The Functional Roles and Regulation of Circular RNAs during Cellular Stresses
Abstract
:1. Introduction
2. Roles of CircRNAs in Cellular Stress
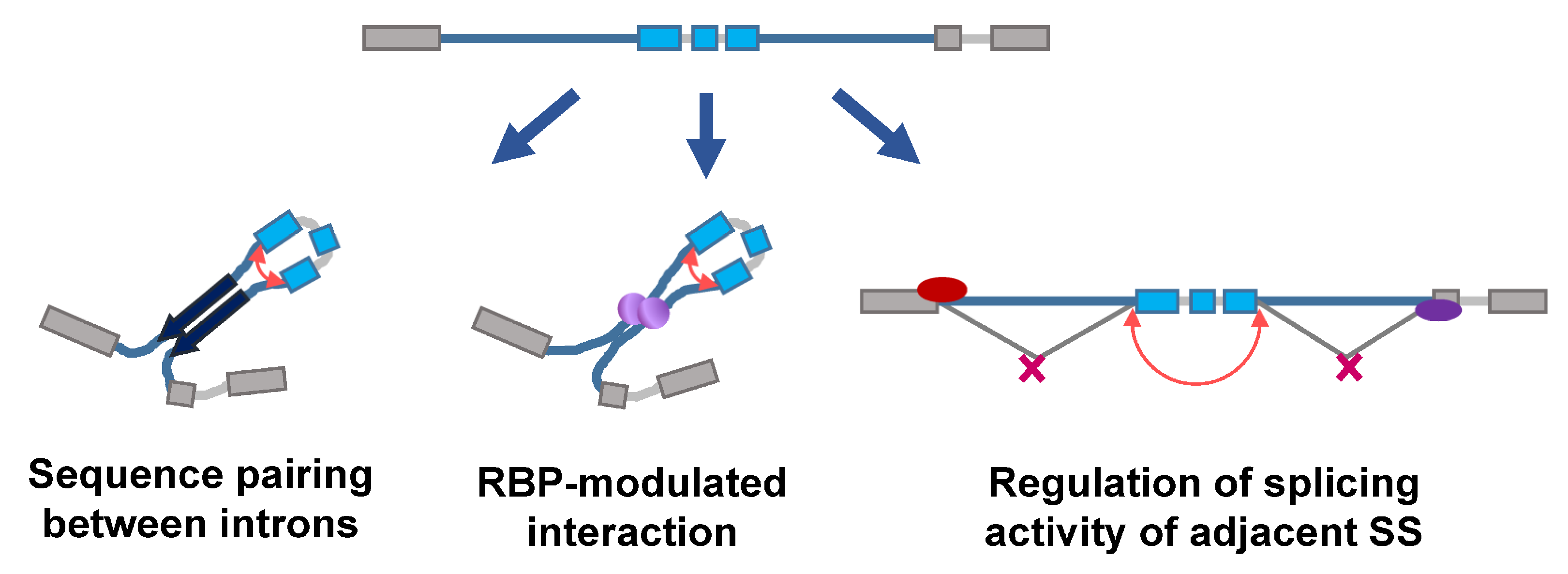
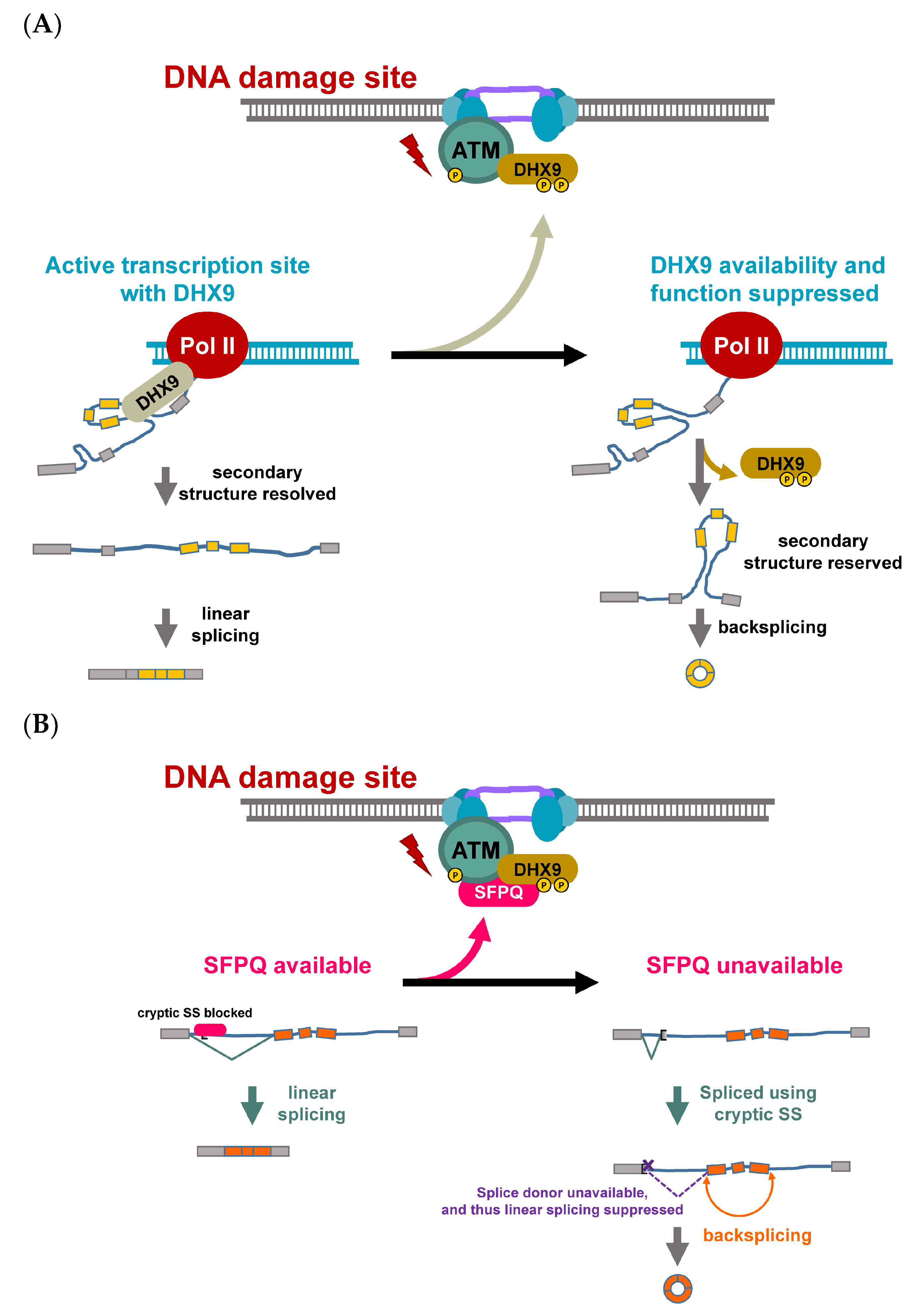
2.1. Roles and Regulation of CircRNAs in Response to Genotoxic Stress
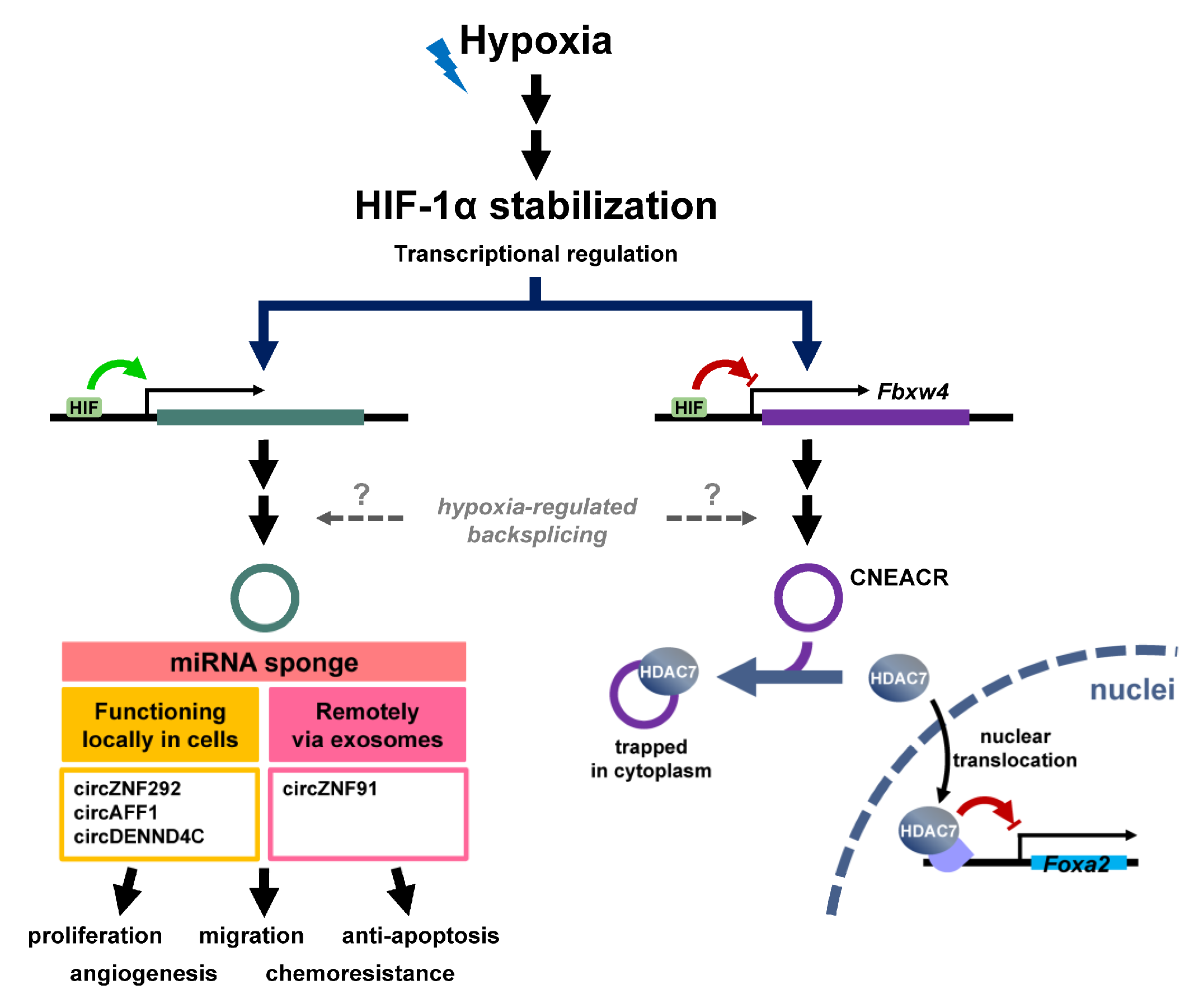
2.2. Modulation of CircRNA under Hypoxic Stress
2.2.1. Hypoxia-Regulated CircRNAs
2.2.2. Players Regulate Function and Abundance of CircRNAs in Hypoxia
3. Heat Shock Stress
4. Closing Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hsu, M.T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Q.; Wang, Y.; Cao, M.; Pantaleo, V.; Burgyan, J.; Li, W.X.; Ding, S.W. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl. Acad. Sci. USA 2012, 109, 3938–3943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Hsiao, K.Y.; Lin, Y.C.; Gupta, S.K.; Chang, N.; Yen, L.; Sun, H.S.; Tsai, S.J. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res. 2017, 77, 2339–2350. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.C.; Lee, Y.C.; Chang, K.L.; Hsiao, K.Y. Analysis of common targets for circular RNAs. BMC Bioinform. 2019, 20, 372. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Chen, N.; Zhao, G.; Yan, X.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z.; et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome. Biol. 2018, 19, 218. [Google Scholar] [CrossRef] [Green Version]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Liu, B.; Shen, D.D.; Yan, P.; Zhang, Y.; Tian, Y.; Hou, L.; Jiang, G.; Zhu, Y.; Liang, Y.; et al. A tumor-suppressive circular RNA mediates uncanonical integrin degradation by the proteasome in liver cancer. Sci. Adv. 2021, 7, eabe5043. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [Green Version]
- Li, X.F.; Lytton, J. A circularized sodium-calcium exchanger exon 2 transcript. J. Biol. Chem. 1999, 274, 8153–8160. [Google Scholar] [CrossRef] [Green Version]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [Green Version]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [Green Version]
- Halbreich, A.; Pajot, P.; Foucher, M.; Grandchamp, C.; Slonimski, P. A pathway of cytochrome b mRNA processing in yeast mitochondria: Specific splicing steps and an intron-derived circular RNA. Cell 1980, 19, 321–329. [Google Scholar] [CrossRef]
