Human-Specific Suppression of Hepatic Fatty Acid Catabolism by RNA-Binding Protein HuR
Abstract
1. Introduction
2. Results
2.1. Human-Specific Suppression of Fatty Acid Catabolism Gene Expression by HuR
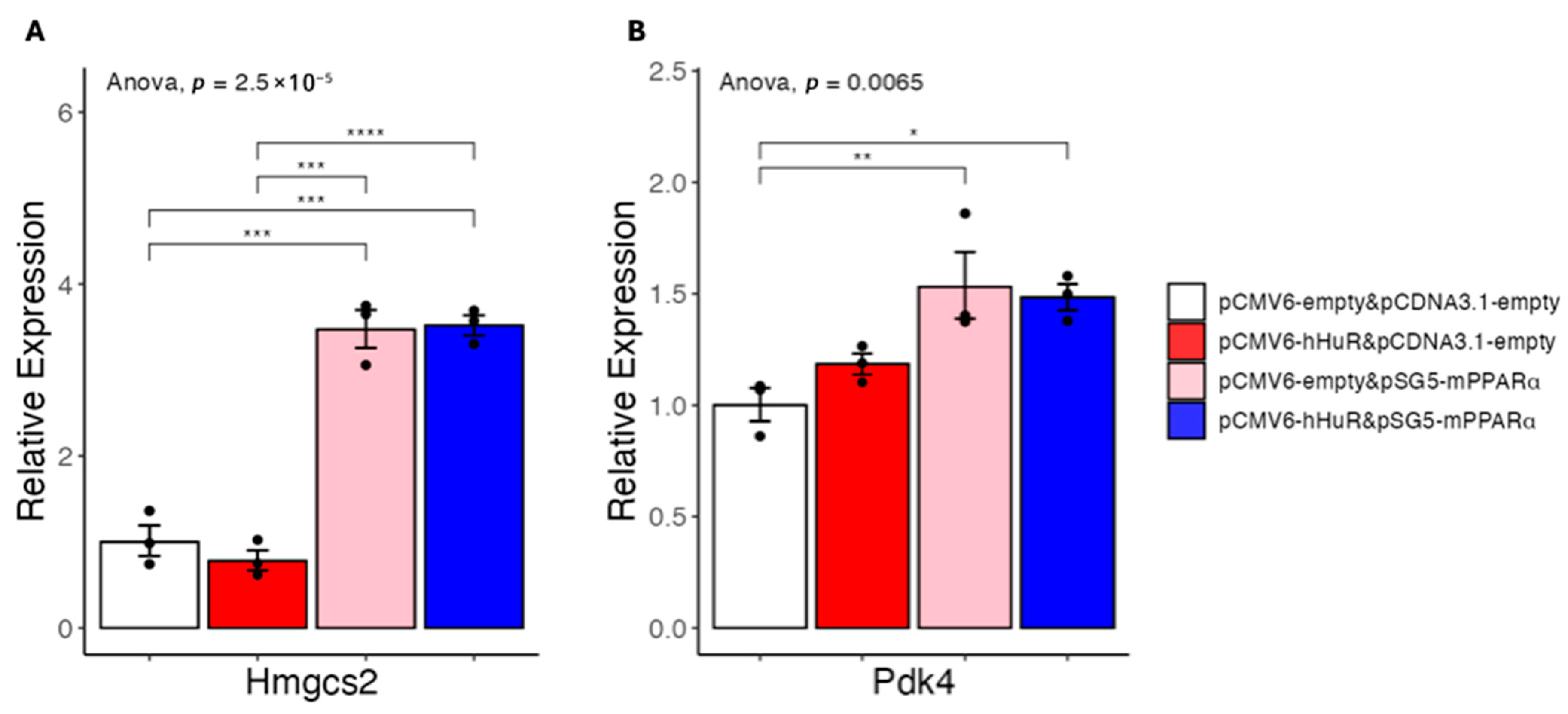
2.2. HuR Suppresses Fatty Acid Catabolism Independent of PPARα
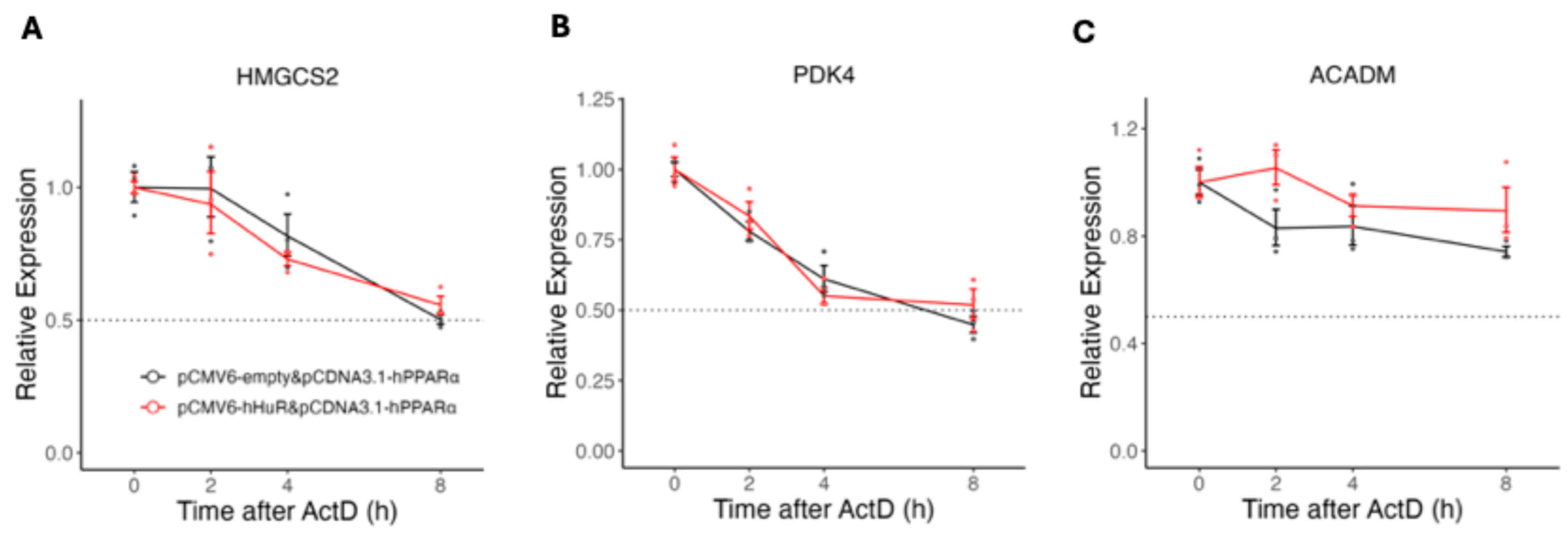
2.3. Human-Specific Role of HuR Is Independent of mRNA Stability or Promoter Activity
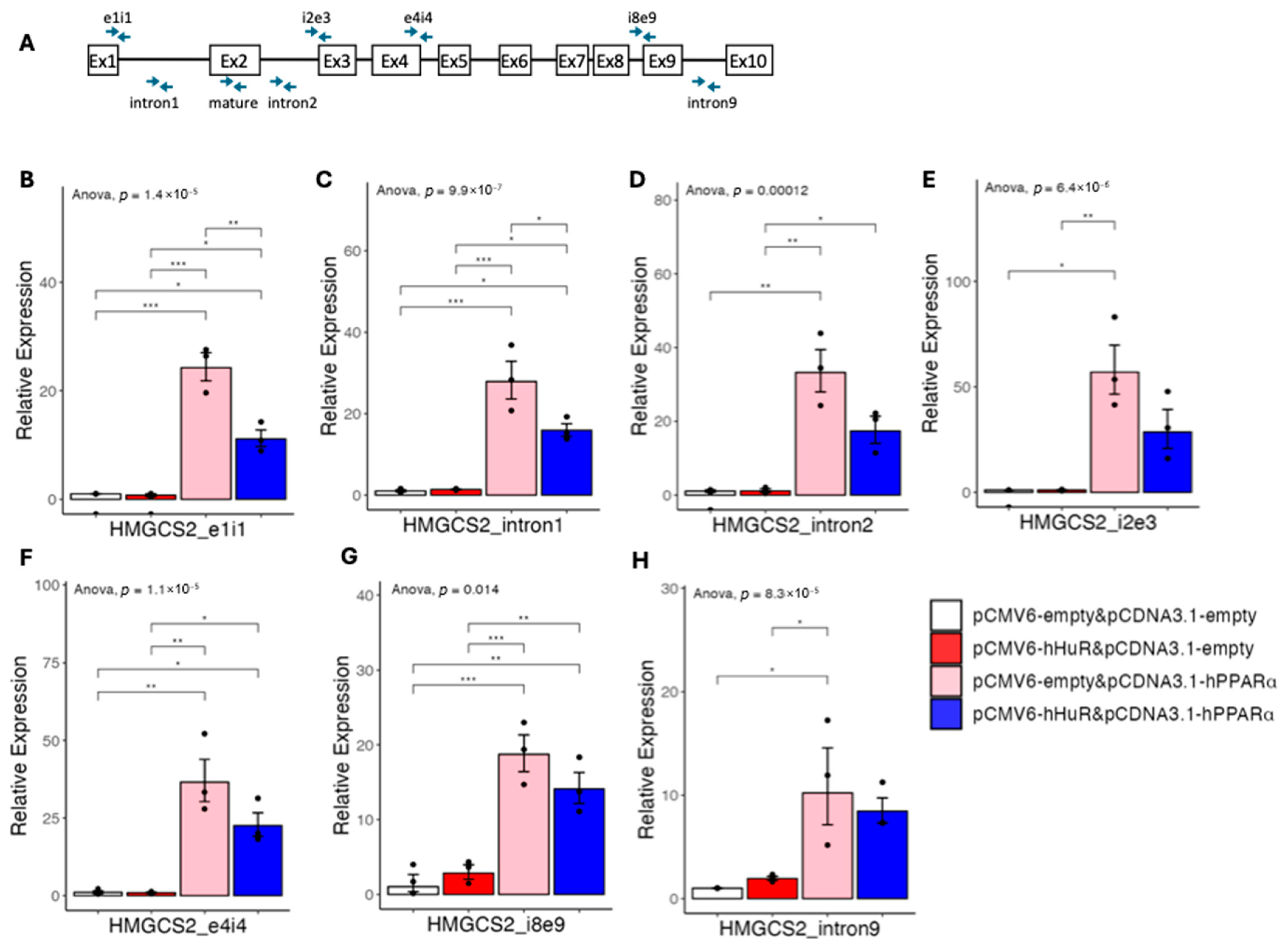
2.4. HuR Blocks Pre-mRNA Processing of PPARα Target Genes
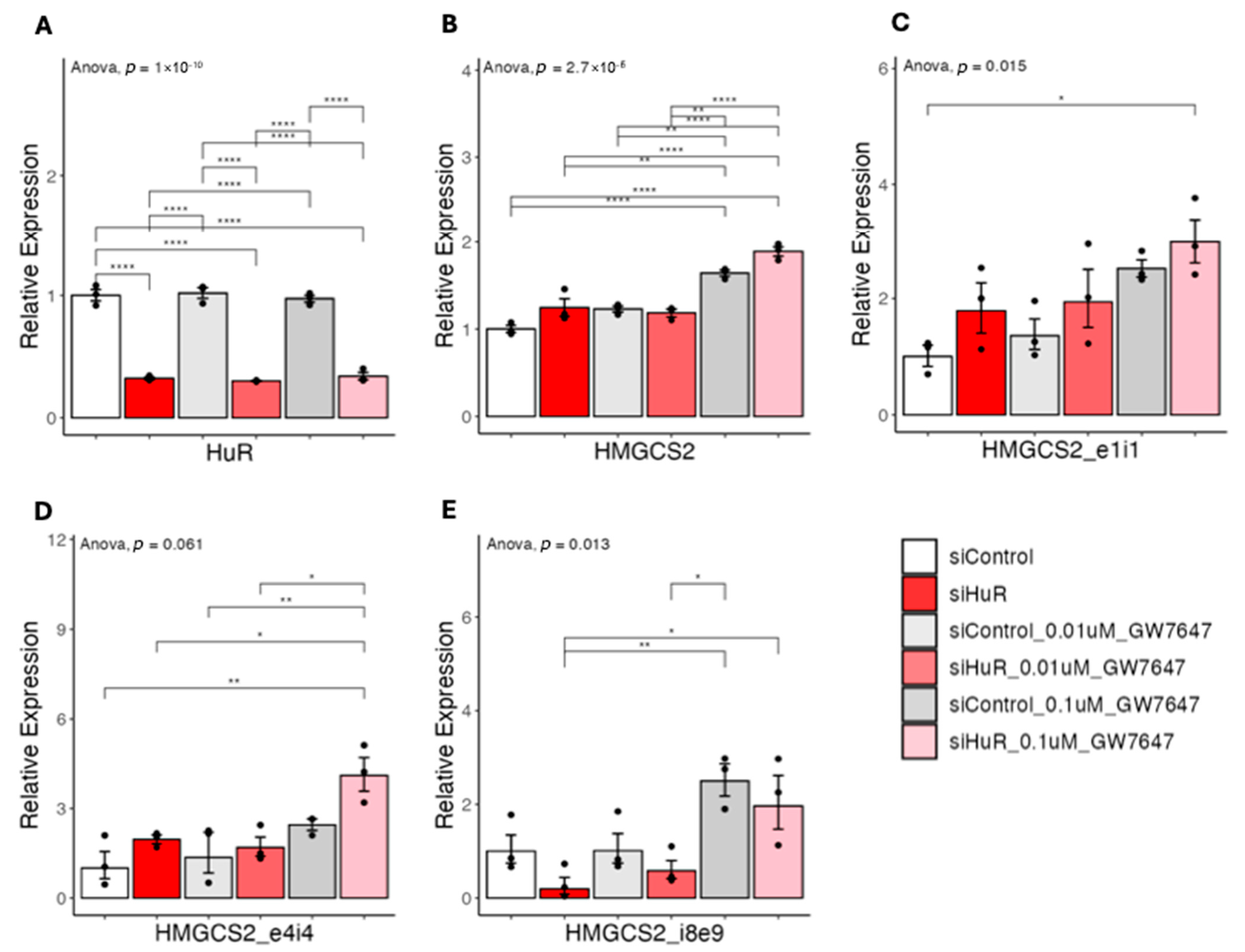
2.5. HuR Suppresses PPARα Agonist-Induced Gene Expression
2.6. Accumulation of HMGCS2/PDK4 Pre-mRNAs in Human Metabolic Dysfunction-Associated Steatohepatitis
3. Discussion
4. Methods
4.1. Cell Culture
4.2. Actinomycin D Treatment
4.3. GW7647 Treatment
4.4. RNA Extraction and qPCR
4.5. Protein Extraction, Cell Fractionation, and Western Blotting
4.6. Luciferase Reporter Assay
