High Throughput FISH Screening Identifies Small Molecules That Modulate Oncogenic lncRNA MALAT1 via GSK3B and hnRNPs
Abstract
1. Introduction
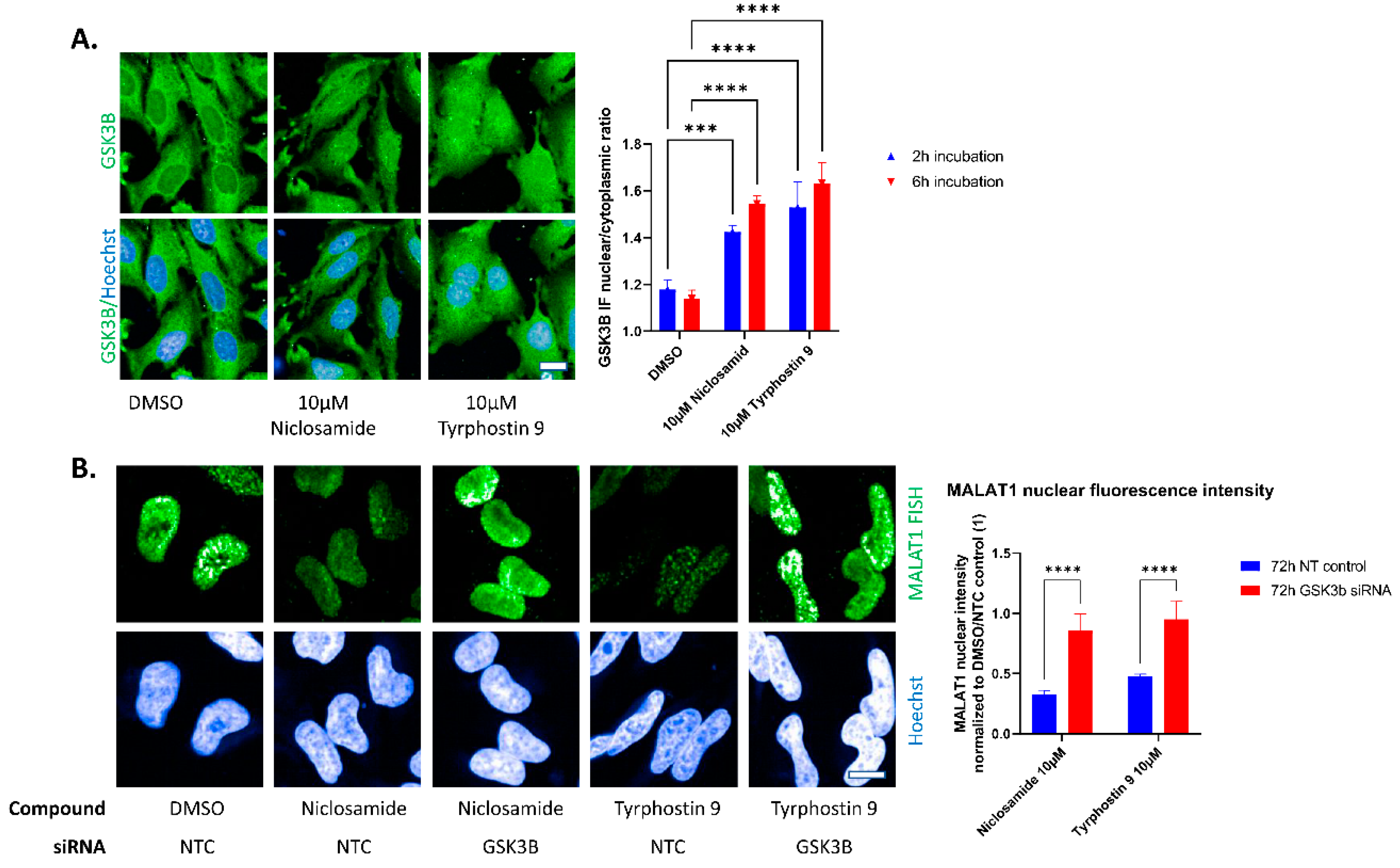
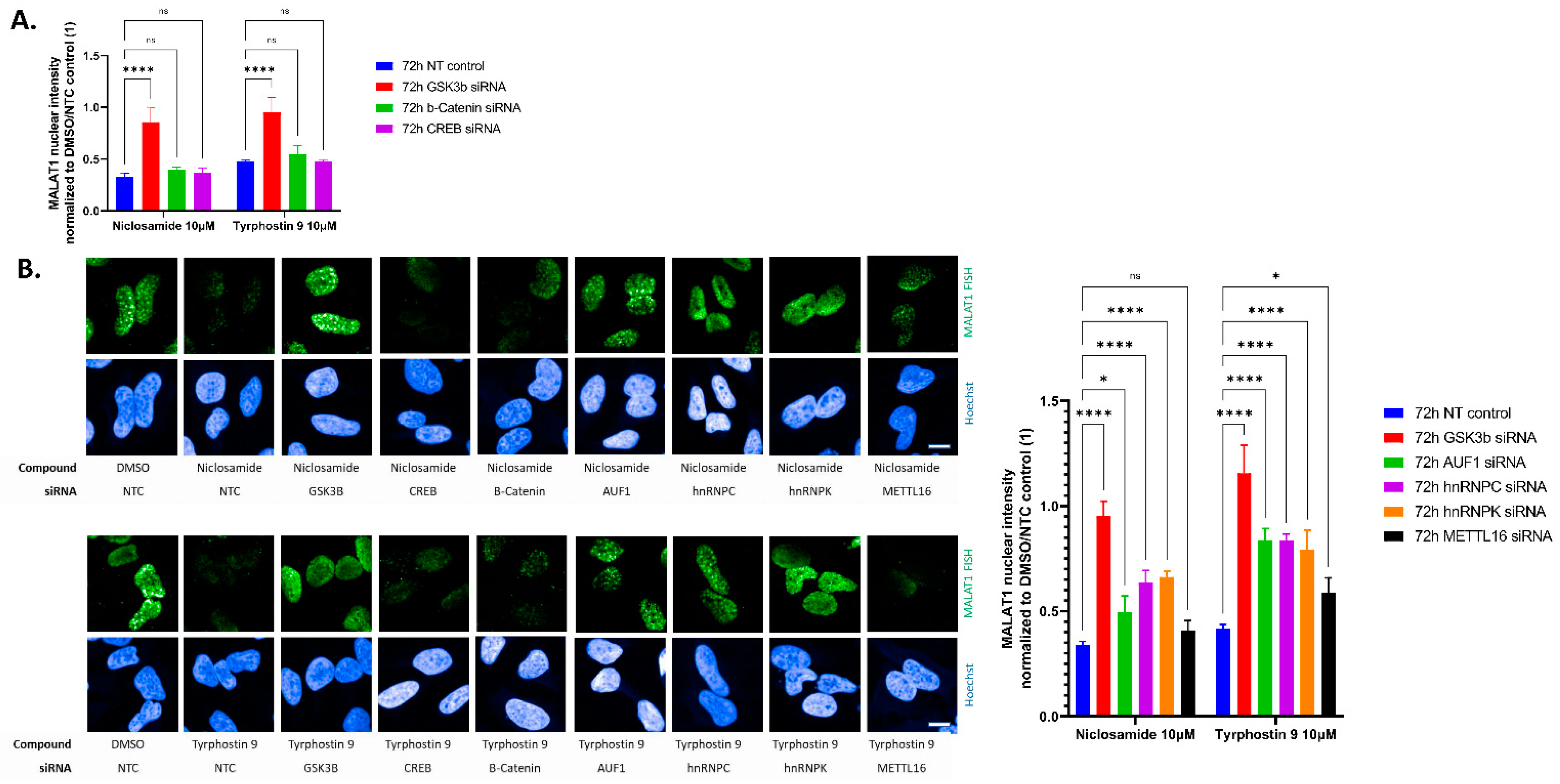
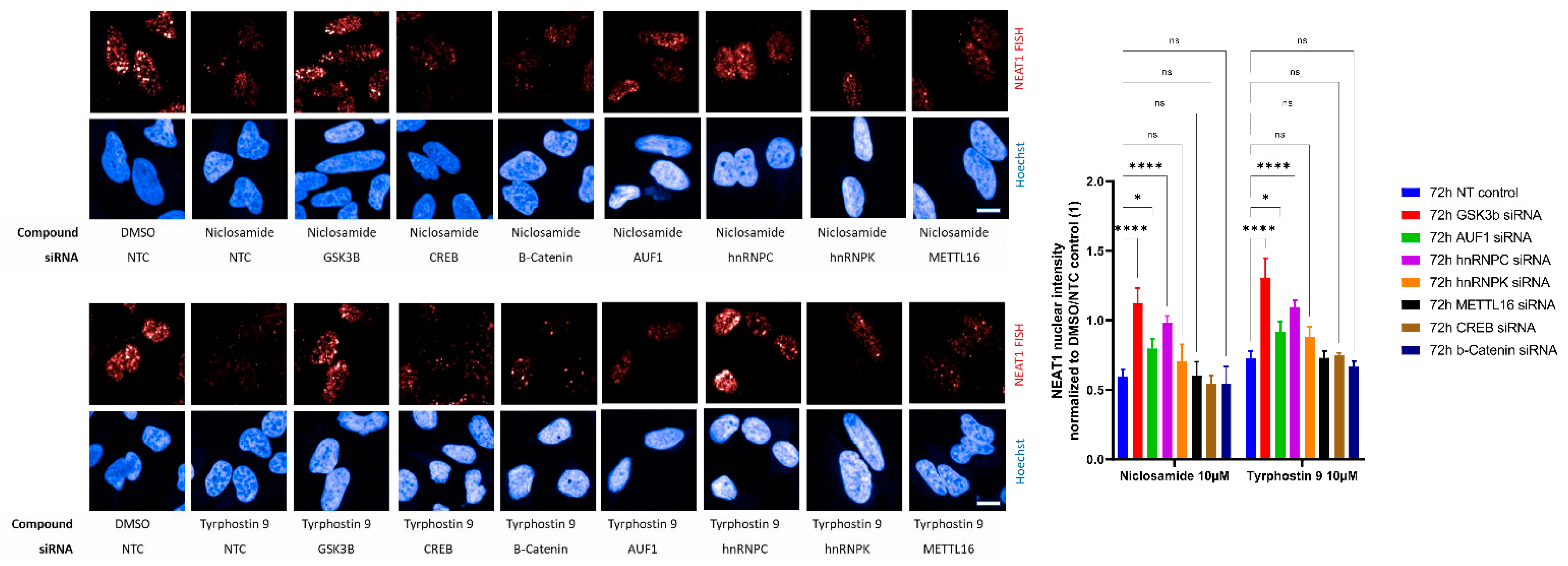
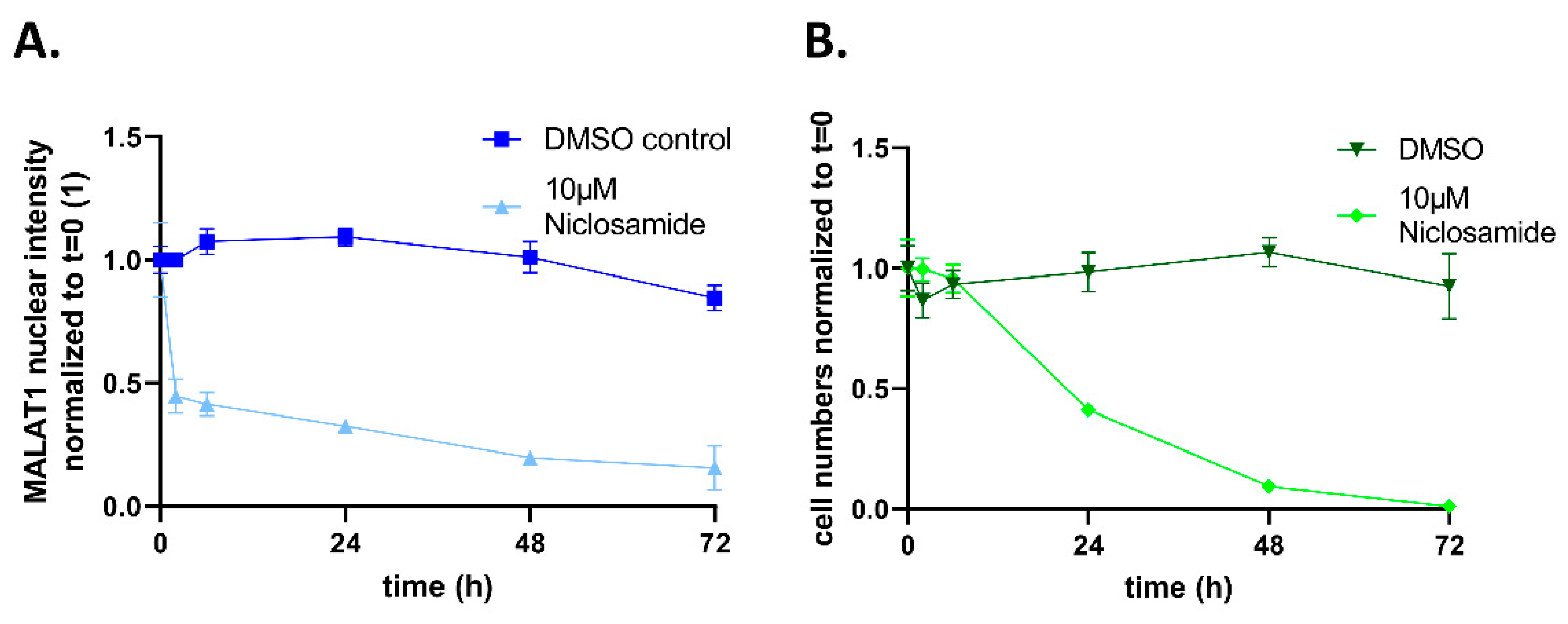
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Fluorescence-In-Situ-Hybridization (FISH)
4.3. Immunofluorescence
4.4. qRT-PCR
4.5. Imaging and Image and Data Analysis
4.6. High Throughput Screen
4.7. siRNA Transfection
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warner, K.D.; Hajdin, C.E.; Weeks, K.M. Principles for Targeting RNA with Drug-like Small Molecules. Nat. Rev. Drug Discov. 2018, 17, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Falese, J.P.; Donlic, A.; Hargrove, A.E. Targeting RNA with Small Molecules: From Fundamental Principles towards the Clinic. Chem. Soc. Rev. 2021, 50, 2224–2243. [Google Scholar] [CrossRef] [PubMed]
- Childs-Disney, J.L.; Yang, X.; Gibaut, Q.M.R.; Tong, Y.; Batey, R.T.; Disney, M.D. Targeting RNA Structures with Small Molecules. Nat. Rev. Drug Discov. 2022, 21, 736–762. [Google Scholar] [CrossRef]
- Zhao, R.; Fu, J.; Zhu, L.; Chen, Y.; Liu, B. Designing Strategies of Small-Molecule Compounds for Modulating Non-Coding RNAs in Cancer Therapy. J. Hematol. Oncol. 2022, 15, 14. [Google Scholar] [CrossRef]
- Raj, A.; Rinn, J.L. Illuminating Genomic Dark Matter with RNA Imaging. Cold Spring Harb. Perspect. Biol. 2019, 11, a032094. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a Novel Noncoding RNA, and Thymosin Beta4 Predict Metastasis and Survival in Early-Stage Non-Small Cell Lung Cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A Druggable Long Non-Coding RNA for Targeted Anti-Cancer Approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A Screen for Nuclear Transcripts Identifies Two Linked Noncoding RNAs Associated with SC35 Splicing Domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef]