- Pasman, Z.; Been, M.D.; Garcia-Blanco, M.A. Exon circularization in mammalian nuclear extracts. RNA 1996, 2, 603–610. [Google Scholar]
- Braun, S.; Domdey, H.; Wiebauer, K. Inverse splicing of a discontinuous pre-mRNA intron generates a circular exon in a HeLa cell nuclear extract. Nucleic Acids Res. 1996, 24, 4152–4157. [Google Scholar] [CrossRef] [PubMed]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Daubersies, P.; Majerus, M.A.; Kerckaert, J.P.; Bailleul, B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992, 11, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Westholm, J.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.; Lai, E.C. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef] [Green Version]
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Das, A.; Sinha, T.; Shyamal, S.; Panda, A.C. Emerging Role of Circular RNA-Protein Interactions. Noncoding RNA 2021, 7, 48. [Google Scholar] [CrossRef]
- Hsiao, K.Y.; Mizzen, C.A. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J. Mol. Cell Biol. 2013, 5, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Munoz, M.J.; Perez Santangelo, M.S.; Paronetto, M.P.; de la Mata, M.; Pelisch, F.; Boireau, S.; Glover-Cutter, K.; Ben-Dov, C.; Blaustein, M.; Lozano, J.J.; et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 2009, 137, 708–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankotai, T.; Bonhomme, C.; Chen, D.; Soutoglou, E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012, 19, 276–282. [Google Scholar] [CrossRef] [Green Version]
- Shanbhag, N.M.; Rafalska-Metcalf, I.U.; Balane-Bolivar, C.; Janicki, S.M.; Greenberg, R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 2010, 141, 970–981. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, T.; Uchida, C.; Anderson, S.F.; Lee, C.G.; Hurwitz, J.; Parvin, J.D.; Montminy, M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 1997, 90, 1107–1112. [Google Scholar] [CrossRef] [Green Version]
- Aktas, T.; Avsar Ilik, I.; Maticzka, D.; Bhardwaj, V.; Pessoa Rodrigues, C.; Mittler, G.; Manke, T.; Backofen, R.; Akhtar, A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 2017, 544, 115–119. [Google Scholar] [CrossRef]
