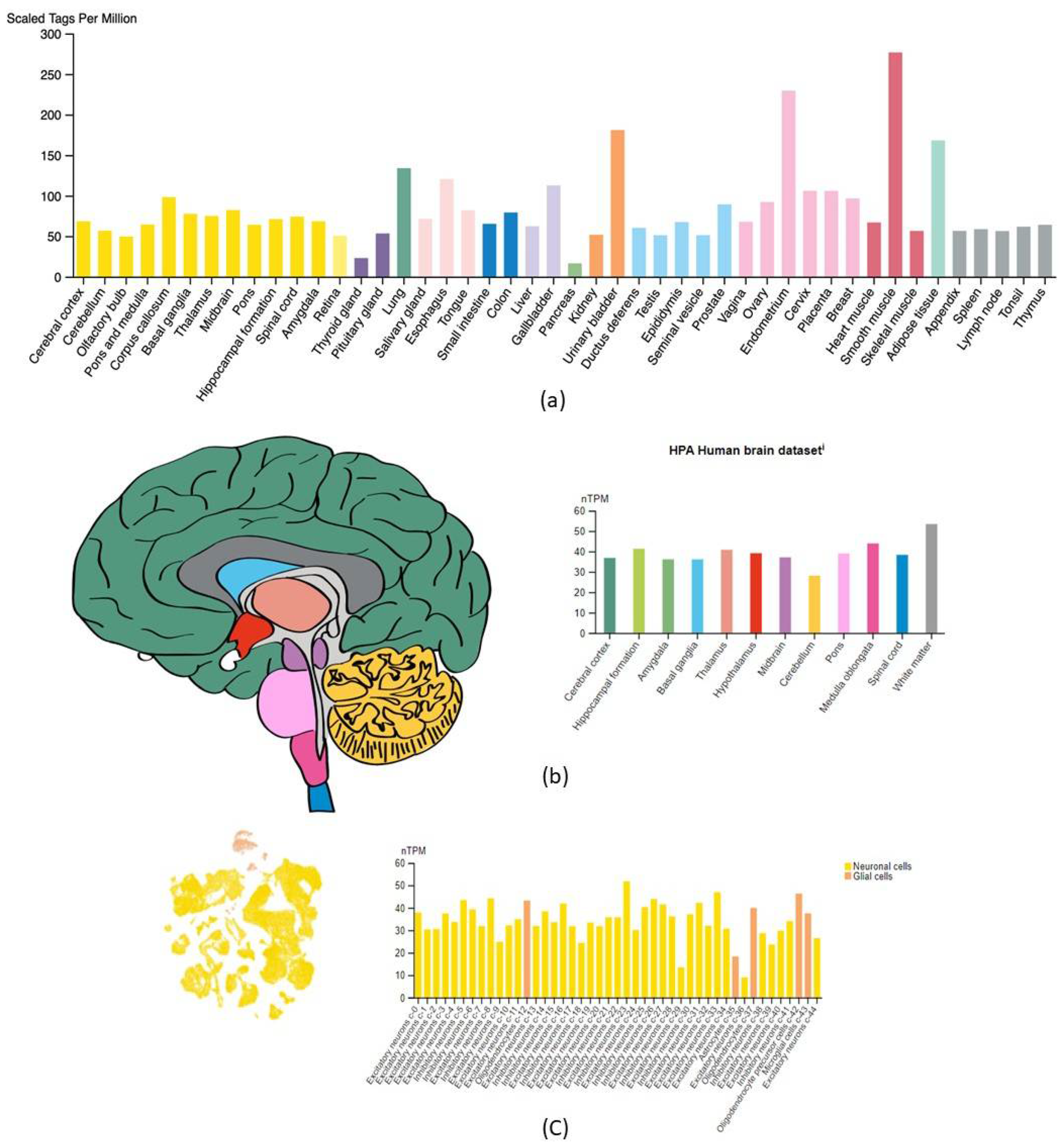
2.1. CORO1C Normal Tissue Distribution
2.2. CORO1C Regional Localization
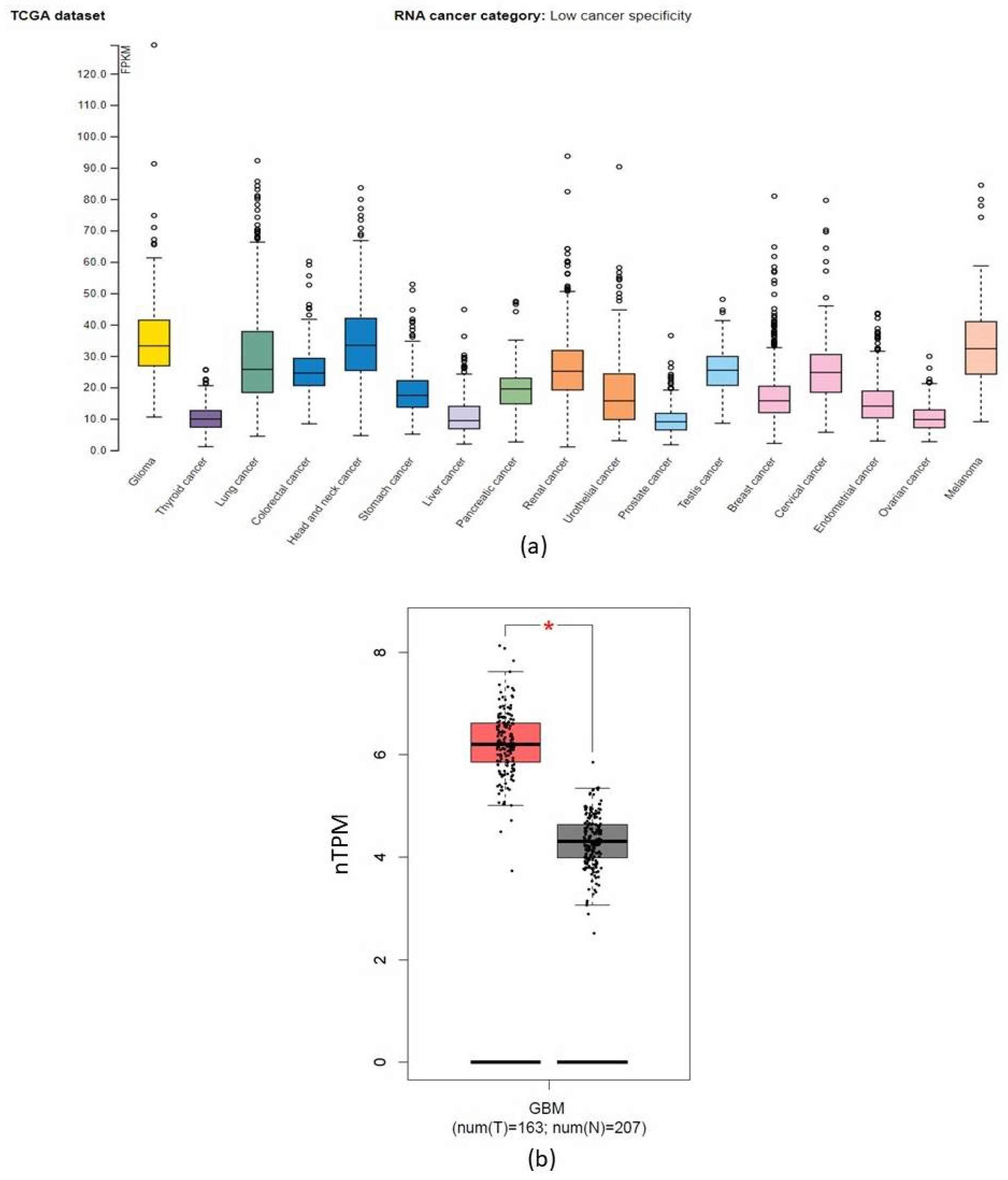
2.3. CORO1C within Various Cancer Types
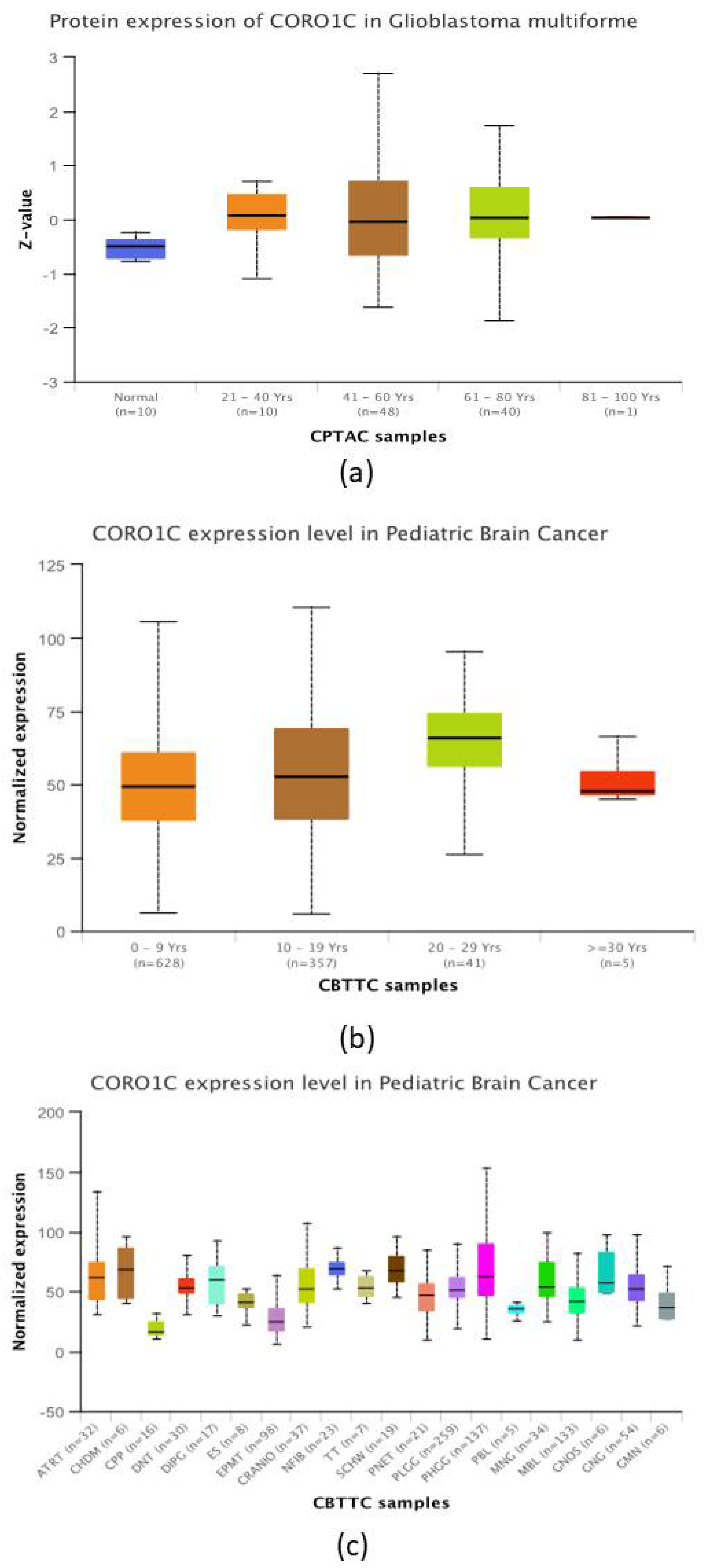
2.4. CORO1C Comparative Analysis of Expression between Paediatric and Adult GBM Cohorts
2.5. CORO1C Single-Cell Expression Analysis in Various GBM Cell Clusters
2.6. miRNA Targets and Physical Interactions of CORO1C
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, X.; Zhang, W.; Mao, X.G.; Cao, W.D.; Zhen, H.N.; Hu, S.J. Malignant intracranial high grade glioma and current treatment strategy. Curr. Cancer Drug Targets 2019, 19, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Davis, M. Glioblastoma: Overview of disease and treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, C.; Bligh, E.; Poon, M.; Solomou, G.; Islim, A.; Mustafa, M.; Rominiyi, O.; Williams, S.; Kalra, N.; Mathew, R.; et al. Imaging timing after glioblastoma surgery (INTERVAL-GB): Protocol for a UK and Ireland, multicentre retrospective cohort study. BMJ Open 2022, 12, e063043. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2014–2018. Neuro-Oncology 2021, 23 (Suppl. 3), iii1–iii105. [Google Scholar] [CrossRef]
- Liu, M.; Thakkar, J.; Garcia, C.; Dolecek, T.; Wagner, L.; Dressler, E.; Villano, J. National cancer database analysis of outcomes in pediatric glioblastoma. Cancer Med. 2018, 7, 1151–1159. [Google Scholar] [CrossRef]
- Gestrich, C.; Jajosky, A.; Elliott, R.; Stearns, D.; Sadri, N.; Cohen, M.; Couce, M. Molecular profiling of pediatric and adult glioblastoma. Am. J. Clin. Pathol. 2020, 155, 606–614. [Google Scholar] [CrossRef]
- Pollack, I.; Hamilton, R.; Sobol, R.; Nikiforova, M.; Lyons-Weiler, M.; La Framboise, B.P.; Brat, D.; Rosenblum, M.; Holmes, E.; Zhou, T.; et al. IDH1 mutations are common in malignant gliomas arising in adolescents: A report from the Children’s Oncology Group. Child’s Nerv. Syst. 2010, 27, 87–94. [Google Scholar] [CrossRef]