- Goyal, B.; Yadav, S.R.M.; Awasthee, N.; Gupta, S.; Kunnumakkara, A.B.; Gupta, S.C. Diagnostic, Prognostic, and Therapeutic Significance of Long Non-Coding RNA MALAT1 in Cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188502. [Google Scholar] [CrossRef]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long Noncoding RNAs in Cancer Metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef]
- Lu, J.; Guo, J.; Liu, J.; Mao, X.; Xu, K. Long Non-Coding RNA MALAT1: A Key Player in Liver Diseases. Front. Med. 2021, 8, 734643. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Banerjee, S.; Guo, S.; Xie, N.; Ge, J.; Jiang, D.; Zörnig, M.; Thannickal, V.J.; Liu, G. Long Noncoding RNA Malat1 Regulates Differential Activation of Macrophages and Response to Lung Injury. JCI Insight 2019, 4, 124522. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Bulkley, D.; Wang, J.; Valenstein, M.L.; Yario, T.A.; Steitz, T.A.; Steitz, J.A. Structural Insights into the Stabilization of MALAT1 Noncoding RNA by a Bipartite Triple Helix. Nat. Struct. Mol. Biol. 2014, 21, 633–640. [Google Scholar] [CrossRef]
- Donlic, A.; Morgan, B.S.; Xu, J.L.; Liu, A.; Roble, C.; Hargrove, A.E. Discovery of Small Molecule Ligands for MALAT1 by Tuning an RNA-Binding Scaffold. Angew. Chem. Int. Ed. Engl. 2018, 57, 13242–13247. [Google Scholar] [CrossRef] [PubMed]
