CARINH, an Interferon-Induced LncRNA in Cancer and Inflammation
Abstract
1. Introduction
2. CARINH: Nomenclature, Genomic Location, and Synteny
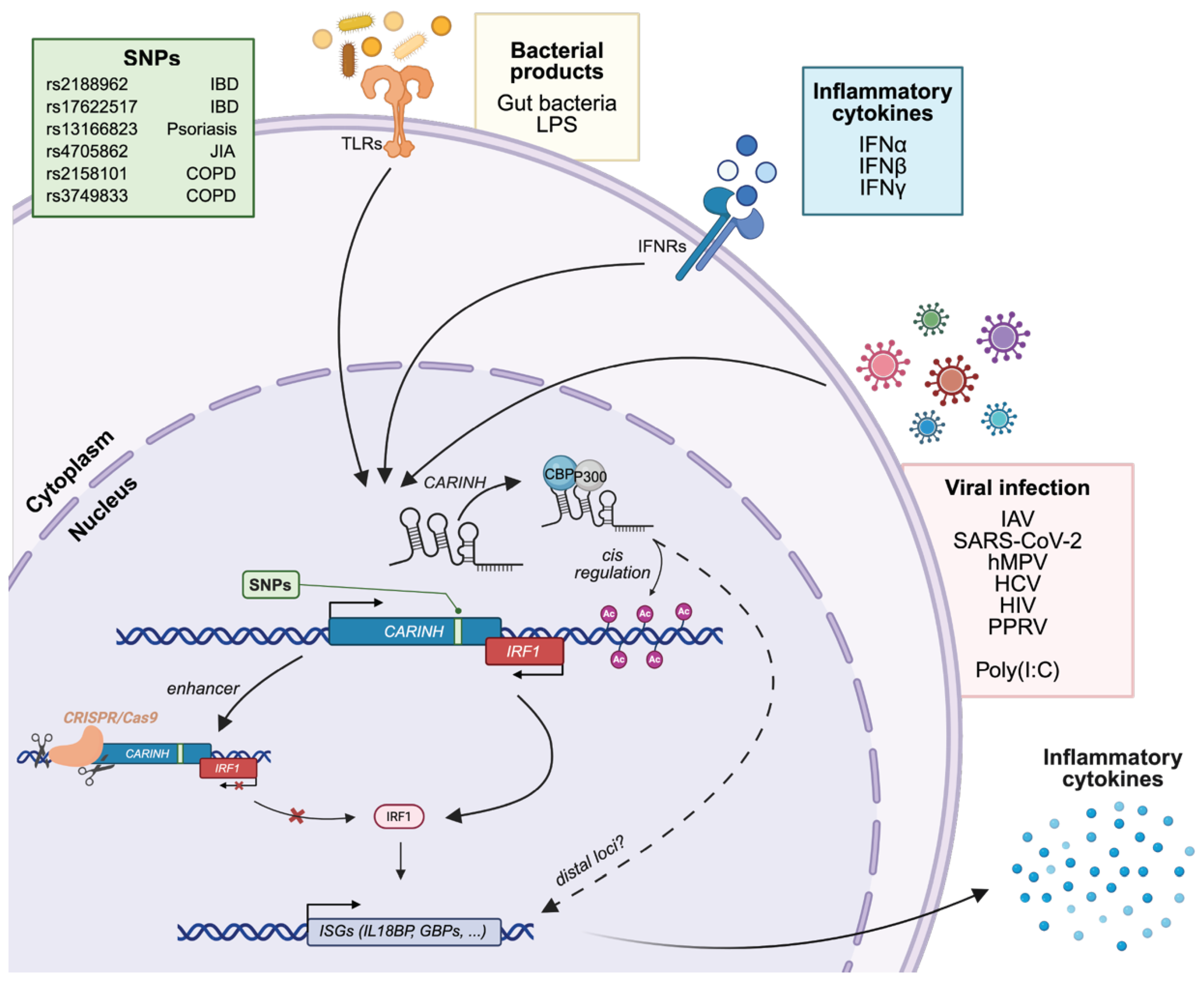
3. CARINH and Innate Immunity
3.1. CARINH in Viral Infection
3.2. CARINH in Autoimmunity and Chronic Inflammatory Diseases
3.2.1. Inflammatory Bowel Disease
3.2.2. Cardiometabolic Disorders
3.2.3. CARINH Variants as Determinants of Inflammatory Disease Risk and Therapeutic Response
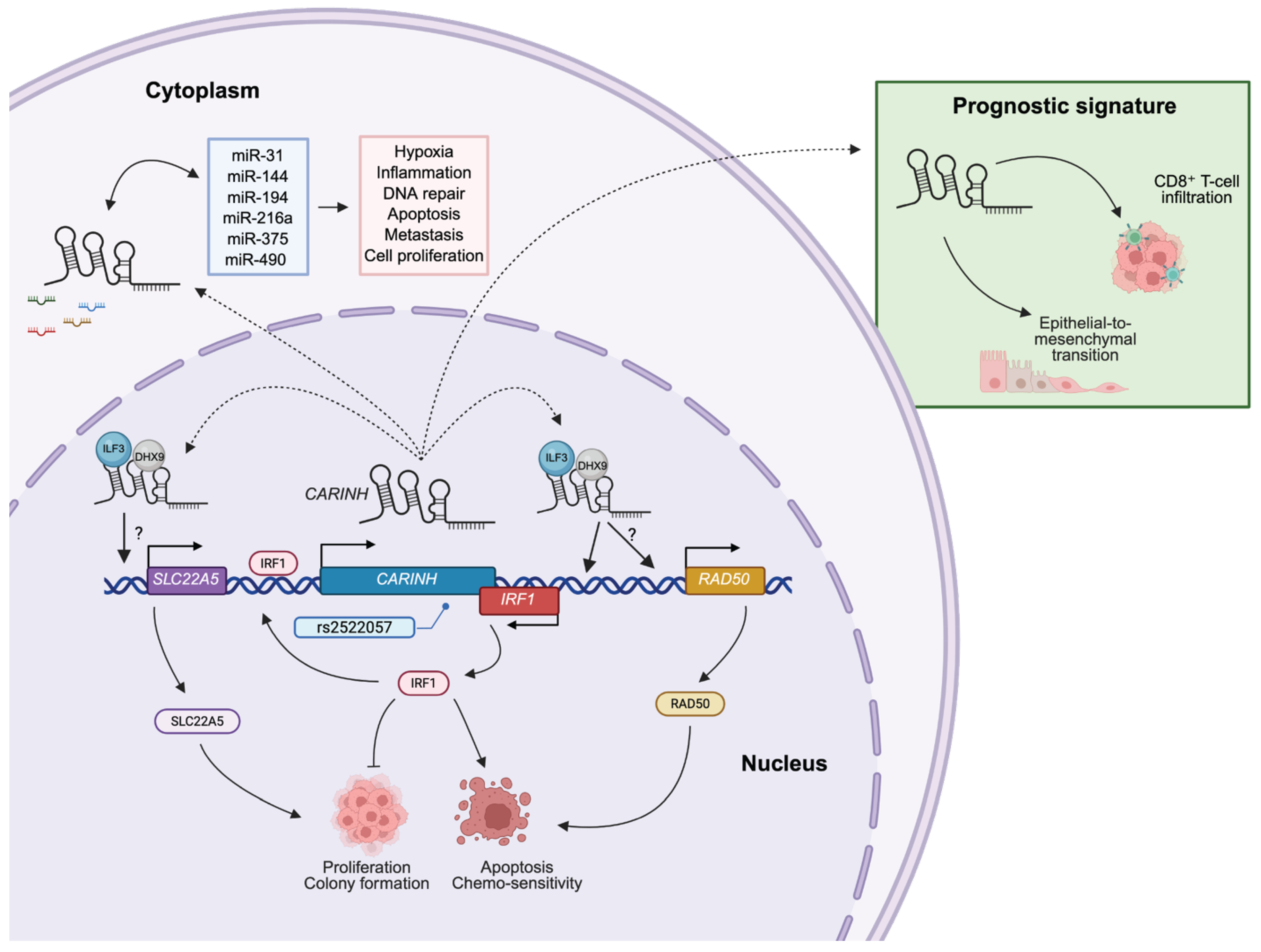
4. CARINH and Cancer
4.1. Esophageal Squamous Cell Carcinoma
4.2. Bladder Cancer
4.3. Acute Lymphoblastic Leukemia
4.4. Breast Cancer
5. Perspective
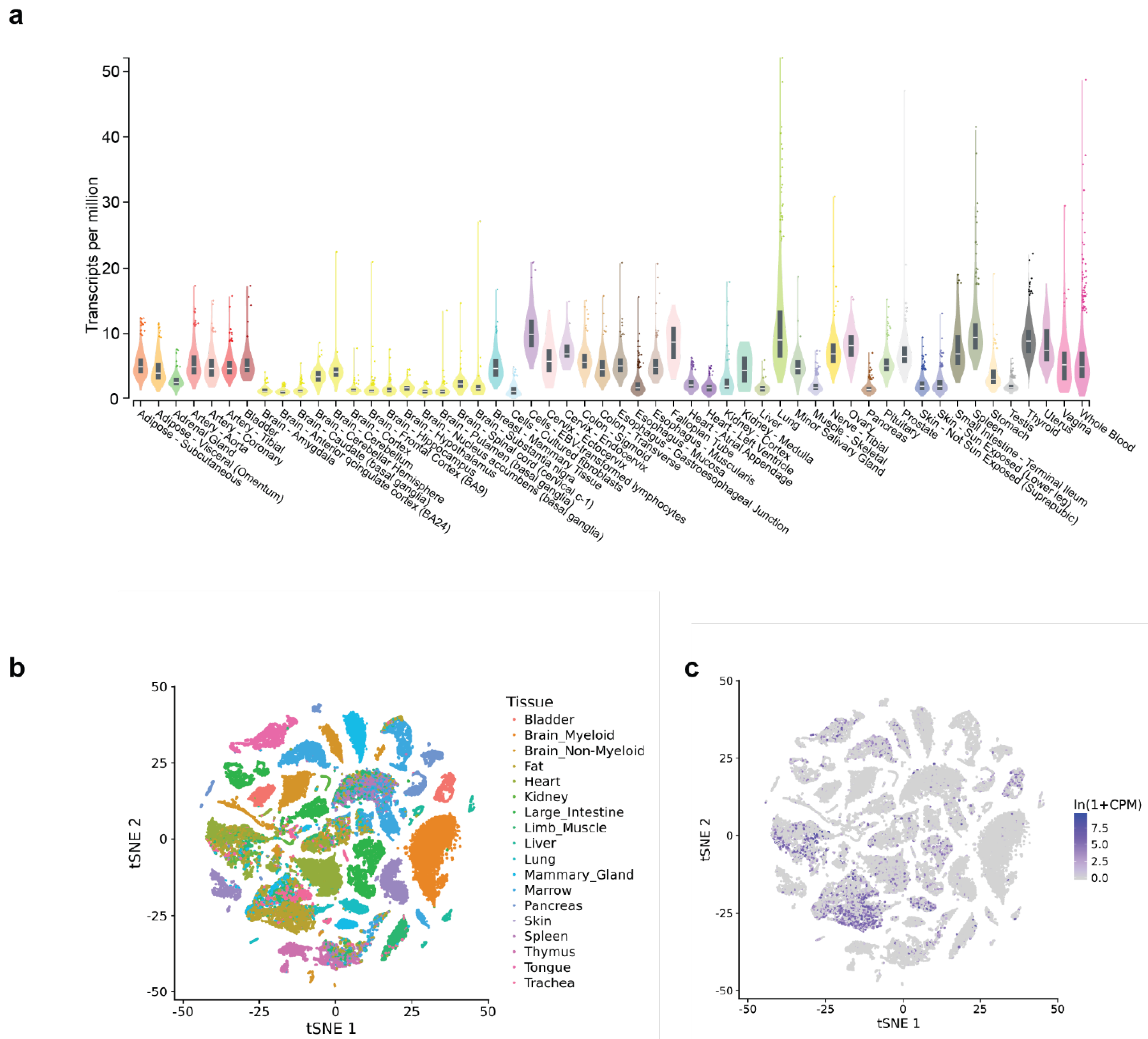
5.1. Expression of CARINH in Other Cells and Cell Types
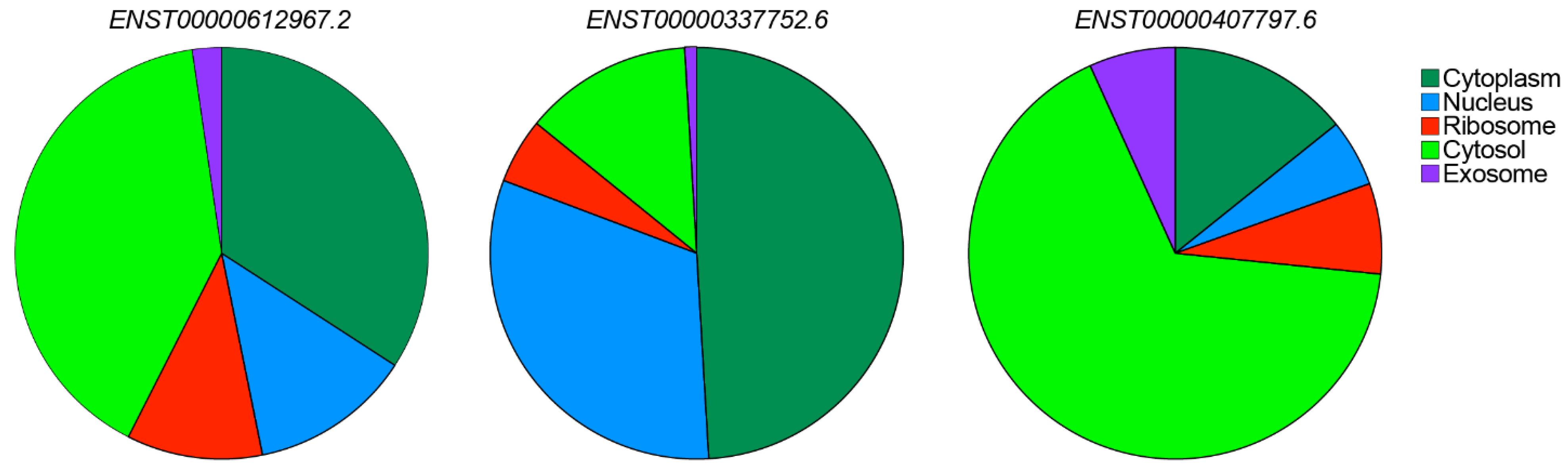
5.2. Subcellular Localization of CARINH
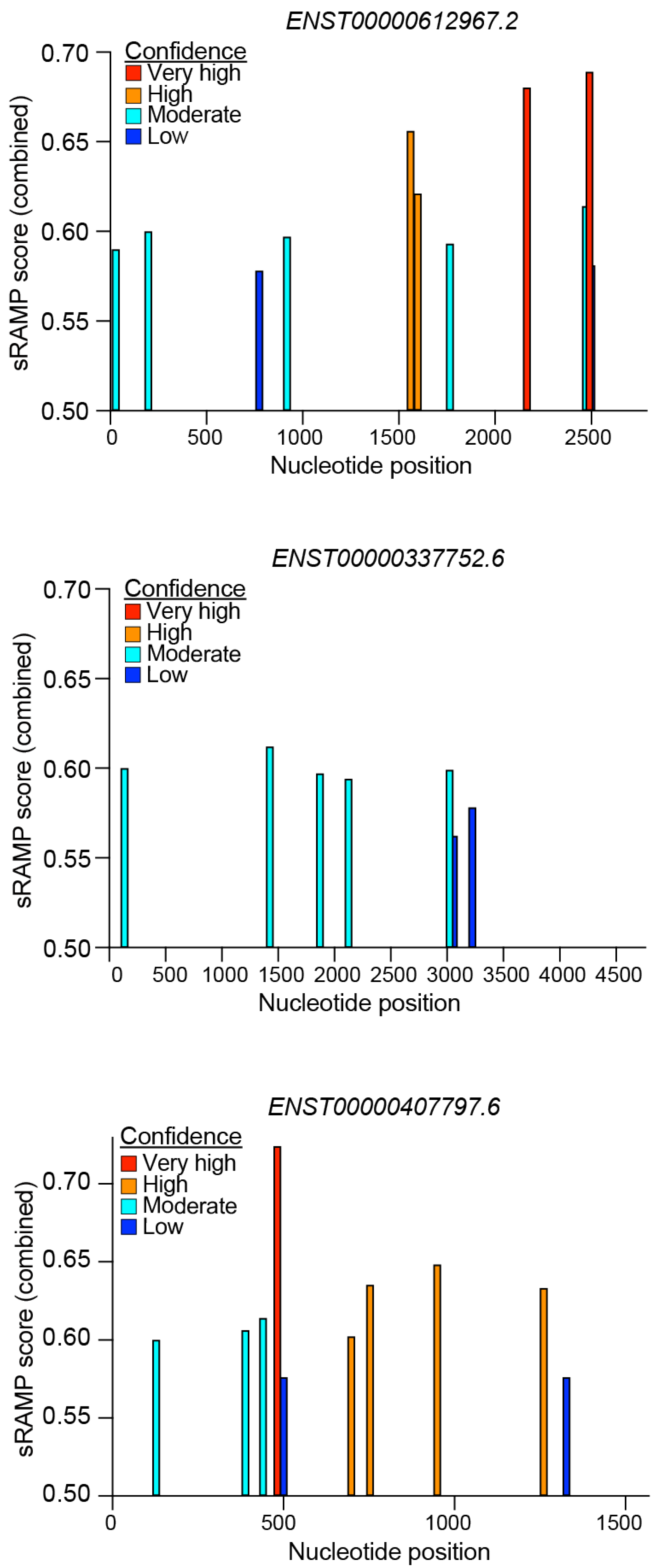
5.3. RNA Modifications Driving CARINH Expression and Function
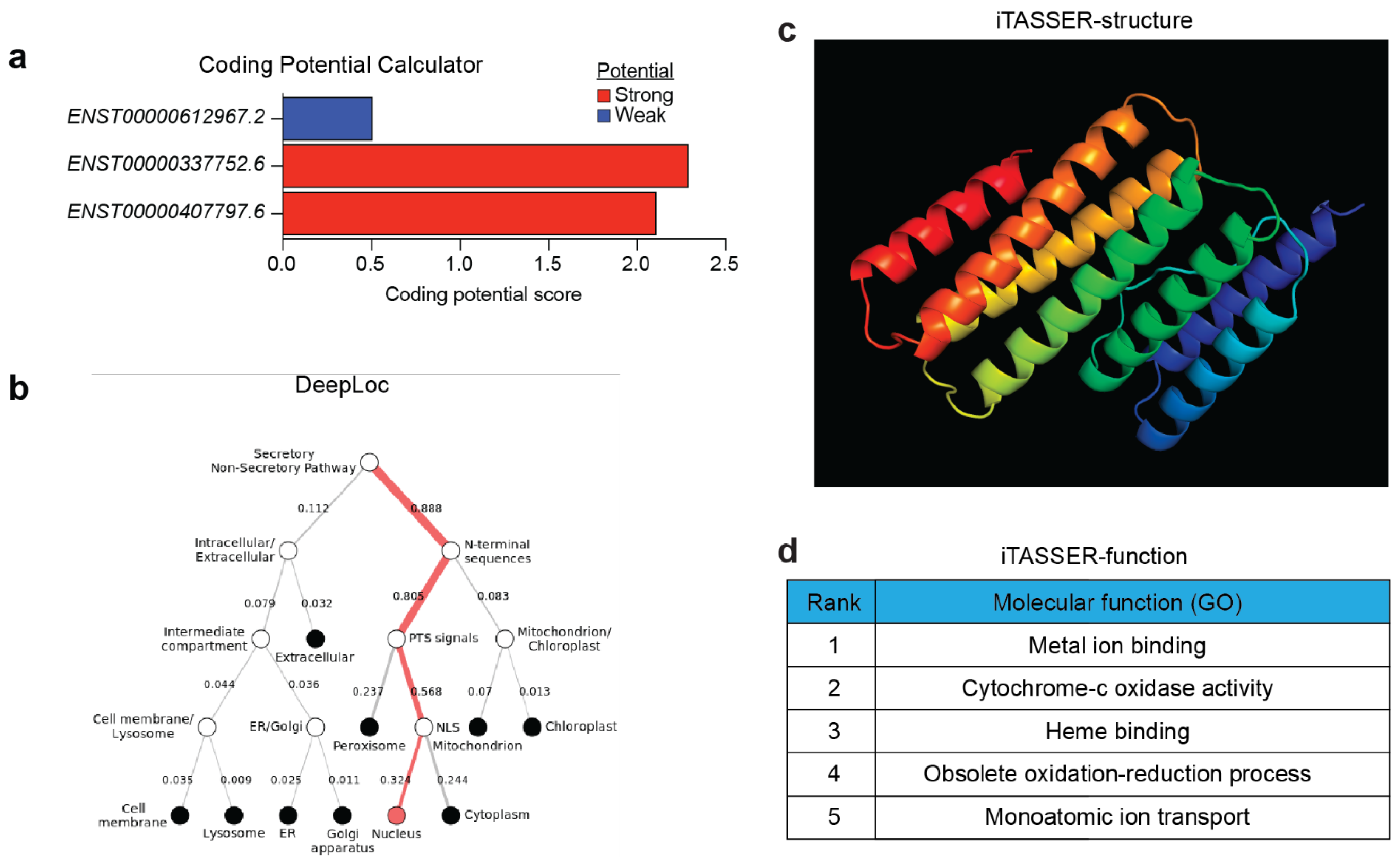
5.4. Coding Potential of CARINH
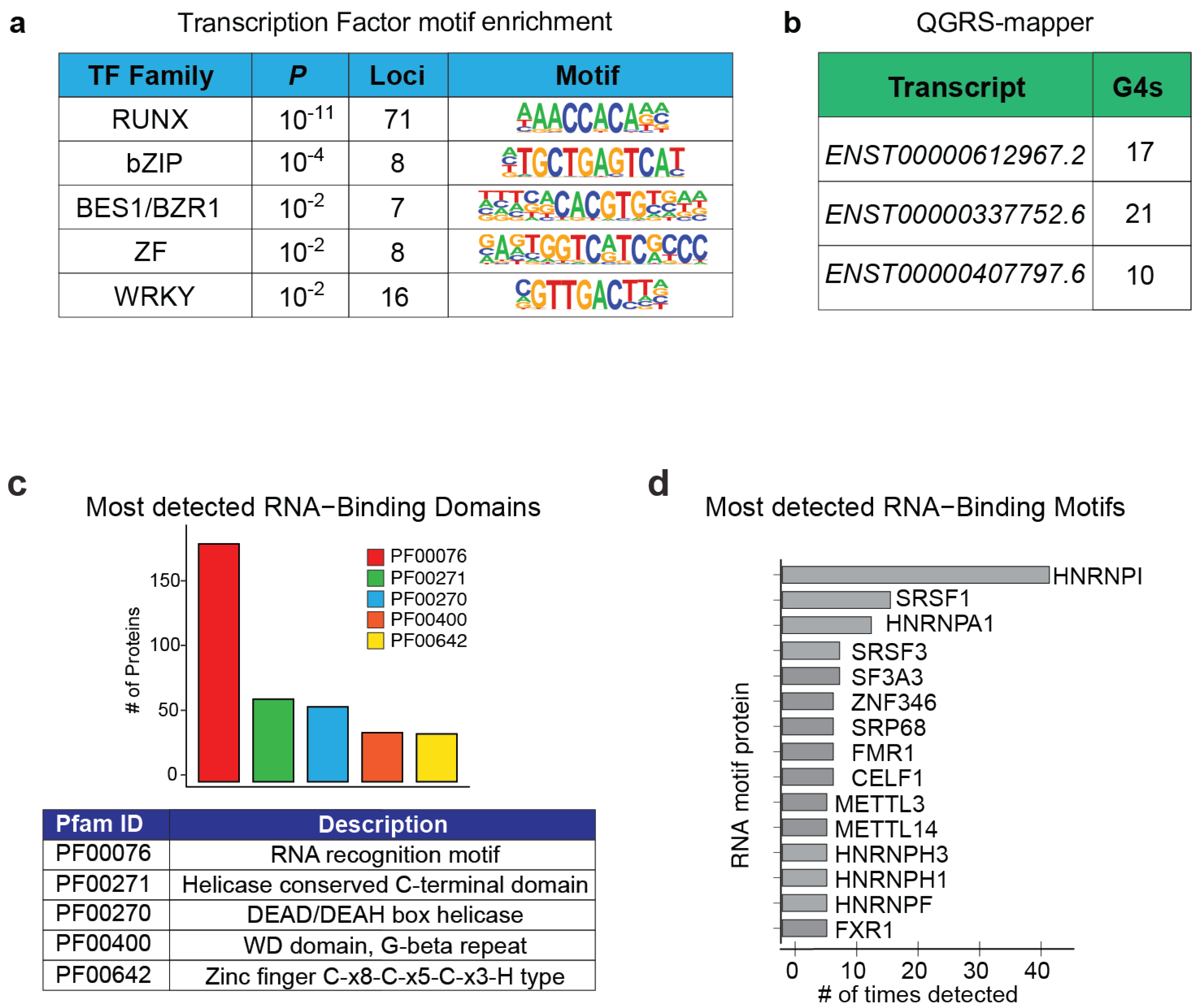
5.5. Chromatin- and Protein-Binding Abilities of CARINH
5.6. CARINH as a Competing Endogenous RNA
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, L.L.; Kim, V.N. Small and Long Non-Coding Rnas: Past, Present, and Future. Cell 2024, 187, 6451–6485. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding Rnas: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Spurlock, C.F., 3rd; Crooke, P.S., 3rd; Aune, T.M. Biogenesis and Transcriptional Regulation of Long Noncoding Rnas in the Human Immune System. J. Immunol. 2016, 197, 4509–4517. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Proudfoot, N.J. Mechanisms of Lncrna Biogenesis as Revealed by Nascent Transcriptomics. Nat. Rev. Mol. Cell Biol. 2022, 23, 389–406. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding Rnas. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-Coding Rna: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of Long Noncoding Rna Function in Development and Disease. Cell Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. Lnccation: Lncrna Localization and Function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Krause, H.M. New and Prospective Roles for Lncrnas in Organelle Formation and Function. Trends Genet. 2018, 34, 736–745. [Google Scholar] [CrossRef]
- Goff, L.A.; Rinn, J.L. Linking Rna Biology to Lncrnas. Genome Res. 2015, 25, 1456–1465. [Google Scholar] [CrossRef]
- Satpathy, A.T.; Chang, H.Y. Long Noncoding Rna in Hematopoiesis and Immunity. Immunity 2015, 42, 792–804. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding Rnas. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Guil, S.; Esteller, M. Cis-Acting Noncoding Rnas: Friends and Foes. Nat. Struct. Mol. Biol. 2012, 19, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F.; Gootenberg, J.S.; Sanjana, N.E.; Wright, J.B.; Fulco, C.P.; et al. Genome-Scale Activation Screen Identifies a Lncrna Locus Regulating a Gene Neighbourhood. Nature 2017, 548, 343–346. [Google Scholar] [CrossRef]
- Hennessy, E.J.; van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The Long Noncoding Rna Chrome Regulates Cholesterol Homeostasis in Primate. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef]
- Ala, U. Competing Endogenous Rnas, Non-Coding Rnas and Diseases: An Intertwined Story. Cells 2020, 9, 1574. [Google Scholar] [CrossRef]