- Zhang, S.; Hemmerich, P.; Grosse, F. Werner syndrome helicase (WRN), nuclear DNA helicase II (NDH II) and histone gammaH2AX are localized to the centrosome. Cell Biol. Int. 2007, 31, 1109–1121. [Google Scholar] [CrossRef]
- Anderson, S.F.; Schlegel, B.P.; Nakajima, T.; Wolpin, E.S.; Parvin, J.D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998, 19, 254–256. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.C.; Yu, Y.S.; Lin, H.H.; Hsiao, K.Y. Oxaliplatin-induced DHX9 phosphorylation promotes oncogenic circular RNA CCDC66 expression and development of chemoresistance. Cancers 2020, 12, 697. [Google Scholar] [CrossRef] [Green Version]
- Xiong, W.; Ai, Y.Q.; Li, Y.F.; Ye, Q.; Chen, Z.T.; Qin, J.Y.; Liu, Q.Y.; Wang, H.; Ju, Y.H.; Li, W.H.; et al. Microarray Analysis of Circular RNA Expression Profile Associated with 5-Fluorouracil-Based Chemoradiation Resistance in Colorectal Cancer Cells. Biomed Res. Int. 2017, 2017, 8421614. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, Z.; Zhang, Q.; Wang, W.; Li, B.; Wang, L.; Xu, Z.; Zeng, A.; Zhang, X.; Zhang, X.; et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer 2019, 18, 71. [Google Scholar] [CrossRef] [PubMed]
- Salton, M.; Lerenthal, Y.; Wang, S.Y.; Chen, D.J.; Shiloh, Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 2010, 9, 1568–1576. [Google Scholar] [CrossRef] [Green Version]
- Stagsted, L.V.W.; O’Leary, E.T.; Ebbesen, K.K.; Hansen, T.B. The RNA-binding protein SFPQ preserves long-intron splicing and regulates circRNA biogenesis in mammals. Elife 2021, 10, e63088. [Google Scholar] [CrossRef] [PubMed]
- Francia, S.; Cabrini, M.; Matti, V.; Oldani, A.; d’Adda di Fagagna, F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J. Cell Sci. 2016, 129, 1468–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; He, Q.; Hu, Z.; Feng, Y.; Fan, L.; Tang, Z.; Yuan, J.; Shan, W.; Li, C.; Hu, X.; et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat. Struct. Mol. Biol. 2016, 23, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Bergstrand, S.; O’Brien, E.M.; Coucoravas, C.; Hrossova, D.; Peirasmaki, D.; Schmidli, S.; Dhanjal, S.; Pederiva, C.; Siggens, L.; Mortusewicz, O.; et al. Small Cajal body-associated RNA 2 (scaRNA2) regulates DNA repair pathway choice by inhibiting DNA-PK. Nat. Commun. 2022, 13, 1015. [Google Scholar] [CrossRef]
