Polymorphism rs2383207 of CDKN2B-AS and Susceptibility to Atherosclerosis: A Mini Review
Abstract
1. Introduction
2. Results
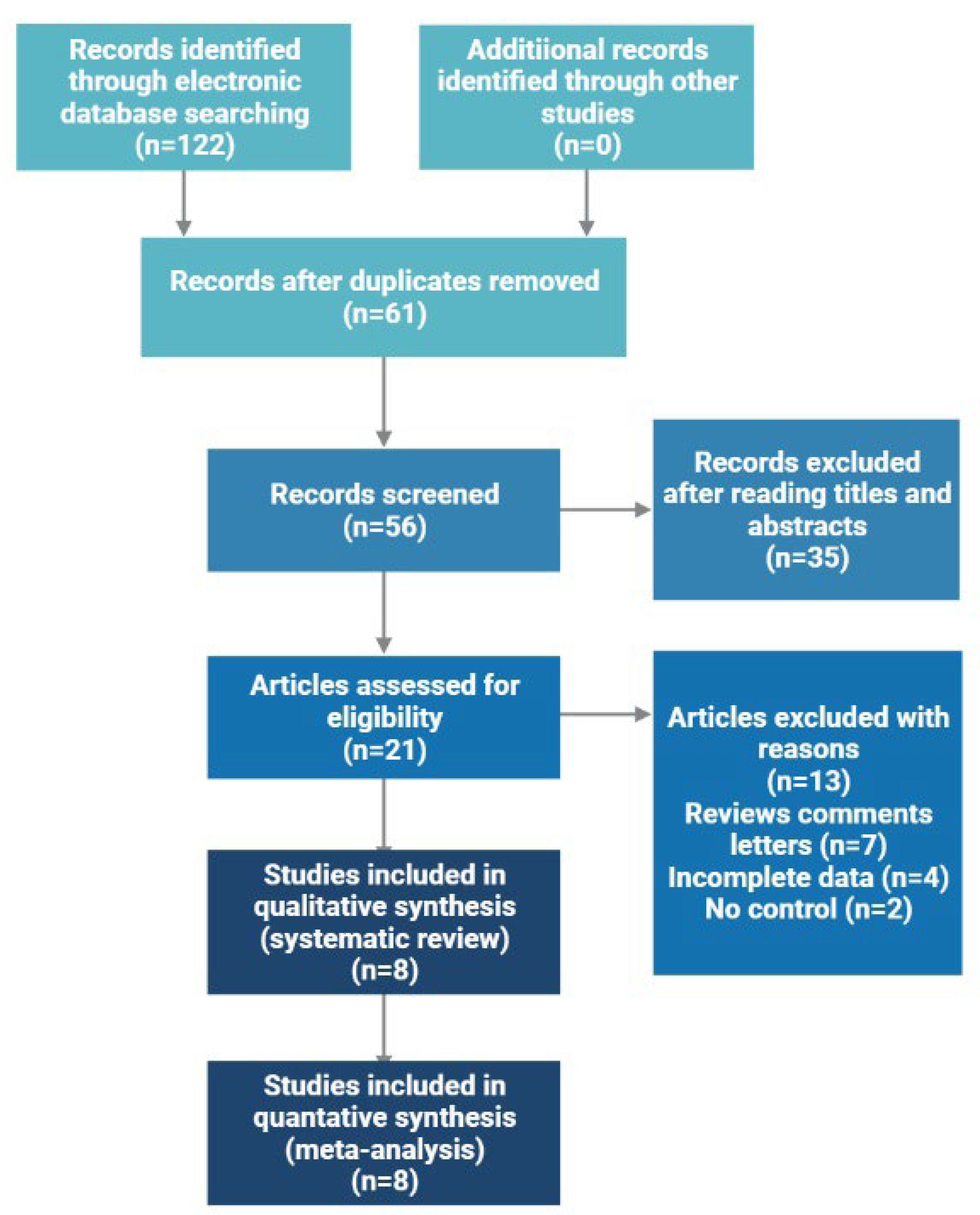
2.1. Characteristics of Included Studies
2.2. Overall and Subgroup Analyses
2.3. Sensitivity Analyses
2.4. Publication Biases
3. Discussion
4. Materials and Methods
4.1. Literature Search and Inclusion Criteria
4.2. Data Extraction and Quality Assessment
4.3. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ballantyne, M.D.; McDonald, R.A.; Baker, A.H. lncRNA/MicroRNA interactions in the vasculature. Clin. Pharmacol. Ther. 2016, 99, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Fasolo, F.; Di Gregoli, K.; Maegdefessel, L.; Johnson, J.L. Non-coding RNAs in cardiovascular cell biology and atherosclerosis. Cardiovasc. Res. 2019, 115, 1732–1756. [Google Scholar] [CrossRef] [PubMed]
- Congrains, A.; Kamide, K.; Oguro, R.; Yasuda, O.; Miyata, K.; Yamamoto, E.; Kawai, T.; Kusunoki, H.; Yamamoto, H.; Takeya, Y.; et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012, 220, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Cunnington, M.S.; Keavney, B. Genetic mechanisms mediating atherosclerosis susceptibility at the chromosome 9p21 locus. Curr. Atheroscler. Rep. 2011, 13, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, J.; Zhang, F.; Li, J.; An, S.; Cheng, M.; Li, J. LncRNA CDKN2B-AS1 hinders the proliferation and facilitates apoptosis of ox-LDL-induced vascular smooth muscle cells via the ceRNA network of CDKN2B-AS1/miR-126-5p/PTPN7. Int. J. Cardiol. 2021, 340, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, K.G.; Li, L.; Shen, G.-Q.; Hu, Y.; Yang, Y.; MacKinlay, K.G.; Topol, E.J.; Wang, Q.K. Four SNPS on Chromosome 9p21 Confer Risk to Premature, Familial CAD and MI in an American Caucasian Population (GeneQuest). Ann. Hum. Genet. 2008, 72, 654–657. [Google Scholar] [CrossRef]
- Cakmak, H.A.; Bayoglu, B.; Durmaz, E.; Can, G.; Karadag, B.; Cengiz, M.; Vural, V.A.; Yuksel, H. Evaluation of association between common genetic variants on chromosome 9p21 and coronary artery disease in Turkish population. Anatol. J. Cardiol. 2015, 15, 196–203. [Google Scholar] [CrossRef]
