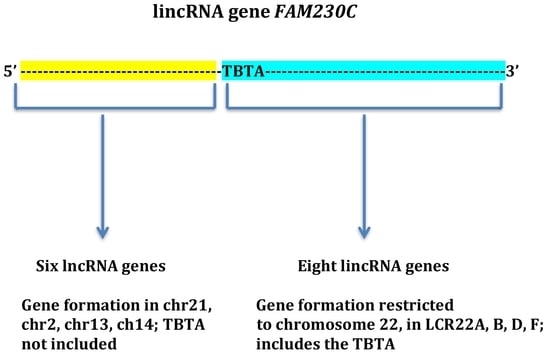
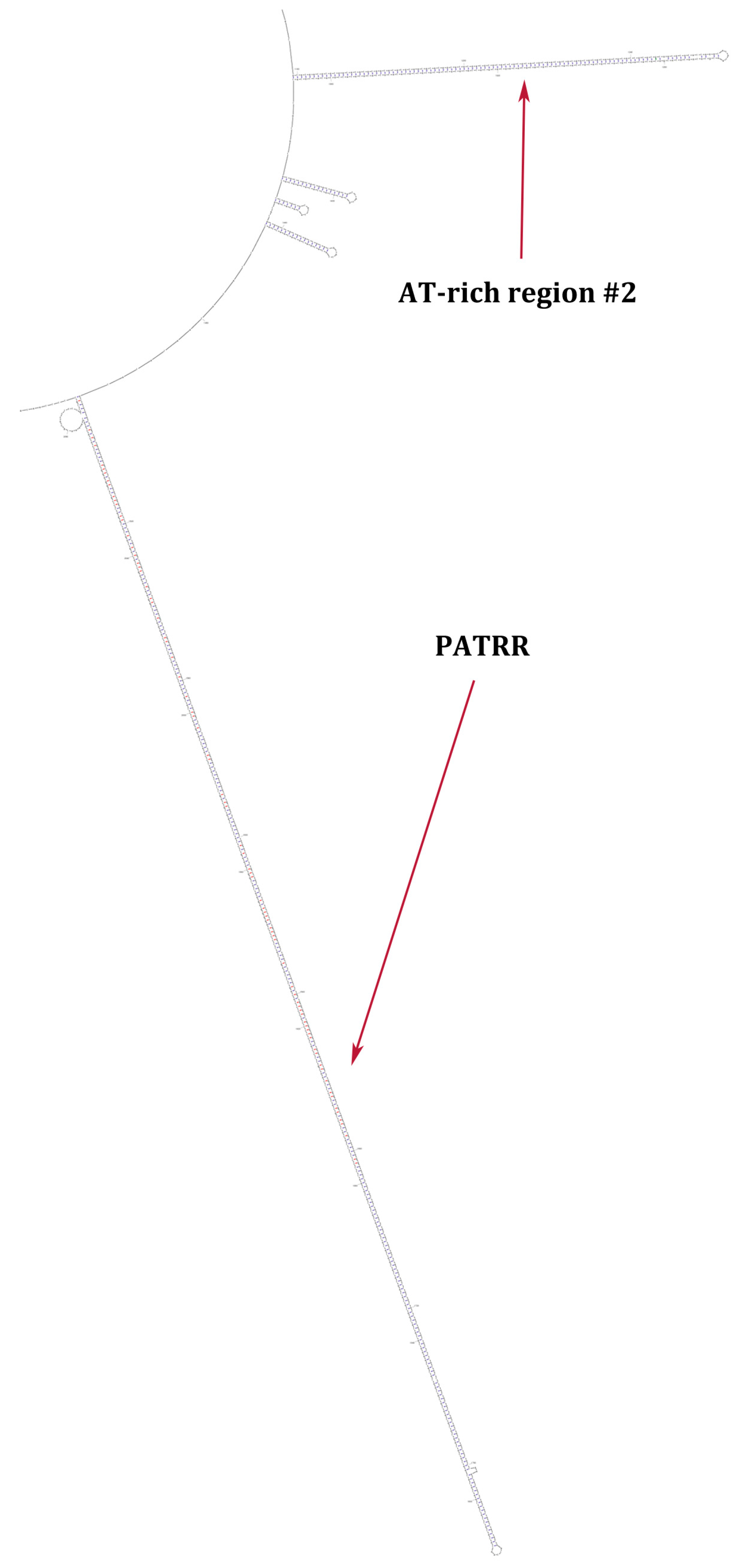
Formation of a Family of Long Intergenic Noncoding RNA Genes with an Embedded Translocation Breakpoint Motif in Human Chromosomal Low Copy Repeats of 22q11.2—Some Surprises and Questions
Abstract
Supplementary Materials
Conflicts of Interest
References
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Goff, L.A.; Rinn, J.L. Linking RNA biology to lncRNAs. Genome Res. 2015, 25, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Feschotte, C. Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lin, M.; Rockowitz, S.; Lachman, H.M.; Zheng, D. Characterization of human pseudogene-derived non-coding RNAs for functional potential. PLoS ONE 2014, 9, e93972. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I. Evolution to the rescue: Using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016, 17, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.H.; Tsai, Z.T.; Tsai, H.K. Comparative genomic analyses highlight the contribution of pseudogenized protein-coding genes to human lincRNAs. BMC Genom. 2017, 18, 786. [Google Scholar] [CrossRef] [PubMed]
- Terracciano, D.; Terreri, S.; de Nigris, F.; Costa, V.; Calin, G.A.; Cimmino, A. The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochim. Biophys. Acta 2017, 1868, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.M. On the Origin of lncRNAs: Missing Link Found. Trends Genet. 2017, 33, 660–662. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Awan, H.M.; Shah, A.; Rashid, F.; Shan, G. Primate-specific Long Non-coding RNAs and MicroRNAs. Genom. Proteom. Bioinform. 2017, 15, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Delihas, N. A family of long intergenic non-coding RNA genes in human chromosomal region 22q11.2 carry a DNA translocation breakpoint/AT-rich sequence. PLoS ONE 2018, 13, e0195702. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, L.; Pandita, R.K.; Morrow, B.E. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 1999, 64, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, T.H.; Kurahashi, H.; Saitta, S.C.; O’Hare, A.M.; Hu, P.; Roe, B.A.; Driscoll, D.A.; McDonald-McGinn, D.M.; Zackai, E.H.; Budarf, M.L.; et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000, 9, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Eichler, E.E. Masquerading repeats: Paralogous pitfalls of the Human Genome. Genome Res. 1998, 8, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Lupski, J.R. Molecular-evolutionary mechanisms for genomic disorders. Curr. Opin. Genet. Dev. 2002, 12, 312–319. [Google Scholar] [CrossRef]