- Pollack, I.; Finkelstein, S.; Woods, J.; Burnham, J.; Holmes, E.; Hamilton, R.; Yates, A.; Boyett, J.; Finlay, J.; Sposto, R. Expression of p53 and prognosis in children with malignant gliomas. N. Engl. J. Med. 2002, 346, 420–427. [Google Scholar] [CrossRef]
- Miklja, Z.; Pasternak, A.; Stallard, S.; Nicolaides, T.; Kline-Nunnally, C.; Cole, B.; Beroukhim, R.; Bandopadhayay, P.; Chi, S.; Ramkissoon, S.; et al. Molecular profiling and targeted therapy in pediatric gliomas: Review and consensus recommendations. Neuro-Oncology 2019, 21, 968–980. [Google Scholar] [CrossRef]
- Cohen, K.; Pollack, I.; Zhou, T.; Buxton, A.; Holmes, E.; Burger, P.; Brat, D.; Rosenblum, M.; Hamilton, R.; Lavey, R.; et al. Temozolomide in the treatment of high-grade gliomas in children: A report from the Children’s Oncology Group. Neuro-Oncology 2011, 13, 317–323. [Google Scholar] [CrossRef]
- Bailey, C.; Figueroa, M.; Mohiuddin, S.; Zaky, W.; Chandra, J. Cutting edge therapeutic insights derived from molecular biology of pediatric high-grade glioma and diffuse intrinsic pontine glioma (DIPG). Bioengineering 2018, 5, 88. [Google Scholar] [CrossRef] [PubMed]
- Huynh-Le, M.; Walker, A.; Burger, P.; Jallo, G.; Cohen, K.; Wharam, M.; Terezakis, S. Management of pediatric intracranial low-grade gliomas: Long-term follow-up after radiation therapy. Child’s Nerv. Syst. 2016, 32, 1425–1430. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.; Baker, S. Capturing the molecular and biological diversity of high-grade astrocytoma in genetically engineered mouse models. Oncotarget 2012, 3, 67–77. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, V.; Ha, M. Regulation of microRNA biogenesis. FASEB J. 2009, 23, 509–524. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, V. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Gatto, L.; Franceschi, E.; Di Nunno, V.; Tosoni, A.; Lodi, R.; Brandes, A. Liquid biopsy in glioblastoma manage-ment: From current research to future perspectives. Oncologist 2021, 26, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Areeb, Z.; Stylli, S.; Koldej, R.; Ritchie, D.; Siegal, T.; Morokoff, A.; Kaye, A.; Luwor, R. microRNA as potential biomarkers in glioblastoma. J. Neuro-Oncol. 2015, 125, 237–248. [Google Scholar] [CrossRef]