- Marsch, E.; Sluimer, J.C.; Daemen, M.J. Hypoxia in atherosclerosis and inflammation. Curr. Opin. Lipidol. 2013, 24, 393–400. [Google Scholar] [CrossRef]
- Maes, C.; Carmeliet, G.; Schipani, E. Hypoxia-driven pathways in bone development, regeneration and disease. Nat. Rev. Rheumatol. 2012, 8, 358–366. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.H.; Hsiao, K.Y.; Tsai, S.J. Hypoxia: The force of endometriosis. J. Obstet. Gynaecol. Res. 2019, 45, 532–541. [Google Scholar] [CrossRef]
- Lin, S.C.; Hsiao, K.Y.; Chang, N.; Hou, P.C.; Tsai, S.J. Loss of dual-specificity phosphatase-2 promotes angiogenesis and metastasis via up-regulation of interleukin-8 in colon cancer. J. Pathol. 2017, 241, 638–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, K.Y.; Chang, N.; Tsai, J.L.; Lin, S.C.; Tsai, S.J.; Wu, M.H. Hypoxia-inhibited DUSP2 expression promotes IL-6/STAT3 signaling in endometriosis. Am. J. Reprod. Immunol. 2017, 78, e12690. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. The Genomics and Genetics of Oxygen Homeostasis. Annu. Rev. Genom. Hum. Genet. 2020, 21, 183–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boeckel, J.N.; Jae, N.; Heumuller, A.W.; Chen, W.; Boon, R.A.; Stellos, K.; Zeiher, A.M.; John, D.; Uchida, S.; Dimmeler, S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ. Res. 2015, 117, 884–890. [Google Scholar] [CrossRef] [Green Version]
- Di Liddo, A.; de Oliveira Freitas Machado, C.; Fischer, S.; Ebersberger, S.; Heumuller, A.W.; Weigand, J.E.; Muller-McNicoll, M.; Zarnack, K. A combined computational pipeline to detect circular RNAs in human cancer cells under hypoxic stress. J. Mol. Cell Biol. 2019, 11, 829–844. [Google Scholar] [CrossRef]
- Gao, X.Q.; Liu, C.Y.; Zhang, Y.H.; Wang, Y.H.; Zhou, L.Y.; Li, X.M.; Wang, K.; Chen, X.Z.; Wang, T.; Ju, J.; et al. The circRNA CNEACR regulates necroptosis of cardiomyocytes through Foxa2 suppression. Cell Death Differ. 2022, 29, 527–539. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Fanale, D.; Taverna, S.; Russo, A.; Bazan, V. Circular RNA in Exosomes. Adv. Exp. Med. Biol. 2018, 1087, 109–117. [Google Scholar]
- Zeng, Z.; Zhao, Y.; Chen, Q.; Zhu, S.; Niu, Y.; Ye, Z.; Hu, P.; Chen, D.; Xu, P.; Chen, J.; et al. Hypoxic exosomal HIF-1alpha-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene 2021, 40, 5505–5517. [Google Scholar] [CrossRef]