- García-González, I.; Pérez-Mendoza, G.; Solís-Cárdenas, A.; Flores-Ocampo, J.; Herrera-Sánchez, L.F.; Mendoza-Alcocer, R.; González-Herrera, L. Genetic variants of PON1, GSTM1, GSTT1, and locus 9p21.3, and the risk for premature coronary artery disease in Yucatan, Mexico. Am. J. Hum. Biol. 2022, 34, e23701. [Google Scholar]
- Zhou, L.; Zhang, X.; He, M.; Cheng, L.; Chen, Y.; Hu, F.B.; Wu, T. Associations Between Single Nucleotide Polymorphisms on Chromosome 9p21 and Risk of Coronary Heart Disease in Chinese Han Population. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 2085–2089. [Google Scholar] [CrossRef]
- Chen, Z.; Qian, Q.; Ma, G.; Wang, J.; Zhang, X.; Feng, Y.; Shen, C.; Yao, Y. A common variant on chromosome 9p21 affects the risk of early-onset coronary artery disease. Mol. Biol. Rep. 2008, 36, 889–893. [Google Scholar] [CrossRef]
- Kumar, J.; Yumnam, S.; Basu, T.; Ghosh, A.; Garg, G.; Karthikeyan, G.; Sengupta, S. Association of polymorphisms in 9p21 region with CAD in North Indian population: Replication of SNPs identified through GWAS. Clin. Genet. 2011, 79, 588–593. [Google Scholar] [CrossRef] [PubMed]
- El-Menyar, A.A.; Rizk, N.M.; Al-Qahtani, A.; AlKindi, F.; Elyas, A.; Farag, F.; Bakhsh, F.D.; Ebrahim, S.; Ahmed, E.; Al-Khinji, M.; et al. The cardiovascular implication of single nucleotide polymorphisms of chromosome 9p21 locus among Arab population. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2015, 20, 346–352. [Google Scholar]
- Yang, J.; Gu, L.; Guo, X.; Huang, J.; Chen, Z.; Huang, G.; Kang, Y.; Zhang, X.; Long, J.; Su, L. LncRNA ANRIL Expression and ANRIL Gene Polymorphisms Contribute to the Risk of Ischemic Stroke in the Chinese Han Population. Cell Mol Neurobiol. 2018, 38, 1253–1269. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Liu, J.; Hu, F.; Bi, Y.; Li, M.; Zhao, L. Genetic variants on chromosome 9p21 confer risks of cerebral infarction in the Chinese population: A meta-analysis. Int. J. Immunopathol. Pharmacol. 2019, 33, 2058738419847852. [Google Scholar] [CrossRef] [PubMed]
- Helgadottir, A.; Thorleifsson, G.; Manolescu, A.; Gretarsdottir, S.; Blondal, T.; Jonasdottir, A.; Jonasdottir, A.; Sigurdsson, A.; Baker, A.; Palsson, A.; et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007, 316, 1491–1493. [Google Scholar] [CrossRef]