- Møller, H.; Rasmussen, A.; Andersen, H.; Johnsen, K.; Henriksen, M.; Duroux, M. A systematic review of mi-croRNA in glioblastoma multiforme: Micro-modulators in the mesenchymal mode of migration and invasion. Mol. Neurobiol. 2012, 47, 131–144. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.; Cohen, S. Principles of microRNA–target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C. The role of microRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 1–9. [Google Scholar] [CrossRef]
- Qu, S.; Guan, J.; Liu, Y. Identification of microRNAs as novel biomarkers for glioma detection: A meta-analysis based on 11 articles. J. Neurol. Sci. 2015, 348, 181–187. [Google Scholar] [CrossRef]
- Kalkan, R.; Atli, E. The impacts of miRNAs in glioblastoma progression. Crit. Rev. Eukaryot. Gene Express. 2016, 26, 137–142. [Google Scholar] [CrossRef]
- Roth, P.; Wischhusen, J.; Happold, C.; Chandran, P.; Hofer, S.; Eisele, G.; Weller, M.; Keller, A. A specific miRNA signature in the peripheral blood of glioblastoma patients. J. Neurochem. 2011, 118, 449–457. [Google Scholar] [CrossRef]
- Tomanek, B.; Iqbal, U.; Blasiak, B.; Abulrob, A.; Albaghdadi, H.; Matyas, J.R.; Ponjevic, D.; Sutherland, G.R. Evaluation of brain tumor vessels specific contrast agents for glioblastoma imaging. Neuro-Oncology 2012, 14, 53–63. [Google Scholar] [CrossRef][Green Version]
- Hadjipanayis, C.G.; Machaidze, R.; Kaluzova, M.; Wang, L.; Schuette, A.J.; Chen, H.; Wu, X.; Mao, H. EGFRvIII Anti-body–Conjugated Iron Oxide Nanoparticles for Magnetic Resonance Imaging–Guided Convection-Enhanced Delivery and Targeted Therapy of GlioblastomaEGFRvIII-Targeted Therapy of GBM by IONPs after CED. Cancer Res. 2010, 70, 6303–6312. [Google Scholar] [CrossRef]
- Kleihues, P.; Cavenee, W.K. Pathology and Genetics: Tumours of the Nervous System; IARC-Press: Lyon, France, 2000. [Google Scholar]