- Zarnack, K.; Konig, J.; Tajnik, M.; Martincorena, I.; Eustermann, S.; Stevant, I.; Reyes, A.; Anders, S.; Luscombe, N.M.; Ule, J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 2013, 152, 453–466. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zhao, W.; Liu, Y.; Tan, X.; Li, X.; Zou, Q.; Xiao, Z.; Xu, H.; Wang, Y.; Yang, X. Function of HNRNPC in breast cancer cells by controlling the dsRNA-induced interferon response. EMBO J. 2018, 37, e99017. [Google Scholar] [CrossRef]
- Hsiao, K.Y.; Wu, M.H.; Chang, N.; Yang, S.H.; Wu, C.W.; Sun, H.S.; Tsai, S.J. Coordination of AUF1 and miR-148a destabilizes DNA methyltransferase 1 mRNA under hypoxia in endometriosis. Mol. Hum. Reprod. 2015, 21, 894–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ripin, N.; Boudet, J.; Duszczyk, M.M.; Hinniger, A.; Faller, M.; Krepl, M.; Gadi, A.; Schneider, R.J.; Sponer, J.; Meisner-Kober, N.C.; et al. Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proc. Natl. Acad. Sci. USA 2019, 116, 2935–2944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e14. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef] [Green Version]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.X.; Chen, X.; Xia, L.P.; Zhang, J.X.; Pan, Z.Z.; Ma, X.D.; Han, K.; Chen, J.W.; Judde, J.G.; Deas, O.; et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef] [Green Version]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507.e8. [Google Scholar] [CrossRef] [PubMed]
- Di Timoteo, G.; Dattilo, D.; Centron-Broco, A.; Colantoni, A.; Guarnacci, M.; Rossi, F.; Incarnato, D.; Oliviero, S.; Fatica, A.; Morlando, M.; et al. Modulation of circRNA Metabolism by m(6)A Modification. Cell Rep. 2020, 31, 107641. [Google Scholar] [CrossRef]
- Li, Y.; Chen, B.; Zhao, J.; Li, Q.; Chen, S.; Guo, T.; Li, Y.; Lai, H.; Chen, Z.; Meng, Z.; et al. HNRNPL Circularizes ARHGAP35 to Produce an Oncogenic Protein. Adv. Sci. 2021, 8, 2001701. [Google Scholar] [CrossRef]
- Ruan, D.Y.; Li, T.; Wang, Y.N.; Meng, Q.; Li, Y.; Yu, K.; Wang, M.; Lin, J.F.; Luo, L.Z.; Wang, D.S.; et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene 2021, 40, 5168–5181. [Google Scholar] [CrossRef]
- Yao, X.; Li, W.; Li, L.; Li, M.; Zhao, Y.; Fang, Z.X.; Luo, Z. YTHDF1 upregulation mediates hypoxia-dependent breast cancer growth and metastasis through regulating PKM2 to affect glycolysis. Cell Death Dis. 2022, 13, 258. [Google Scholar] [CrossRef]