- Liu, X.H.; Sun, M.; Nie, F.Q.; Ge, Y.B.; Zhang, E.B.; Yin, D.D.; Kong, R.; Xia, R.; Lu, K.H.; Li, J.H.; et al. Lnc Rna Hotair Functions as a Competing Endogenous Rna to Regulate Her2 Expression by Sponging Mir-331-3p in Gastric Cancer. Mol. Cancer 2014, 13, 92. [Google Scholar] [CrossRef]
- Monroy-Eklund, A.; Taylor, C.; Weidmann, C.A.; Burch, C.; Laederach, A. Structural Analysis of Malat1 Long Noncoding Rna in Cells and in Evolution. RNA 2023, 29, 691–704. [Google Scholar] [CrossRef]
- Quinn, J.J.; Zhang, Q.C.; Georgiev, P.; Ilik, I.A.; Akhtar, A.; Chang, H.Y. Rapid Evolutionary Turnover Underlies Conserved Lncrna-Genome Interactions. Genes. Dev. 2016, 30, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Barriocanal, M.; Prats-Mari, L.; Razquin, N.; Prior, C.; Unfried, J.P.; Fortes, P. Isr8/Irf1-As1 Is Relevant for Ifnalpha and Nf-Kappab Responses. Front. Immunol. 2022, 13, 829335. [Google Scholar] [CrossRef]
- Carnero, E.; Barriocanal, M.; Segura, V.; Guruceaga, E.; Prior, C.; Borner, K.; Grimm, D.; Fortes, P. Type I Interferon Regulates the Expression of Long Non-Coding Rnas. Front. Immunol. 2014, 5, 548. [Google Scholar] [CrossRef]
- Cyr, Y.; Gourvest, M.; Ciabattoni, G.O.; Zhang, T.; Newman, A.A.; Zahr, T.; Delbare, S.; Schlamp, F.; Dittmann, M.; Moore, K.J.; et al. Lncrna Carinh Regulates Expression and Function of Innate Immune Transcription Factor Irf1 in Macrophages. Life Sci. Alliance 2025, 8, e2024003021. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Hu, T.; Tao, W.; Tong, J.; Han, Z.; Herndler-Brandstetter, D.; Wei, Z.; Liu, R.; Zhou, T.; Liu, Q.; et al. A Lncrna from an Inflammatory Bowel Disease Risk Locus Maintains Intestinal Host-Commensal Homeostasis. Cell Res. 2023, 33, 372–388. [Google Scholar] [CrossRef]
- Johnson, J.L.; Sargsyan, D.; Neiman, E.M.; Hart, A.; Stojmirovic, A.; Kosoy, R.; Irizar, H.; Suarez-Farinas, M.; Song, W.M.; Argmann, C.; et al. Gene Coexpression Networks Reveal a Broad Role for Lncrnas in Inflammatory Bowel Disease. JCI Insight 2024, 9, e168988. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, J.; Li, Y.; Lu, Z.; Che, Y.; Mao, S.; Lei, Y.; Zang, R.; Zheng, S.; Liu, C.; et al. Interferon-Inducible Lncrna Irf1-as Represses Esophageal Squamous Cell Carcinoma by Promoting Interferon Response. Cancer Lett. 2019, 459, 86–99. [Google Scholar] [CrossRef]
- Nekoeian, S.; Rostami, T.; Norouzy, A.; Hussein, S.; Tavoosidana, G.; Chahardouli, B.; Rostami, S.; Asgari, Y.; Azizi, Z. Identification of Lncrnas Associated with the Progression of Acute Lymphoblastic Leukemia Using a Competing Endogenous Rnas Network. Oncol. Res. 2022, 30, 259–268. [Google Scholar] [CrossRef]
- Tong, H.; Li, T.; Gao, S.; Yin, H.; Cao, H.; He, W. An Epithelial-Mesenchymal Transition-Related Long Noncoding Rna Signature Correlates with the Prognosis and Progression in Patients with Bladder Cancer. Biosci. Rep. 2021, 41, BSR20203944. [Google Scholar] [CrossRef] [PubMed]
- Adedokun, B.; Du, Z.; Gao, G.; Ahearn, T.U.; Lunetta, K.L.; Zirpoli, G.; Figueroa, J.; John, E.M.; Bernstein, L.; Zheng, W.; et al. Cross-Ancestry Gwas Meta-Analysis Identifies Six Breast Cancer Loci in African and European Ancestry Women. Nat. Commun. 2021, 12, 4198. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Wang, X.; Yang, Q. Identification of a Six-Immune-Related Long Non-Coding Rna Signature for Predicting Survival and Immune Infiltrating Status in Breast Cancer. Front. Genet. 2020, 11, 680. [Google Scholar] [CrossRef]