- Abulwerdi, F.A.; Xu, W.; Ageeli, A.A.; Yonkunas, M.J.; Arun, G.; Nam, H.; Schneekloth, J.S.; Dayie, T.K.; Spector, D.; Baird, N.; et al. Selective Small-Molecule Targeting of a Triple Helix Encoded by the Long Noncoding RNA, MALAT1. ACS Chem. Biol. 2019, 14, 223–235. [Google Scholar] [CrossRef]
- Soares, R.J.; Maglieri, G.; Gutschner, T.; Diederichs, S.; Lund, A.H.; Nielsen, B.S.; Holmstrøm, K. Evaluation of Fluorescence in Situ Hybridization Techniques to Study Long Non-Coding RNA Expression in Cultured Cells. Nucleic Acids Res. 2018, 46, e4. [Google Scholar] [CrossRef]
- Tani, H.; Nakamura, Y.; Ijiri, K.; Akimitsu, N. Stability of MALAT-1, a Nuclear Long Non-Coding RNA in Mammalian Cells, Varies in Various Cancer Cells. Drug Discov. 2010, 4, 235–239. [Google Scholar]
- Coassin, S.R.; Orjalo, A.V.; Semaan, S.J.; Johansson, H.E. Simultaneous Detection of Nuclear and Cytoplasmic RNA Variants Utilizing Stellaris® RNA Fluorescence in Situ Hybridization in Adherent Cells. Methods Mol. Biol. 2014, 1211, 189–199. [Google Scholar] [CrossRef]
- Orjalo, A.; Johansson, H.E. Duplex Imaging of Pre-LncRNAs and Mature LncRNAs by Stellaris® RNA Fluorescence in Situ Hybridization (RNA FISH). Available online: https://biosearchassets.blob.core.windows.net/assets/poster_duplex_imaging_prelncRNA_matureIncRNAs.pdf (accessed on 6 October 2022).
- Querido, E.; Dekakra-Bellili, L.; Chartrand, P. RNA Fluorescence in Situ Hybridization for High-Content Screening. Methods 2017, 126, 149–155. [Google Scholar] [CrossRef]
- Bensaude, O. Inhibiting Eukaryotic Transcription: Which Compound to Choose? How to Evaluate Its Activity? Transcription 2011, 2, 103–108. [Google Scholar] [CrossRef]