- Edelmann, L.; Spiteri, E.; Koren, K.; Pulijaal, V.; Bialer, M.G.; Shanske, A.; Goldberg, R.; Morrow, B.E. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kurahashi, H.; Emanuel, B.S. Chromosomal translocations and palindromic AT-rich repeats. Curr. Opin. Genet. Dev. 2012, 22, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Kurahashi, H.; Inagaki, H.; Hosoba, E.; Kato, T.; Ohye, T.; Kogo, H.; Emanuel, B.S. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007, 17, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Babcock, M.; Yatsenko, S.; Stankiewicz, P.; Lupski, J.R.; Morrow, B.E. AT-rich repeats associated with chromosome 22q11.2 rearrangement disorders shape human genome architecture on Yq12. Genome Res. 2007, 17, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2017, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
- Samonte, R.V.; Eichler, E.E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002, 3, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Shaw, C.J.; Withers, M.; Inoue, K.; Lupski, J.R. Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res. 2004, 14, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Dennehey, B.K.; Gutches, D.G.; McConkey, E.H.; Krauter, K.S. Inversion, duplication, and changes in gene context are associated with human chromosome 18 evolution. Genomics 2004, 83, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Dennis, M.Y.; Eichler, E.E. Human adaptation and evolution by segmental duplication. Curr. Opin. Genet. Dev. 2016, 41, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Freyer, L.; Morrow, B.; Zheng, D. Characterization of the past and current duplication activities in the human 22q11.2 region. BMC Genom. 2011, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Harel T Pehlivan, D.; Caskey, C.T.; Lupski, J.R. Mendelian, Non-Mendelian, Multigenic Inheritance, and Epigenetics. In Rosenberg’s Molecular and Genetic Basis of Neurological and Psychiatric Disease, 5th ed.; Academic Press: Waltham, MA, USA, 2015; Chapter 1; pp. 3–27. [Google Scholar]
- Shaikh, T.H.; Kurahashi, H.; Emanuel, B.S. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: An update and literature review. Genet. Med. 2001, 3, 6–13. [Google Scholar] [CrossRef] [PubMed]
- McDonald-McGinn, D.M.; Sullivan, K.E.; Marino, B.; Philip, N.; Swillen, A.; Vorstman, J.A.; Zackai, E.H.; Emanuel, B.S.; Vermeesch, J.R.; Morrow, B.E.; et al. 22q11.2 deletion syndrome. Nat. Rev. Dis. Primers 2015, 1, 15071. [Google Scholar] [CrossRef] [PubMed]
- Kobrynski, L.J.; Sullivan, K.E. Velocardiofacial syndrome, DiGeorge syndrome: The chromosome 22q11.2 deletion syndromes. Lancet 2007, 370, 1443–1452. [Google Scholar] [CrossRef]