- Seal, R.L.; Tweedie, S.; Bruford, E.A. A Standardised Nomenclature for Long Non-Coding Rnas. IUBMB Life 2023, 75, 380–389. [Google Scholar] [CrossRef]
- Wright, M.W. A Short Guide to Long Non-Coding Rna Gene Nomenclature. Hum. Genom. 2014, 8, 7. [Google Scholar] [CrossRef]
- Weinstock, A.; Rahman, K.; Yaacov, O.; Nishi, H.; Menon, P.; Nikain, C.A.; Garabedian, M.L.; Pena, S.; Akbar, N.; Sansbury, B.E.; et al. Wnt Signaling Enhances Macrophage Responses to Il-4 and Promotes Resolution of Atherosclerosis. Elife 2021, 10, e67932. [Google Scholar] [CrossRef]
- Zhao, W.; Lei, T.; Li, H.; Sun, D.; Mo, X.; Wang, Z.; Zhang, K.; Ou, H. Macrophage-Specific Overexpression of Interleukin-5 Attenuates Atherosclerosis in Ldl Receptor-Deficient Mice. Gene Ther. 2015, 22, 645–652. [Google Scholar] [CrossRef]
- Kissling, V.M.; Reginato, G.; Bianco, E.; Kasaciunaite, K.; Tilma, J.; Cereghetti, G.; Schindler, N.; Lee, S.S.; Guerois, R.; Luke, B.; et al. Mre11-Rad50 Oligomerization Promotes DNA Double-Strand Break Repair. Nat. Commun. 2022, 13, 2374. [Google Scholar] [CrossRef] [PubMed]
- Trinchieri, G. Type I Interferon: Friend or Foe? J. Exp. Med. 2010, 207, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Navajas, J.M.; Lee, J.; David, M.; Raz, E. Immunomodulatory Functions of Type I Interferons. Nat. Rev. Immunol. 2012, 12, 125–135. [Google Scholar] [CrossRef]
- Kopitar-Jerala, N. The Role of Interferons in Inflammation and Inflammasome Activation. Front. Immunol. 2017, 8, 873. [Google Scholar] [CrossRef]
- Wen, B.; Qi, X.; Lv, D.; Yang, L.; Tang, P.; Chang, W.; Han, S.; Yu, S.; Wei, S.; Xue, Q.; et al. Long Noncoding Rna Irf1-as Is Associated with Peste Des Petits Ruminants Infection. Vet. Res. 2022, 53, 89. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yan, Z.; Lan, H.; Wu, Y.; Chen, S.; Qiu, G.; Wu, Y. Genetic Comorbidity of Psoriasis and Four Cardiovascular Diseases: Uncovering Shared Mechanisms and Potential Therapeutic Targets. Exp. Dermatol. 2025, 34, e70158. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, L.; Dai, H.; Qiu, G.; Liu, J.; Yu, D.; Liu, J.; Lyu, C.Z.; Liu, L.; Zheng, M. Genome-Wide Association Analysis of Anti-Tnf-Alpha Treatment Response in Chinese Patients with Psoriasis. Front. Pharmacol. 2022, 13, 968935. [Google Scholar] [CrossRef]
- John, C.; Guyatt, A.L.; Shrine, N.; Packer, R.; Olafsdottir, T.A.; Liu, J.; Hayden, L.P.; Chu, S.H.; Koskela, J.T.; Luan, J.; et al. Genetic Associations and Architecture of Asthma-Copd Overlap. Chest 2022, 161, 1155–1166. [Google Scholar] [CrossRef]
- Joo, J.; Himes, B. Gene-Based Analysis Reveals Sex-Specific Genetic Risk Factors of Copd. AMIA Annu. Symp. Proc. 2021, 2021, 601–610. [Google Scholar] [PubMed]
- Park, J.H.; Park, K.J. Genetic Variants Associated with Metabolic Dysfunction-Associated Fatty Liver Diseases in a Korean Population. Eur. J. Med. Res. 2025, 30, 318. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Z.; Peng, R.; Zhang, L.; Liu, H.; Wang, X.; Tian, Y.; Sun, Y. Rna-Seq Analysis Reveals Critical Transcriptome Changes Caused by Sodium Butyrate in Dn Mouse Models. Biosci. Rep. 2021, 41, BSR20203005. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhu, C. Identification of Hub Lncrnas Correlated with Tetralogy of Fallot Based on Weighted Gene Co-Expression Network Analysis. Biochem. Biophys. Rep. 2024, 39, 101786. [Google Scholar] [CrossRef]