- Ahn, S.Y.; Yang, J.H.; Kim, N.H.; Lee, K.; Cha, Y.H.; Yun, J.S.; Kang, H.E.; Lee, Y.; Choi, J.; Kim, H.S.; et al. Anti-Helminthic Niclosamide Inhibits Ras-Driven Oncogenic Transformation via Activation of GSK-3. Oncotarget 2017, 8, 31856–31863. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.Y.; Kim, N.H.; Lee, K.; Cha, Y.H.; Yang, J.H.; Cha, S.Y.; Cho, E.S.; Lee, Y.; Cha, J.S.; Cho, H.S.; et al. Niclosamide Is a Potential Therapeutic for Familial Adenomatosis Polyposis by Disrupting Axin-GSK3 Interaction. Oncotarget 2017, 8, 31842–31855. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, M.A.; Raheja, R.; Murugaiyan, G.; Rajabi, H.; Kumar, D.; Pertel, T.; Regev, K.; Griffin, R.; Aly, L.; Kivisakk, P.; et al. Identification of a Novel Mechanism of Action of Fingolimod (FTY720) on Human Effector T Cell Function through TCF-1 Upregulation. J. Neuroinflamm. 2015, 12, 245. [Google Scholar] [CrossRef]
- Scarpa, M.; Singh, P.; Bailey, C.M.; Lee, J.K.; Kapoor, S.; Lapidus, R.G.; Niyongere, S.; Sangodkar, J.; Wang, Y.; Perrotti, D.; et al. PP2A-Activating Drugs Enhance FLT3 Inhibitor Efficacy through AKT Inhibition-Dependent GSK-3β-Mediated c-Myc and Pim-1 Proteasomal Degradation. Mol. Cancer 2021, 20, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhu, H.; Xiao, Y.; Guo, H.; Lin, M.; Yuan, Z.; Yang, X.; Huang, Y.; Zhang, Q.; Bai, Y. The Anthelmintic Drug Niclosamide Induces GSK-β-Mediated β-Catenin Degradation to Potentiate Gemcitabine Activity, Reduce Immune Evasion Ability and Suppress Pancreatic Cancer Progression. Cell Death Dis. 2022, 13, 112. [Google Scholar] [CrossRef]
- Stamos, J.L.; Chu, M.L.-H.; Enos, M.D.; Shah, N.; Weis, W.I. Structural Basis of GSK-3 Inhibition by N-Terminal Phosphorylation and by the Wnt Receptor LRP6. eLife 2014, 3, e01998. [Google Scholar] [CrossRef]