- Kurahashi, H.; Inagaki, H.; Ohye, T.; Kogo, H.; Kato, T.; Emanuel, B.S. Chromosomal translocations mediated by palindromic DNA. Cell Cycle 2006, 5, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H.; Kato, T.; Tsutsumi, M.; Ouchi, Y.; Ohye, T.; Kurahashi, H. Palindrome-Mediated Translocations in Humans: A New Mechanistic Model for Gross Chromosomal Rearrangements. Front. Genet. 2016, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Gotter, A.L.; Shaikh, T.H.; Budarf, M.L.; Rhodes, C.H.; Emanuel, B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004, 13, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Tong, M.; Kato, T.; Yamada, K.; Inagaki, H.; Kogo, H.; Ohye, T.; Tsutsumi, M.; Wang, J.; Emanuel, B.S.; Kurahashi, H. Polymorphisms of the 22q11.2 breakpoint region influence the frequency of de novo constitutional t(11;22)s in sperm. Hum. Mol. Genet. 2010, 19, 2630–2637. [Google Scholar] [CrossRef] [PubMed]
- Frommer, M.; Prosser, J.; Vincent, P.C. Human satellite I sequences include a male specific 2.47 kb tandemly repeated unit containing one Alu family member per repeat. Nucleic Acids Res. 1984, 12, 2887–2900. [Google Scholar] [CrossRef] [PubMed]
- Delihas, N. Complexity of a small non-protein coding sequence in chromosomal region 22q11.2: Presence of specialized DNA secondary structures and RNA exon/intron motifs. BMC Genom 2015, 16, 785. [Google Scholar] [CrossRef] [PubMed]
- Bacolla, A.; Collins, J.R.; Gold, B.; Chuzhanova, N.; Yi, M.; Stephens, R.M.; Stefanov, S.; Olsh, A.; Jakupciak, J.P.; Dean, M.; et al. Long homopurine·homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 2006, 34, 2663–2675. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H.; Ohye, T.; Kogo, H.; Yamada, K.; Kowa, H.; Shaikh, T.H.; Emanuel, B.S. Kurahashi Palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved in primates. Hum. Mutat. 2005, 26, 332–342. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delihas, N. Formation of a Family of Long Intergenic Noncoding RNA Genes with an Embedded Translocation Breakpoint Motif in Human Chromosomal Low Copy Repeats of 22q11.2—Some Surprises and Questions. Non-Coding RNA 2018, 4, 16. https://doi.org/10.3390/ncrna4030016
Delihas N. Formation of a Family of Long Intergenic Noncoding RNA Genes with an Embedded Translocation Breakpoint Motif in Human Chromosomal Low Copy Repeats of 22q11.2—Some Surprises and Questions. Non-Coding RNA. 2018; 4(3):16. https://doi.org/10.3390/ncrna4030016
Chicago/Turabian StyleDelihas, Nicholas. 2018. "Formation of a Family of Long Intergenic Noncoding RNA Genes with an Embedded Translocation Breakpoint Motif in Human Chromosomal Low Copy Repeats of 22q11.2—Some Surprises and Questions" Non-Coding RNA 4, no. 3: 16. https://doi.org/10.3390/ncrna4030016
APA StyleDelihas, N. (2018). Formation of a Family of Long Intergenic Noncoding RNA Genes with an Embedded Translocation Breakpoint Motif in Human Chromosomal Low Copy Repeats of 22q11.2—Some Surprises and Questions. Non-Coding RNA, 4(3), 16. https://doi.org/10.3390/ncrna4030016