- Chiaroni-Clarke, R.C.; Munro, J.E.; Chavez, R.A.; Pezic, A.; Allen, R.C.; Akikusa, J.D.; Piper, S.E.; Saffery, R.; Ponsonby, A.L.; Ellis, J.A. Independent Confirmation of Juvenile Idiopathic Arthritis Genetic Risk Loci Previously Identified by Immunochip Array Analysis. Pediatr. Rheumatol. Online J. 2014, 12, 53. [Google Scholar] [CrossRef]
- Weber, B.; Merola, J.F.; Husni, M.E.; Di Carli, M.; Berger, J.S.; Garshick, M.S. Psoriasis and Cardiovascular Disease: Novel Mechanisms and Evolving Therapeutics. Curr. Atheroscler. Rep. 2021, 23, 67. [Google Scholar] [CrossRef]
- Kumar, V.; Westra, H.J.; Karjalainen, J.; Zhernakova, D.V.; Esko, T.; Hrdlickova, B.; Almeida, R.; Zhernakova, A.; Reinmaa, E.; Vosa, U.; et al. Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding Rna Expression. PLoS Genet. 2013, 9, e1003201. [Google Scholar] [CrossRef]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. Magma: Generalized Gene-Set Analysis of Gwas Data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional Mapping and Annotation of Genetic Associations with Fuma. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef]
- Grunert, M.; Dorn, C.; Schueler, M.; Dunkel, I.; Schlesinger, J.; Mebus, S.; Alexi-Meskishvili, V.; Perrot, A.; Wassilew, K.; Timmermann, B.; et al. Rare and Private Variations in Neural Crest, Apoptosis and Sarcomere Genes Define the Polygenic Background of Isolated Tetralogy of Fallot. Hum. Mol. Genet. 2014, 23, 3115–3128. [Google Scholar] [CrossRef] [PubMed]
- Grunert, M.; Dorn, C.; Cui, H.; Dunkel, I.; Schulz, K.; Schoenhals, S.; Sun, W.; Berger, F.; Chen, W.; Sperling, S.R. Comparative DNA Methylation and Gene Expression Analysis Identifies Novel Genes for Structural Congenital Heart Diseases. Cardiovasc. Res. 2016, 112, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Hinks, A.; Cobb, J.; Marion, M.C.; Prahalad, S.; Sudman, M.; Bowes, J.; Martin, P.; Comeau, M.E.; Sajuthi, S.; Andrews, R.; et al. Dense Genotyping of Immune-Related Disease Regions Identifies 14 New Susceptibility Loci for Juvenile Idiopathic Arthritis. Nat. Genet. 2013, 45, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Brandt, M.; Kim-Hellmuth, S.; Ziosi, M.; Gokden, A.; Wolman, A.; Lam, N.; Recinos, Y.; Daniloski, Z.; Morris, J.A.; Hornung, V.; et al. An Autoimmune Disease Risk Variant: A Trans Master Regulatory Effect Mediated by Irf1 under Immune Stimulation? PLoS Genet. 2021, 17, e1009684. [Google Scholar] [CrossRef]
- Baek, M.; Chai, J.C.; Choi, H.I.; Yoo, E.; Binas, B.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. Analysis of Differentially Expressed Long Non-Coding Rnas in Lps-Induced Human Hmc3 Microglial Cells. BMC Genom. 2022, 23, 853. [Google Scholar] [CrossRef]
- Parker, B.S.; Rautela, J.; Hertzog, P.J. Antitumour Actions of Interferons: Implications for Cancer Therapy. Nat. Rev. Cancer 2016, 16, 131–144. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Li, J.; Han, L.; Roebuck, P.; Diao, L.; Liu, L.; Yuan, Y.; Weinstein, J.N.; Liang, H. Tanric: An Interactive Open Platform to Explore the Function of Lncrnas in Cancer. Cancer Res. 2015, 75, 3728–3737. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- De Clara, E.; Gourvest, M.; Ma, H.; Vergez, F.; Tosolini, M.; Dejean, S.; Demur, C.; Delabesse, E.; Recher, C.; Touriol, C.; et al. Long Non-Coding Rna Expression Profile in Cytogenetically Normal Acute Myeloid Leukemia Identifies a Distinct Signature and a New Biomarker in Npm1-Mutated Patients. Haematologica 2017, 102, 1718–1726. [Google Scholar] [CrossRef] [PubMed]