- Yang, B.; Wang, S.; Zeng, J.; Zhang, Y.; Ruan, X.; Han, W.; Yin, B.; Yuan, J.; Qiang, B.; Ying, W.; et al. Proteomic screening and identification of microRNA-128 targets in glioma cells. Proteomics 2015, 15, 2602–2617. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. Publ. Protein Soc. 2018, 27, 233–244. [Google Scholar] [CrossRef]
- Hawrylycz, M.J.; Lein, E.S.; Guillozet-Bongaarts, A.L.; Shen, E.H.; Ng, L.; Miller, J.A.; van de Lagemaat, L.N.; Smith, K.A.; Ebbert, A.; Riley, Z.L.; et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012, 489, 391–399. [Google Scholar] [CrossRef]
- Molinie, N.; Rubtsova, S.N.; Fokin, A.; Visweshwaran, S.P.; Rocques, N.; Polesskaya, A.; Schnitzler, A.; Vacher, S.; Den-isov, E.V.; Tashireva, L.A.; et al. Cortical branched actin determines cell cycle progression. Cell Res. 2019, 29, 432–445. [Google Scholar] [CrossRef]
- Wang, Z.; Jia, L.; Sun, Y.; Li, C.; Zhang, L.; Wang, X.; Chen, H. CORO1C is associated with poor prognosis and promotes metastasis through PI3K/AKT pathway in colorectal cancer. Front. Mol. Biosci. 2021, 8, 682594. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Kong, S.; Ma, S.; Shen, L.; Zheng, M.; Qin, S.; Qi, J.; Wang, Q.; Cui, X.; Ju, S. Hsa_circ_0000437 promotes pathogenesis of gastric cancer and lymph node metastasis. Oncogene 2022, 41, 4724–4735. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Sheng, S.; Ye, L.; Xu, X.; Ma, Y.; Feng, X.; Qiu, L.; Fan, Z.; Wang, Y.; Xia, X.; et al. Extracellular vesicles derived from glioblastoma promote proliferation and migration of neural progenitor cells via PI3K-Akt pathway. Cell Commun. Signal. CCS 2022, 20, 7. [Google Scholar] [CrossRef]
- Rosentreter, A.; Hofmann, A.; Xavier, C.P.; Stumpf, M.; Noegel, A.A.; Clemen, C.S. Coronin 3 involvement in F-actin-dependent processes at the cell cortex. Exp. Cell Res. 2007, 313, 878–895. [Google Scholar] [CrossRef] [PubMed]
- Thal, D.; Xavier, C.P.; Rosentreter, A.; Linder, S.; Friedrichs, B.; Waha, A.; Pietsch, T.; Stumpf, M.; Noegel, A.; Clemen, C. Expression of coronin-3 (coronin-1C) in diffuse gliomas is related to malignancy. J. Pathol. 2008, 214, 415–424. [Google Scholar] [CrossRef]