- Akinyemi, R.; Arnett, D.K.; Tiwari, H.K.; Ovbiagele, B.; Sarfo, F.; Srinivasasainagendra, V.; Irvin, M.R.; Adeoye, A.; Perry, R.T.; Akpalu, A.; et al. Interleukin-6 (IL-6) rs1800796 and cyclin dependent kinase inhibitor (CDKN2A/CDKN2B) rs2383207 are associated with ischemic stroke in indigenous West African Men. J. Neurol. Sci. 2017, 379, 229–235. [Google Scholar] [CrossRef]
- Dichgans, M.; Malik, R.; König, I.R.; Rosand, J.; Clarke, R.; Gretarsdottir, S.; Thorleifsson, G.; Mitchell, B.D.; Assimes, T.L.; Levi, C.; et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: A genome-wide analysis of common variants. Stroke 2014, 45, 24–36. [Google Scholar] [CrossRef]
- Dehghan, A.; Bis, J.C.; White, C.C.; Smith, A.V.; Morrison, A.C.; Cupples, L.A.; Trompet, S.; Chasman, D.I.; Lumley, T.; Völker, U.; et al. Genome-Wide Association Study for Incident Myocardial Infarction and Coronary Heart Disease in Prospective Cohort Studies: The CHARGE Consortium. PLoS ONE 2016, 11, e0144997. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred Reporting Items for Systematic Reviews and MetaAnalyses: The PRISMA statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef]
- Lo, C.K.; Mertz, D.; Loeb, M. Newcastle-Ottawa Scale: Comparing reviewers’ to authors’ assessments. BMC Med. Res. Methodol. 2014, 14, 45. [Google Scholar] [CrossRef]
- Zhang, T.; Xu, C.; Zhao, R.; Cao, Z. Diagnostic Value of sST2 in Cardiovascular Diseases: A Systematic Review and Meta-Analysis. Front. Cardiovasc. Med. 2021, 8, 697837. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Chu, H. Quantifying publication bias in meta-analysis. Biometrics 2018, 74, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Shi, G. Meta-Analysis of SNP-Environment Interaction with Heterogeneity. Hum. Hered. 2019, 84, 117–126. [Google Scholar] [CrossRef] [PubMed]
| First Author, Year | Country | Ethnicity | Sample Size | Genotype Distribution (MM/Mm/mm) | p Value for HWE | NOS Score | |
|---|---|---|---|---|---|---|---|
| Cases Control | |||||||
| Abdulah, 2008 | United States | Caucasian | 310/560 | 139/121/50 | 147/277/136 | 0.88 | 8 |
| Zhou, 2008 | China | Asia | 1360/1360 | 702/520/138 | 592/605/163 | 0.75 | 8 |
| Chen, 2009 | China | Asia | 212/232 | 107/69/36 | 71/114/47 | 0.96 | 8 |
| Kumar, 2011 | India | Asia | 301/424 | 137/124/40 | 174/190/60 | 0.63 | 7 |
| Cakmak, 2015 | Turkey | Caucasian | 220/240 | 83/101/36 | 102/118/20 | 0.21 | 7 |