- Gourvest, M.; Brousset, P.; Bousquet, M. Long Noncoding Rnas in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance. Cancers 2019, 11, 1638. [Google Scholar] [CrossRef]
- Gourvest, M.; De Clara, E.; Wu, H.C.; Touriol, C.; Meggetto, F.; De The, H.; Pyronnet, S.; Brousset, P.; Bousquet, M. A Novel Leukemic Route of Mutant Npm1 through Nuclear Import of the Overexpressed Long Noncoding Rna Lona. Leukemia 2021, 35, 2784–2798. [Google Scholar] [CrossRef]
- Outhwaite, J.E.; McGuire, V.; Simmons, D.G. Genetic Ablation of Placental Sinusoidal Trophoblast Giant Cells Causes Fetal Growth Restriction and Embryonic Lethality. Placenta 2015, 36, 951–955. [Google Scholar] [CrossRef]
- Chaim, I.A.; Nagel, Z.D.; Jordan, J.J.; Mazzucato, P.; Ngo, L.P.; Samson, L.D. In Vivo Measurements of Interindividual Differences in DNA Glycosylases and Ape1 Activities. Proc. Natl. Acad. Sci. USA 2017, 114, E10379–E10388. [Google Scholar] [CrossRef]
- Cruz-Miranda, G.M.; Hidalgo-Miranda, A.; Barcenas-Lopez, D.A.; Nunez-Enriquez, J.C.; Ramirez-Bello, J.; Mejia-Arangure, J.M.; Jimenez-Morales, S. Long Non-Coding Rna and Acute Leukemia. Int. J. Mol. Sci. 2019, 20, 735. [Google Scholar] [CrossRef]
- Yeoh, A.E.; Li, Z.; Dong, D.; Lu, Y.; Jiang, N.; Trka, J.; Tan, A.M.; Lin, H.P.; Quah, T.C.; Ariffin, H.; et al. Effective Response Metric: A Novel Tool to Predict Relapse in Childhood Acute Lymphoblastic Leukaemia Using Time-Series Gene Expression Profiling. Br. J. Haematol. 2018, 181, 653–663. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Jeggari, A.; Marks, D.S.; Larsson, E. Mircode: A Map of Putative Microrna Target Sites in the Long Non-Coding Transcriptome. Bioinformatics 2012, 28, 2062–2063. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Uray, I.P.; Mazumdar, A.; Mayer, J.A.; Brown, P.H. Slc22a5/Octn2 Expression in Breast Cancer Is Induced by Estrogen Via a Novel Intronic Estrogen-Response Element (Ere). Breast Cancer Res. Treat. 2012, 134, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Chen, C.F.; Li, S.; Chen, Y.; Wang, C.C.; Xiao, J.; Chen, P.L.; Sharp, Z.D.; Lee, W.H. Association of Brca1 with the Hrad50-Hmre11-P95 Complex and the DNA Damage Response. Science 1999, 285, 747–750. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Stang, M.T.; Liu, Y.; Yan, J.; Pizzoferrato, E.; Yim, J.H. Irf-1 Inhibits Nf-Kappab Activity, Suppresses Traf2 and Ciap1 and Induces Breast Cancer Cell Specific Growth Inhibition. Cancer Biol. Ther. 2015, 16, 1029–1041. [Google Scholar] [CrossRef]
- Bouker, K.B.; Skaar, T.C.; Riggins, R.B.; Harburger, D.S.; Fernandez, D.R.; Zwart, A.; Wang, A.; Clarke, R. Interferon Regulatory Factor-1 (Irf-1) Exhibits Tumor Suppressor Activities in Breast Cancer Associated with Caspase Activation and Induction of Apoptosis. Carcinogenesis 2005, 26, 1527–1535. [Google Scholar] [CrossRef]
- Kao, K.J.; Chang, K.M.; Hsu, H.C.; Huang, A.T. Correlation of Microarray-Based Breast Cancer Molecular Subtypes and Clinical Outcomes: Implications for Treatment Optimization. BMC Cancer 2011, 11, 143. [Google Scholar] [CrossRef]