- Martorella, M.; Barford, K.; Winkler, B.; Deppmann, C.D. Emergent role of Coronin-1a in neuronal signaling. Vitam. Horm. 2017, 104, 113–131. [Google Scholar]
- Hasse, A.; Rosentreter, A.; Spoerl, Z.; Stumpf, M.; Noegel, A.A.; Clemen, C.S. Coronin 3 and its role in murine brain morphogenesis. Eur. J. Neurosci. 2005, 21, 1155–1168. [Google Scholar] [CrossRef]
- Wang, M.; Li, Q.; Yu, S.; Zhang, Z.; Qiu, P.; Zhang, Y.; Yang, W.; Xu, G.; Xu, T. Coronin 3 promotes the devel-opment of oncogenic properties in glioma through the Wnt/β-catenin signaling pathway. OncoTargets Ther. 2020, 13, 6661–6673. [Google Scholar] [CrossRef]
- Campbell, P.J.; Getz, G.; Stuart, J.M.; Korbel, J.O.; Stein, L.D. ICGC/TCGA Pan-cancer analysis of whole genomes consortium Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar]
- Michaeli, O.; Tabori, U. Pediatric high grade gliomas in the context of cancer predisposition syndromes. J. Korean Neurosurg. Soc. 2018, 61, 319–332. [Google Scholar] [CrossRef]
- Tabori, U.; Hansford, J.R.; Achatz, M.I.; Kratz, C.P.; Plon, S.E.; Frebourg, T.; Brugières, L. Clinical management and tumor surveillance recommendations of inherited mismatch repair deficiency in childhood. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, e32–e37. [Google Scholar] [CrossRef]
- Goldblum, J.R.; Weiss, S.W.; Folpe, A.L. Enzinger and Weiss’s Soft Tissue Tumors E-Book; Elsevier Health Sciences: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Costa, A.D.A.; Gutmann, D.H. Brain tumors in neurofibromatosis type 1. Neuro-Oncol. Adv. 2020, 2 (Suppl. 1), i85–i97. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An integrative model of cellular states, plasticity, and genetics for G glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef]
- Borst, K.; Dumas, A.A.; Prinz, M. Microglia: Immune and non-immune functions. Immunity 2021, 54, 2194–2208. [Google Scholar] [CrossRef]
- Broekman, M.L.; Maas, S.L.; Abels, E.R.; Mempel, T.R.; Krichevsky, A.M.; Breakefield, X.O. Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 2018, 14, 482–495. [Google Scholar] [CrossRef]