| El-Menyar, 2015 | Egypt | Asia | 236/152 | 146/77/12 | 84/58/10 | 1 | 7 |
| Yang, 2018 | China | Asia | 540/548 | 247/236/57 | 244/251/53 | 0.49 | 8 |
| García-González, 2021 | Yucatan | Caucasian | 98/101 | 23/53/22 | 26/51/24 | 0.94 | 8 |
| Genetic Model | Population | Number of Studies | Test of Association | Test of Heterogeneity | |||
|---|---|---|---|---|---|---|---|
| OR (95%CI) | p | Ph | I2, % | Model | |||
| C vs. G | Total | 8 | 0.91 [0.79, 1.05] | 0.20 | 0.002 | 70 | R |
| Asian | 5 | 0.82 [0.71, 0.95] | 0.007 | 0.05 | 57 | R | |
| Caucasian | 3 | 1.12 [0.96, 1.30] | 0.16 | 0.33 | 9 | F | |
| CG vs. CC | Total | 8 | 0.86 [0.78, 0.96] | 0.007 | 0.0009 | 72 | R |
| Asian | 5 | 0.85 [0.75, 0.95] | 0.005 | 0.001 | 65 | R | |
| Caucasian | 3 | 0.91 [0.70, 1.19] | 0.48 | 0.02 | 85 | R | |
| GG vs. CC | Total | 8 | 0.80 [0.57, 1.12] | 0.19 | 0.0002 | 75 | R |
| Asian | 5 | 0.76 [0.61, 0.91] | 0.002 | 0.26 | 24 | F | |
| Caucasian | 3 | 0.94 [0.30, 2.97] | 0.92 | <0.0001 | 91 | R | |
| GG + CGvs.CC | Total | 8 | 0.78 [0.71, 0.86] | 0.13 | <0.00001 | 37 | F |
| Asian | 5 | 0.79 [0.70, 0.88] | 0.12 | <0.0001 | 46 | F | |
| Caucasian | 3 | 0.78 [0.63, 0.96] | 0.15 | 0.02 | 47 | F | |
| GGvs.CC + CG | Total | 8 | 0.92 [0.73, 1.16] | 0.47 | 0.03 | 54 | R |
| Asian | 5 | 0.88 [0.75, 1.05] | 0.15 | 0.77 | 0 | F | |
| Caucasian | 3 | 1.04 [0.47, 2.29] | 0.92 | 0.001 | 85 | R | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timofeeva, S.V.; Sherchkova, T.A.; Shkurat, T.P. Polymorphism rs2383207 of CDKN2B-AS and Susceptibility to Atherosclerosis: A Mini Review. Non-Coding RNA 2022, 8, 78. https://doi.org/10.3390/ncrna8060078
Timofeeva SV, Sherchkova TA, Shkurat TP. Polymorphism rs2383207 of CDKN2B-AS and Susceptibility to Atherosclerosis: A Mini Review. Non-Coding RNA. 2022; 8(6):78. https://doi.org/10.3390/ncrna8060078
Chicago/Turabian StyleTimofeeva, Sofia Vladimorovna, Tatiana Alexandrovna Sherchkova, and Tatiana Pavlovna Shkurat. 2022. "Polymorphism rs2383207 of CDKN2B-AS and Susceptibility to Atherosclerosis: A Mini Review" Non-Coding RNA 8, no. 6: 78. https://doi.org/10.3390/ncrna8060078
APA StyleTimofeeva, S. V., Sherchkova, T. A., & Shkurat, T. P. (2022). Polymorphism rs2383207 of CDKN2B-AS and Susceptibility to Atherosclerosis: A Mini Review. Non-Coding RNA, 8(6), 78. https://doi.org/10.3390/ncrna8060078