- Consortium, G. TEx. The Genotype-Tissue Expression (Gtex) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators. Single-Cell Transcriptomics of 20 Mouse Organs Creates a Tabula Muris. Nature 2018, 562, 367–372. [Google Scholar] [CrossRef]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and Abundance Analysis of Human Lncrnas at Single-Cell and Single-Molecule Resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Pan, X.; Yang, Y.; Huang, Y.; Shen, H.B. The Lnclocator: A Subcellular Localization Predictor for Long Non-Coding Rnas Based on a Stacked Ensemble Classifier. Bioinformatics 2018, 34, 2185–2194. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, L.; Hu, S.; Tang, G.; Chen, J.; Yi, Y.; Xie, H.; Lin, J.; Wang, M.; Wang, D.; et al. Rnalocate V3.0: Advancing the Repository of Rna Subcellular Localization with Dynamic Analysis and Prediction. Nucleic Acids Res. 2025, 53, D284–D292. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic Transcriptomic M(6)a Decoration: Writers, Erasers, Readers and Functions in Rna Metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, P.; Li, Y.H.; Zhang, Z.; Cui, Q. Sramp: Prediction of Mammalian N6-Methyladenosine (M6a) Sites Based on Sequence-Derived Features. Nucleic Acids Res. 2016, 44, e91. [Google Scholar] [CrossRef]
- Andrews, S.J.; Rothnagel, J.A. Emerging Evidence for Functional Peptides Encoded by Short Open Reading Frames. Nat. Rev. Genet. 2014, 15, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Johnstone, T.G.; Christiano, R.; Mackowiak, S.D.; Obermayer, B.; Fleming, E.S.; Vejnar, C.E.; Lee, M.T.; Rajewsky, N.; Walther, T.C.; et al. Identification of Small Orfs in Vertebrates Using Ribosome Footprinting and Evolutionary Conservation. EMBO J. 2014, 33, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Hofman, D.A.; Ruiz-Orera, J.; Yannuzzi, I.; Murugesan, R.; Brown, A.; Clauser, K.R.; Condurat, A.L.; van Dinter, J.T.; Engels, S.A.G.; Goodale, A.; et al. Translation of Non-Canonical Open Reading Frames as a Cancer Cell Survival Mechanism in Childhood Medulloblastoma. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sandmann, C.L.; Schulz, J.F.; Ruiz-Orera, J.; Kirchner, M.; Ziehm, M.; Adami, E.; Marczenke, M.; Christ, A.; Liebe, N.; Greiner, J.; et al. Evolutionary Origins and Interactomes of Human, Young Microproteins and Small Peptides Translated from Short Open Reading Frames. Mol. Cell 2023, 83, 994–1011.e18. [Google Scholar] [CrossRef]
- Prensner, J.R.; Enache, O.M.; Luria, V.; Krug, K.; Clauser, K.R.; Dempster, J.M.; Karger, A.; Wang, L.; Stumbraite, K.; Wang, V.M.; et al. Noncanonical Open Reading Frames Encode Functional Proteins Essential for Cancer Cell Survival. Nat. Biotechnol. 2021, 39, 697–704. [Google Scholar] [CrossRef]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e29. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Abelin, J.G.; Kok, L.W.; Clauser, K.R.; Mudge, J.M.; Ruiz-Orera, J.; Bassani-Sternberg, M.; Moritz, R.L.; Deutsch, E.W.; van Heesch, S. What Can Ribo-Seq, Immunopeptidomics, and Proteomics Tell Us About the Noncanonical Proteome? Mol. Cell Proteom. 2023, 22, 100631. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. Cpc: Assess the Protein-Coding Potential of Transcripts Using Sequence Features and Support Vector Machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-Tasser: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sonderby, C.K.; Sonderby, S.K.; Nielsen, H.; Winther, O. Deeploc: Prediction of Protein Subcellular Localization Using Deep Learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Bencze, D.; Fekete, T.; Pazmandi, K. Type I Interferon Production of Plasmacytoid Dendritic Cells under Control. Int. J. Mol. Sci. 2021, 22, 4190. [Google Scholar] [CrossRef]
- Hu, Y.; Pan, Q.; Zhou, K.; Ling, Y.; Wang, H.; Li, Y. Runx1 Inhibits the Antiviral Immune Response against Influenza a Virus through Attenuating Type I Interferon Signaling. Virol. J. 2022, 19, 39. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The Regulation and Functions of DNA and Rna G-Quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. Qgrs Mapper: A Web-Based Server for Predicting G-Quadruplexes in Nucleotide Sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef] [PubMed]
- van Solingen, C.; Cyr, Y.; Scacalossi, K.R.; de Vries, M.; Barrett, T.J.; de Jong, A.; Gourvest, M.; Zhang, T.; Peled, D.; Kher, R.; et al. Long Noncoding Rna Chromr Regulates Antiviral Immunity in Humans. Proc. Natl. Acad. Sci. USA 2022, 119, e2210321119. [Google Scholar] [CrossRef]
- Shukla, C.; Datta, B. G-Quadruplexes in Long Non-Coding Rnas and Their Interactions with Proteins. Int. J. Biol. Macromol. 2024, 278, 134946. [Google Scholar] [CrossRef]
- Armaos, A.; Colantoni, A.; Proietti, G.; Rupert, J.; Tartaglia, G.G. Catrapid Omics V2.0: Going Deeper and Wider in the Prediction of Protein-Rna Interactions. Nucleic Acids Res. 2021, 49, W72–W79. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The Hnrnp Family: Insights into Their Role in Health and Disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Atianand, M.K.; Hu, W.; Satpathy, A.T.; Shen, Y.; Ricci, E.P.; Alvarez-Dominguez, J.R.; Bhatta, A.; Schattgen, S.A.; McGowan, J.D.; Blin, J.; et al. A Long Noncoding Rna Lincrna-Eps Acts as a Transcriptional Brake to Restrain Inflammation. Cell 2016, 165, 1672–1685. [Google Scholar] [CrossRef]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B.; et al. A Long Noncoding Rna Mediates Both Activation and Repression of Immune Response Genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y. The Novel Regulatory Role of Lncrna-Mirna-Mrna Axis in Cardiovascular Diseases. J. Cell Mol. Med. 2018, 22, 5768–5775. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of Functional Microrna Targets by Integrative Modeling of Microrna Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Wang, C.; Li, X.; Zhang, L.; Chen, Y.; Dong, R.; Zhang, J.; Zhao, J.; Guo, X.; Yang, G.; Li, Y.; et al. Mir-194-5p down-Regulates Tumor Cell Pd-L1 Expression and Promotes Anti-Tumor Immunity in Pancreatic Cancer. Int. Immunopharmacol. 2021, 97, 107822. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, X.; Yang, X.; Zhang, R.; Chen, J.; Wang, X. Mirna Sequencing Identifies Immune-Associated Mirnas and Highlights the Role of Mir-193b-5p in Sepsis and Septic Shock Progression. Sci. Rep. 2025, 15, 5323. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Lai, H.F.; Huang, T.C.; Chen, Y.G.; Ye, R.H.; Chang, P.Y.; Lai, S.W.; Chen, Y.C.; Lee, C.H.; Liu, W.N.; et al. Aberrantly Reduced Expression of Mir-342-5p Contributes to Ccnd1-Associated Chronic Myeloid Leukemia Progression and Imatinib Resistance. Cell Death Dis. 2021, 12, 908. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ji, L.; Yu, C.; Chen, Q.; Ge, Q.; Lu, Y. Mir-423-5p Regulates Cells Apoptosis and Extracellular Matrix Degradation Via Nucleotide-Binding, Leucine-Rich Repeat Containing X1 (Nlrx1) in Interleukin 1 Beta (Il-1beta)-Induced Human Nucleus Pulposus Cells. Med. Sci. Monit. 2020, 26, e922497. [Google Scholar] [CrossRef]