- Wang, J.; Liu, J.; Sun, G.; Meng, H.; Wang, J.; Guan, Y.; Yin, Y.; Zhao, Z.; Dong, X.; Yin, S.; et al. Glioblastoma extracellular vesicles induce the tumour-promoting transformation of neural stem cells. Cancer Lett. 2019, 466, 1–12. [Google Scholar] [CrossRef]
- Zhang, Y.; Chao, T.; Li, R.; Liu, W.; Chen, Y.; Yan, X.; Gong, Y.; Yin, B.; Liu, W.; Qiang, B.; et al. microRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J. Mol. Med. 2009, 87, 43–51. [Google Scholar] [CrossRef]
- Hersi, H.M.; Raulf, N.; Gaken, J.; Folarin, N.; Tavassoli, M. MicroRNA-9 inhibits growth and invasion of head and neck cancer cells and is a predictive biomarker of response to plerixafor, an inhibitor of its target CXCR4. Mol. Oncol. 2018, 12, 2023–2041. [Google Scholar] [CrossRef]
- Xu, F.; Li, F.; Zhang, W.; Jia, P. Growth of glioblastoma is inhibited by miR-133-mediated EGFR suppression. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 9553–9558. [Google Scholar] [CrossRef]
- Zhou, F.; Cao, W.; Xu, R.; Zhang, J.; Yu, T.; Xu, X.; Zhi, T.; Yin, J.; Cao, S.; Liu, N.; et al. Mi-croRNA-206 attenuates glioma cell proliferation, migration, and invasion by blocking the WNT/β-catenin pathway via direct targeting of Frizzled 7 mRNA. Am. J. Transl. Res. 2019, 11, 4584–4601. [Google Scholar]
- Luo, G.; Luo, W.; Sun, X.; Lin, J.; Wang, M.; Zhang, Y.; Luo, W.; Zhang, Y. MicroRNA-21 promotes migration and invasion of glioma cells via activation of Sox2 and β-catenin signaling. Mol. Med. Rep. 2017, 15, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Pang, B.; Xin, T.; Guo, H.; Xing, Y.; Xu, S.; Feng, B.; Liu, B.; Pang, Q. Plasma miR-221/222 family as novel descriptive and prognostic biomarkers for glioma. Mol. Neurobiol. 2016, 53, 1452–1460. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Qi, L.; Tong, S.; Cui, Y.; Chen, J.; Huang, T.; Chen, Z.; Zu, X. miR-128 downregulation promotes growth and metastasis of bladder cancer cells and involves VEGF-C upregulation. Oncol. Lett. 2015, 10, 3183–3190. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Shen, X.; Han, X. miR-128 Functions as an OncomiR for the Downregulation of HIC1 in Breast Cancer. Front. Pharmacol. 2019, 10, 1202. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.; Chinnaiyan, A. The role of non-coding RNAs in oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Zhang, W.; Kim, P.J.; Chen, Z.; Lokman, H.; Qiu, L.; Zhang, K.; Rozen, S.G.; Tan, E.K.; Je, H.S.; Zeng, L. MiRNA-128 regulates the proliferation and neurogenesis of neural precursors by targeting PCM1 in the developing cortex. eLife 2016, 5, e11324. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, H.; Xia, M.; Sun, C.; Li, N.; Guo, E.; Guo, L.; Shan, W.; Lu, H.; Wu, Y.; et al. Overexpressed miR-9 promotes tumor metastasis via targeting E-cadherin in serous ovarian cancer. Front. Med. 2017, 11, 214–222. [Google Scholar] [CrossRef]
- Ben-Hamo, R.; Zilberberg, A.; Cohen, H.; Efroni, S. hsa-miR-9 controls the mobility behavior of glioblastoma cells via regulation of MAPK14 signaling elements. Oncotarget 2016, 7, 23170–23181. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Li, Y.; Tan, Y.; Liu, Q.; Jiang, S.; Liu, D.; Chen, Q.; Zhang, S. MiR-9-5p Inhibits Glioblastoma Cells Proliferation Through Directly Targeting FOXP2 (Forkhead Box P2). Front. Oncol. 2019, 9, 1176. [Google Scholar] [CrossRef]
- Luo, T.; Yi, X.; Si, W. Identification of miRNA and genes involving in osteosarcoma by comprehensive analysis of microRNA and copy number variation data. Oncol. Lett. 2017, 14, 5427–5433. [Google Scholar] [CrossRef]
- Han, S.; Ding, X.; Wang, S.; Xu, L.; Li, W.; Sun, W. miR-133a-3p regulates hepatocellular carcinoma progression through targeting CORO1C. Cancer Manag. Res. 2020, 12, 8685. [Google Scholar] [CrossRef]
- Liao, M.; Peng, L. Mir-206 may suppress non-small lung cancer metastasis by targeting Coro1c. Cell. Mol. Biol. Lett. 2020, 25, 1–13. [Google Scholar] [CrossRef]
- Zhang, C.Z.; Zhang, J.X.; Zhang, A.L.; Shi, Z.D.; Han, L.; Jia, Z.F.; Yang, W.D.; Wang, G.X.; Jiang, T.; You, Y.P.; et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer 2010, 9, 229. [Google Scholar] [CrossRef]