- Imai, Y.; Matsuo, N.; Ogawa, S.; Tohyama, M.; Takagi, T. Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation. Mol. Brain Res. 1998, 53, 33–40. [Google Scholar] [CrossRef]
- Thalhammer, A.; Bencokova, Z.; Poole, R.; Loenarz, C.; Adam, J.; O’Flaherty, L.; Schodel, J.; Mole, D.; Giaslakiotis, K.; Schofield, C.J.; et al. Human AlkB homologue 5 is a nuclear 2-oxoglutarate dependent oxygenase and a direct target of hypoxia-inducible factor 1alpha (HIF-1alpha). PLoS ONE 2011, 6, e16210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, E.A.; Weber, J.D.; Spiegelman, V.S. Involvement of the mRNA binding protein CRD-BP in the regulation of metastatic melanoma cell proliferation and invasion by hypoxia. J. Cell Sci. 2012, 125, 5950–5954. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Shao, Y.L.; Wang, L.L.; Lin, J.; Zhang, J.B.; Ding, Y.; Gao, B.B.; Liu, D.H.; Gao, X.N. YTHDF2 is a potential target of AML1/ETO-HIF1alpha loop-mediated cell proliferation in t(8;21) AML. Oncogene 2021, 40, 3786–3798. [Google Scholar] [CrossRef]
- Chen, L.Y.; Wang, L.; Ren, Y.X.; Pang, Z.; Liu, Y.; Sun, X.D.; Tu, J.; Zhi, Z.; Qin, Y.; Sun, L.N.; et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1alpha translation. Mol. Cancer 2020, 19, 164. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-C.; Wang, W.-Y.; Lin, H.-H.; Huang, Y.-R.; Lin, Y.-C.; Hsiao, K.-Y. The Functional Roles and Regulation of Circular RNAs during Cellular Stresses. Non-Coding RNA 2022, 8, 38. https://doi.org/10.3390/ncrna8030038
Lee Y-C, Wang W-Y, Lin H-H, Huang Y-R, Lin Y-C, Hsiao K-Y. The Functional Roles and Regulation of Circular RNAs during Cellular Stresses. Non-Coding RNA. 2022; 8(3):38. https://doi.org/10.3390/ncrna8030038
Chicago/Turabian StyleLee, Yueh-Chun, Wei-Yu Wang, Hui-Hsuan Lin, Yi-Ren Huang, Ya-Chi Lin, and Kuei-Yang Hsiao. 2022. "The Functional Roles and Regulation of Circular RNAs during Cellular Stresses" Non-Coding RNA 8, no. 3: 38. https://doi.org/10.3390/ncrna8030038
APA StyleLee, Y.-C., Wang, W.-Y., Lin, H.-H., Huang, Y.-R., Lin, Y.-C., & Hsiao, K.-Y. (2022). The Functional Roles and Regulation of Circular RNAs during Cellular Stresses. Non-Coding RNA, 8(3), 38. https://doi.org/10.3390/ncrna8030038