- Yao, J.; Wang, X.-Q.; Li, Y.-J.; Shan, K.; Yang, H.; Wang, Y.-N.-Z.; Yao, M.-D.; Liu, C.; Li, X.-M.; Shen, Y.; et al. Long Non-Coding RNA MALAT1 Regulates Retinal Neurodegeneration through CREB Signaling. EMBO Mol. Med. 2016, 8, 346–362. [Google Scholar] [CrossRef]
- Fan, X.; Xiong, H.; Wei, J.; Gao, X.; Feng, Y.; Liu, X.; Zhang, G.; He, Q.-Y.; Xu, J.; Liu, L. Cytoplasmic HnRNPK Interacts with GSK3β and Is Essential for the Osteoclast Differentiation. Sci. Rep. 2015, 5, 17732. [Google Scholar] [CrossRef]
- Tolnay, M.; Juang, Y.-T.; Tsokos, G.C. Protein Kinase A Enhances, Whereas Glycogen Synthase Kinase-3 Beta Inhibits, the Activity of the Exon 2-Encoded Transactivator Domain of Heterogeneous Nuclear Ribonucleoprotein D in a Hierarchical Fashion. Biochem. J. 2002, 363, 127–136. [Google Scholar] [CrossRef]
- Wilson, G.M.; Lu, J.; Sutphen, K.; Suarez, Y.; Sinha, S.; Brewer, B.; Villanueva-Feliciano, E.C.; Ysla, R.M.; Charles, S.; Brewer, G. Phosphorylation of P40AUF1 Regulates Binding to A + U-Rich MRNA-Destabilizing Elements and Protein-Induced Changes in Ribonucleoprotein Structure. J. Biol. Chem. 2003, 278, 33039–33048. [Google Scholar] [CrossRef]
- Yoon, J.-H.; De, S.; Srikantan, S.; Abdelmohsen, K.; Grammatikakis, I.; Kim, J.; Kim, K.M.; Noh, J.H.; White, E.J.F.; Martindale, J.L.; et al. PAR-CLIP Analysis Uncovers AUF1 Impact on Target RNA Fate and Genome Integrity. Nat. Commun. 2014, 5, 5248. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.; Levin, M.; Butter, F.; Scheibe, M. Quantitative Proteomics to Identify Nuclear RNA-Binding Proteins of Malat1. Int. J. Mol. Sci. 2020, 21, 1166. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Liu, Y.; Zhuang, H.; Yang, B.; Hei, K.; Xiao, M.; Hou, C.; Gao, H.; Zhang, X.; Jia, C.; et al. Quantitative Proteomics Reveals That Long Non-Coding RNA MALAT1 Interacts with DBC1 to Regulate P53 Acetylation. Nucleic Acids Res. 2017, 45, 9947–9959. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-Methyladenosine-Dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-Methyladenosine Alters RNA Structure to Regulate Binding of a Low-Complexity Protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef]
- Yang, F.; Yi, F.; Han, X.; Du, Q.; Liang, Z. MALAT-1 Interacts with HnRNP C in Cell Cycle Regulation. FEBS Lett. 2013, 587, 3175–3181. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Kabotyanski, E.B.; Reineke, L.C.; Shao, J.; Xiong, F.; Lee, J.-H.; Dubrulle, J.; Johnson, H.; Stossi, F.; Tsoi, P.S.; et al. The SINEB1 Element in the Long Non-Coding RNA Malat1 Is Necessary for TDP-43 Proteostasis. Nucleic Acids Res. 2020, 48, 2621–2642. [Google Scholar] [CrossRef]
- Brown, J.A.; Kinzig, C.G.; DeGregorio, S.J.; Steitz, J.A. Methyltransferase-like Protein 16 Binds the 3′-Terminal Triple Helix of MALAT1 Long Noncoding RNA. Proc. Natl. Acad. Sci. USA 2016, 113, 14013–14018. [Google Scholar] [CrossRef]
- Wilusz, J.E.; JnBaptiste, C.K.; Lu, L.Y.; Kuhn, C.-D.; Joshua-Tor, L.; Sharp, P.A. A Triple Helix Stabilizes the 3′ Ends of Long Noncoding RNAs That Lack Poly(A) Tails. Genes Dev. 2012, 26, 2392–2407. [Google Scholar] [CrossRef]
- Ageeli, A.A.; McGovern-Gooch, K.R.; Kaminska, M.M.; Baird, N.J. Finely Tuned Conformational Dynamics Regulate the Protective Function of the LncRNA MALAT1 Triple Helix. Nucleic Acids Res. 2019, 47, 1468–1481. [Google Scholar] [CrossRef]
- Liu, X.; Klein, P.S. Glycogen Synthase Kinase-3 and Alternative Splicing. WIREs RNA 2018, 9, e1501. [Google Scholar] [CrossRef] [PubMed]