- Tan, X.; Tang, H.; Bi, J.; Li, N.; Jia, Y. MicroRNA-222-3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion, and inhibits apoptosis in gastric cancer. J. Cell. Biochem. 2018, 119, 5153–5162. [Google Scholar] [CrossRef]
- Zhou, H.; Zhu, X. MicroRNA-21 and microRNA-30c as diagnostic biomarkers for prostate cancer: A meta-analysis. Cancer Manag. Res. 2019, 11, 2039–2050. [Google Scholar] [CrossRef]
- Bica-Pop, C.; Cojocneanu-Petric, R.; Magdo, L.; Raduly, L.; Gulei, D.; Berindan-Neagoe, I. Overview upon miR-21 in lung cancer: Focus on NSCLC. Cell. Mol. Life Sci. CMLS 2018, 75, 3539–3551. [Google Scholar] [CrossRef]
- Zeng, L.P.; Hu, Z.M.; Li, K.; Xia, K. miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR Axis in bladder cancer cells. J. Cell. Mol. Med. 2016, 20, 559–567. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, T.; Yang, S.; Xia, M.; Xu, J.; An, H.; Yang, Y.; Li, S. High-mobility group A1 proteins enhance the expression of the oncogenic miR-222 in lung cancer cells. Mol. Cell. Biochem. 2011, 357, 363–371. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mustafov, D.; Karteris, E.; Braoudaki, M. Deciphering the Role of microRNA Mediated Regulation of Coronin 1C in Glioblastoma Development and Metastasis. Non-Coding RNA 2023, 9, 4. https://doi.org/10.3390/ncrna9010004
Mustafov D, Karteris E, Braoudaki M. Deciphering the Role of microRNA Mediated Regulation of Coronin 1C in Glioblastoma Development and Metastasis. Non-Coding RNA. 2023; 9(1):4. https://doi.org/10.3390/ncrna9010004
Chicago/Turabian StyleMustafov, Denis, Emmanouil Karteris, and Maria Braoudaki. 2023. "Deciphering the Role of microRNA Mediated Regulation of Coronin 1C in Glioblastoma Development and Metastasis" Non-Coding RNA 9, no. 1: 4. https://doi.org/10.3390/ncrna9010004
APA StyleMustafov, D., Karteris, E., & Braoudaki, M. (2023). Deciphering the Role of microRNA Mediated Regulation of Coronin 1C in Glioblastoma Development and Metastasis. Non-Coding RNA, 9(1), 4. https://doi.org/10.3390/ncrna9010004