- Tang, X.; Zeng, X.; Huang, Y.; Chen, S.; Lin, F.; Yang, G.; Yang, N. Mir-423-5p Serves as a Diagnostic Indicator and Inhibits the Proliferation and Invasion of Ovarian Cancer. Exp. Ther. Med. 2018, 15, 4723–4730. [Google Scholar] [CrossRef]
- Hayakawa, K.; Kawasaki, M.; Hirai, T.; Yoshida, Y.; Tsushima, H.; Fujishiro, M.; Ikeda, K.; Morimoto, S.; Takamori, K.; Sekigawa, I. Microrna-766-3p Contributes to Anti-Inflammatory Responses through the Indirect Inhibition of Nf-Kappab Signaling. Int. J. Mol. Sci. 2019, 20, 809. [Google Scholar] [CrossRef]
- Gao, J.; Fei, L.; Wu, X.; Li, H. Mir-766-3p Suppresses Malignant Behaviors and Stimulates Apoptosis of Colon Cancer Cells Via Targeting Tgfbi. Can. J. Gastroenterol. Hepatol. 2022, 2022, 7234704. [Google Scholar] [CrossRef] [PubMed]
- Chowdhari, S.; Saini, N. Hsa-Mir-4516 Mediated Downregulation of Stat3/Cdk6/Ube2n Plays a Role in Puva Induced Apoptosis in Keratinocytes. J. Cell Physiol. 2014, 229, 1630–1638. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. Rna-Based Therapeutics: An Overview and Prospectus. Cell Death Dis. 2022, 13, 644. [Google Scholar] [CrossRef]
- Collotta, D.; Bertocchi, I.; Chiapello, E.; Collino, M. Antisense Oligonucleotides: A Novel Frontier in Pharmacological Strategy. Front. Pharmacol. 2023, 14, 1304342. [Google Scholar] [CrossRef]
- Scharner, J.; Aznarez, I. Clinical Applications of Single-Stranded Oligonucleotides: Current Landscape of Approved and in-Development Therapeutics. Mol. Ther. 2021, 29, 540–554. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The Current Landscape of Nucleic Acid Therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Kasuya, T.; Hori, S.; Watanabe, A.; Nakajima, M.; Gahara, Y.; Rokushima, M.; Yanagimoto, T.; Kugimiya, A. Ribonuclease H1-Dependent Hepatotoxicity Caused by Locked Nucleic Acid-Modified Gapmer Antisense Oligonucleotides. Sci. Rep. 2016, 6, 30377. [Google Scholar] [CrossRef]
- Crooke, S.T.; Baker, B.F.; Crooke, R.M.; Liang, X.H. Antisense Technology: An Overview and Prospectus. Nat. Rev. Drug Discov. 2021, 20, 427–453. [Google Scholar] [CrossRef] [PubMed]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gourvest, M.; van Solingen, C. CARINH, an Interferon-Induced LncRNA in Cancer and Inflammation. Non-Coding RNA 2025, 11, 79. https://doi.org/10.3390/ncrna11060079
Gourvest M, van Solingen C. CARINH, an Interferon-Induced LncRNA in Cancer and Inflammation. Non-Coding RNA. 2025; 11(6):79. https://doi.org/10.3390/ncrna11060079
Chicago/Turabian StyleGourvest, Morgane, and Coen van Solingen. 2025. "CARINH, an Interferon-Induced LncRNA in Cancer and Inflammation" Non-Coding RNA 11, no. 6: 79. https://doi.org/10.3390/ncrna11060079
APA StyleGourvest, M., & van Solingen, C. (2025). CARINH, an Interferon-Induced LncRNA in Cancer and Inflammation. Non-Coding RNA, 11(6), 79. https://doi.org/10.3390/ncrna11060079