- Piñol-Roma, S.; Dreyfuss, G. HnRNP Proteins:Localization and Transport between the Nucleus and the Cytoplasm. Trends Cell Biol. 1993, 3, 151–155. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The HnRNP Family: Insights into Their Role in Health and Disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Faulds, K.J.; Egelston, J.N.; Sedivy, L.J.; Mitchell, M.K.; Garimella, S.; Kozlowski, H.; D’Alessandro, A.; Hansen, K.C.; Balsbaugh, J.L.; Phiel, C.J. Glycogen Synthase Kinase-3 (GSK-3) Activity Regulates MRNA Methylation in Mouse Embryonic Stem Cells. J. Biol. Chem. 2018, 293, 10731–10743. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The Dynamic Epitranscriptome: N6-Methyladenosine and Gene Expression Control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhu, C.; Jin, Y. The Oncogenic and Tumor Suppressive Functions of the Long Noncoding RNA MALAT1: An Emerging Controversy. Front. Genet. 2020, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Mook, R.A.; Premont, R.T.; Wang, J. Niclosamide: Beyond an Antihelminthic Drug. Cell. Signal. 2018, 41, 89–96. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Ball, C.A.; Berman, J.J.; Bova, G.S.; Brazma, A.; Bumgarner, R.E.; Campbell, D.; Causton, H.C.; Christiansen, J.H.; Daian, F.; et al. Minimum Information Specification for in Situ Hybridization and Immunohistochemistry Experiments (MISFISHIE). Nat. Biotechnol. 2008, 26, 305–312. [Google Scholar] [CrossRef]
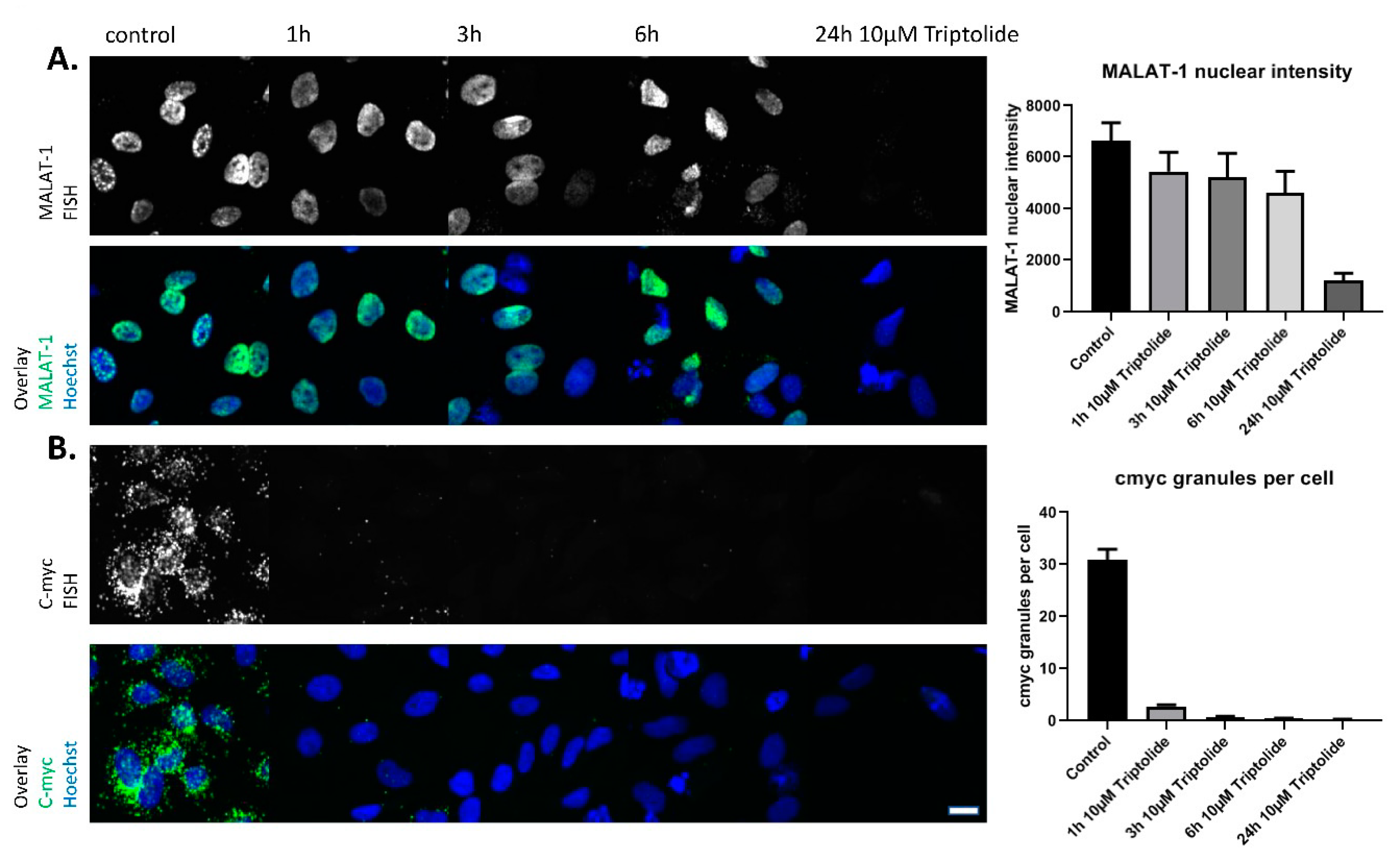
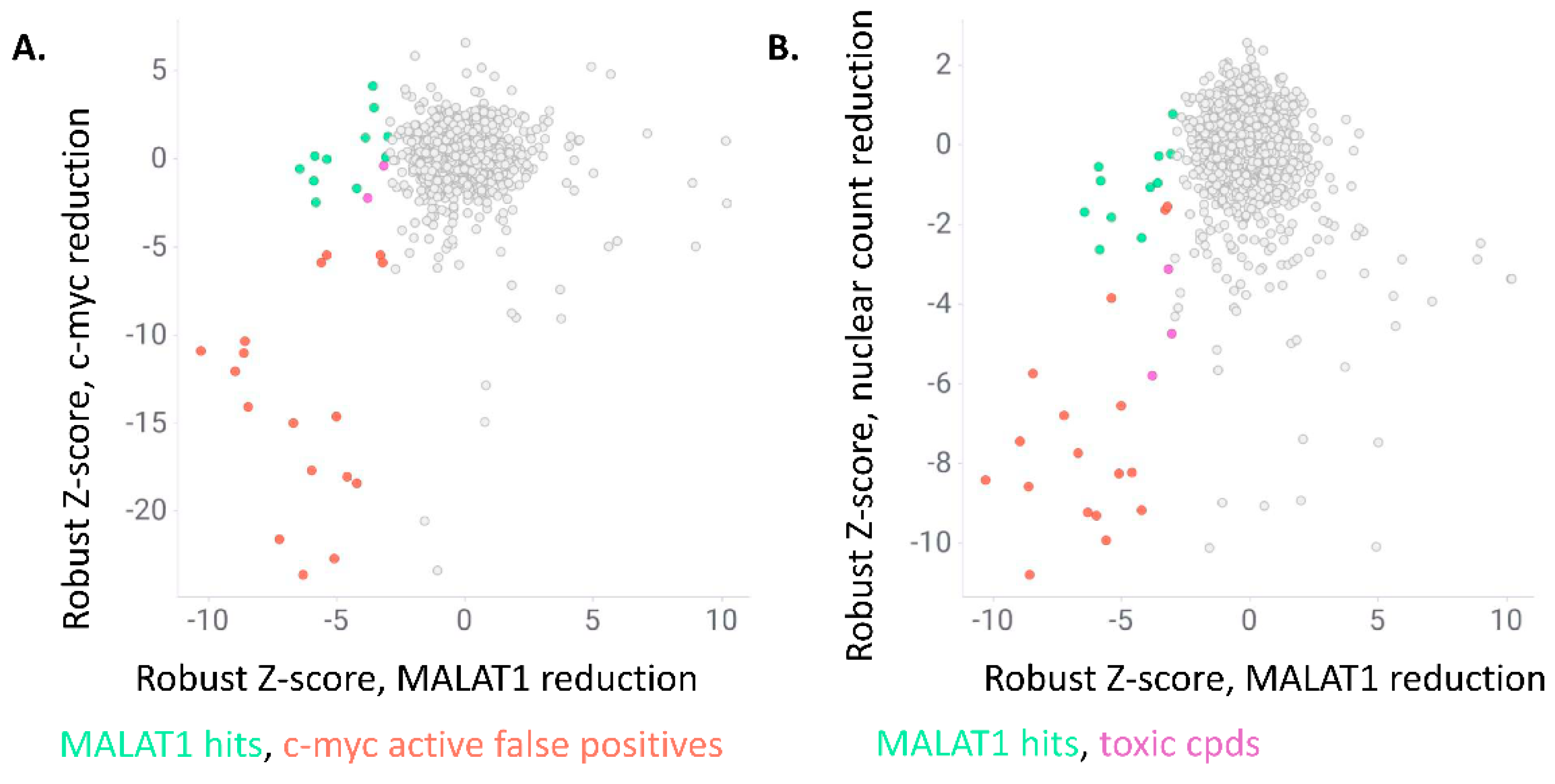
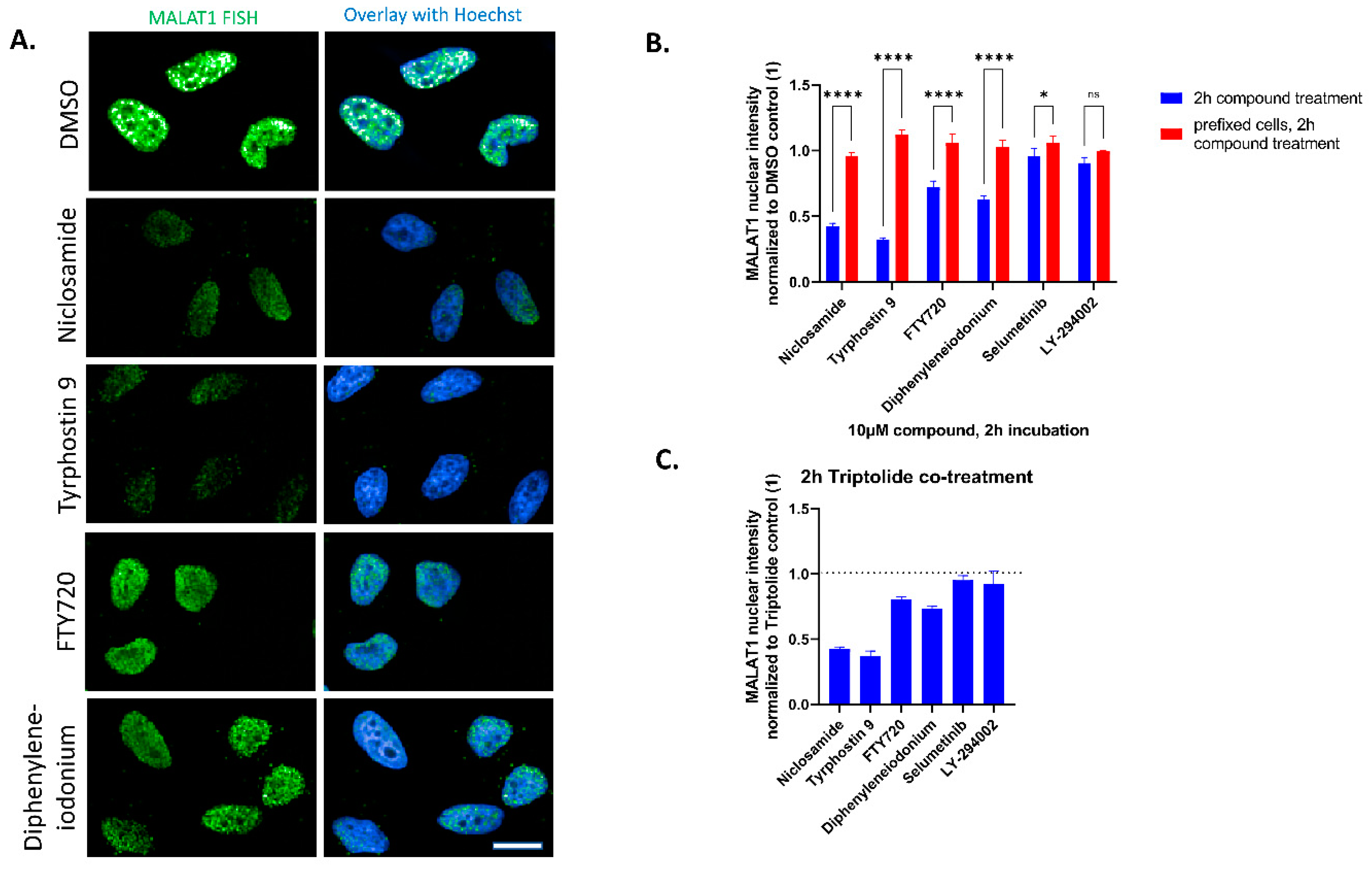
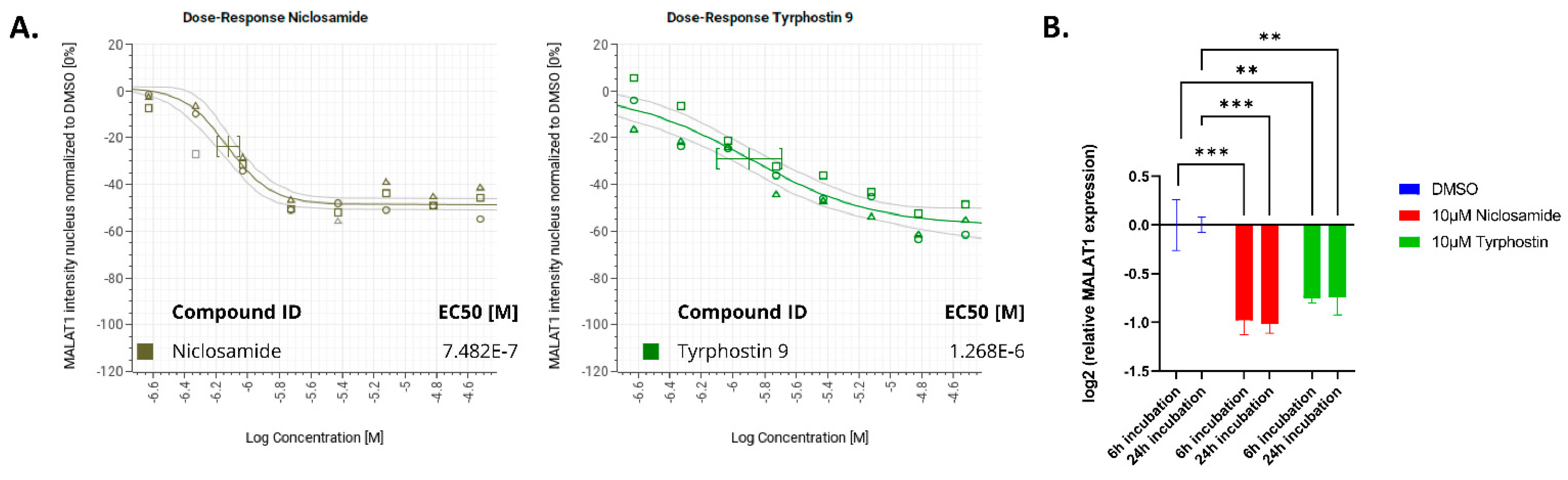
| Compound ID | Robust Z-Score: MALAT1 Nuclear Intensity | Robust Z-Score: c-myc Granules per Cell | Robust Z-Score: Nuclear Count | Retest @ 10 µM and 2 h | EC50 Determination |
|---|---|---|---|---|---|
| Niclosamide | −5.906422 | 0.1783263 | −2.623441 | Strongly active; >50% reduction | 851 nM +/− 193 nM |
| Tyrphostin 9 | −3.908122 | 1.203798 | −1.069688 | Strongly active; >50% reduction | 2.16 µM +/− 1.1 µM |
| FTY720 | −5.875497 | −2.437235 | −0.8817972 | active | |
| Diphenyleneiodonium | −4.243739 | −1.65379 | −2.316559 | active | |
| Selumetinib (AZD6244) | −6.467096 | −0.541567 | −1.678847 | inactive | |
| LY-294002 | −3.060566 | 1.285709 | 0.7897846 | inactive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zablowsky, N.; Farack, L.; Rofall, S.; Kramer, J.; Meyer, H.; Nguyen, D.; Ulrich, A.K.C.; Bader, B.; Steigemann, P. High Throughput FISH Screening Identifies Small Molecules That Modulate Oncogenic lncRNA MALAT1 via GSK3B and hnRNPs. Non-Coding RNA 2023, 9, 2. https://doi.org/10.3390/ncrna9010002
Zablowsky N, Farack L, Rofall S, Kramer J, Meyer H, Nguyen D, Ulrich AKC, Bader B, Steigemann P. High Throughput FISH Screening Identifies Small Molecules That Modulate Oncogenic lncRNA MALAT1 via GSK3B and hnRNPs. Non-Coding RNA. 2023; 9(1):2. https://doi.org/10.3390/ncrna9010002
Chicago/Turabian StyleZablowsky, Nina, Lydia Farack, Sven Rofall, Jan Kramer, Hanna Meyer, Duy Nguyen, Alexander K. C. Ulrich, Benjamin Bader, and Patrick Steigemann. 2023. "High Throughput FISH Screening Identifies Small Molecules That Modulate Oncogenic lncRNA MALAT1 via GSK3B and hnRNPs" Non-Coding RNA 9, no. 1: 2. https://doi.org/10.3390/ncrna9010002
APA StyleZablowsky, N., Farack, L., Rofall, S., Kramer, J., Meyer, H., Nguyen, D., Ulrich, A. K. C., Bader, B., & Steigemann, P. (2023). High Throughput FISH Screening Identifies Small Molecules That Modulate Oncogenic lncRNA MALAT1 via GSK3B and hnRNPs. Non-Coding RNA, 9(1), 2. https://doi.org/10.3390/ncrna9010002