4.7. Bioinformatics & Bulk RNA-Seq Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Brennan, C.M.; Steitz, J.A. HuR and mRNA stability. Cell Mol. Life Sci. 2001, 58, 266–277. [Google Scholar] [CrossRef]
- Lebedeva, S.; Jens, M.; Theil, K.; Schwanhausser, B.; Selbach, M.; Landthaler, M.; Rajewsky, N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 2011, 43, 340–352. [Google Scholar] [CrossRef]
- Zhang, Z.; Zong, C.; Jiang, M.; Hu, H.; Cheng, X.; Ni, J.; Yi, X.; Jiang, B.; Tian, F.; Chang, M.W.; et al. Hepatic HuR modulates lipid homeostasis in response to high-fat diet. Nat. Commun. 2020, 11, 3067. [Google Scholar] [CrossRef]
- Subramanian, P.; Gargani, S.; Palladini, A.; Chatzimike, M.; Grzybek, M.; Peitzsch, M.; Papanastasiou, A.D.; Pyrina, I.; Ntafis, V.; Gercken, B.; et al. The RNA binding protein human antigen R is a gatekeeper of liver homeostasis. Hepatology 2022, 75, 881–897. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tai, Y.L.; Way, G.; Zeng, J.; Zhao, D.; Su, L.; Jiang, X.; Jackson, K.G.; Wang, X.; Gurley, E.C.; et al. RNA binding protein HuR protects against NAFLD by suppressing long noncoding RNA H19 expression. Cell Biosci. 2022, 12, 172. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Wang, J.; Liu, S.; Li, X.; Li, J.; Yang, J.; Zhang, C.; Zhang, W. Hepatic HuR protects against the pathogenesis of non-alcoholic fatty liver disease by targeting PTEN. Cell Death Dis. 2021, 12, 236. [Google Scholar] [CrossRef]
- Jiang, C.; Li, P.; Ruan, X.; Ma, Y.; Kawai, K.; Suemizu, H.; Cao, H. Comparative Transcriptomics Analyses in Livers of Mice, Humans, and Humanized Mice Define Human-Specific Gene Networks. Cells 2020, 9, 2566. [Google Scholar] [CrossRef]
- Hsu, M.H.; Savas, U.; Griffin, K.J.; Johnson, E.F. Identification of peroxisome proliferator-responsive human genes by elevated expression of the peroxisome proliferator-activated receptor alpha in HepG2 cells. J. Biol. Chem. 2001, 276, 27950–27958. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; He, X.; Gao, X.; Liu, Q.; Zhao, Y.; Hong, Y.; Zhu, W.; Yan, J.; Li, Y.; Li, Y.; et al. Hyodeoxycholic acid ameliorates nonalcoholic fatty liver disease by inhibiting RAN-mediated PPARalpha nucleus-cytoplasm shuttling. Nat. Commun. 2023, 14, 5451. [Google Scholar] [CrossRef]
- Fan, X.C.; Steitz, J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 1998, 95, 15293–15298. [Google Scholar] [CrossRef]
- Kim, J.B.; Wright, H.M.; Wright, M.; Spiegelman, B.M. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. USA 1998, 95, 4333–4337. [Google Scholar] [CrossRef]
- Rosonina, E.; Blencowe, B.J. Gene expression: The close coupling of transcription and splicing. Curr. Biol. 2002, 12, R319–R321. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Tarn, W.Y. Coupling pre-mRNA processing to transcription on the RNA factory assembly line. RNA Biol. 2013, 10, 380–390. [Google Scholar] [CrossRef]
- Zhu, H.; Berkova, Z.; Mathur, R.; Sehgal, L.; Khashab, T.; Tao, R.H.; Ao, X.; Feng, L.; Sabichi, A.L.; Blechacz, B.; et al. HuR Suppresses Fas Expression and Correlates with Patient Outcome in Liver Cancer. Mol. Cancer Res. 2015, 13, 809–818. [Google Scholar] [CrossRef]
- Suppli, M.P.; Rigbolt, K.T.G.; Veidal, S.S.; Heeboll, S.; Eriksen, P.L.; Demant, M.; Bagger, J.I.; Nielsen, J.C.; Oro, D.; Thrane, S.W.; et al. Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 316, G462–G472. [Google Scholar] [CrossRef]
- Mooli, R.G.R.; Ramakrishnan, S.K. Emerging Role of Hepatic Ketogenesis in Fatty Liver Disease. Front. Physiol. 2022, 13, 946474. [Google Scholar] [CrossRef]
- Pradas-Juni, M.; Hansmeier, N.R.; Link, J.C.; Schmidt, E.; Larsen, B.D.; Klemm, P.; Meola, N.; Topel, H.; Loureiro, R.; Dhaouadi, I.; et al. A MAFG-lncRNA axis links systemic nutrient abundance to hepatic glucose metabolism. Nat. Commun. 2020, 11, 644. [Google Scholar] [CrossRef]
- Tateno, C.; Yamamoto, T.; Utoh, R.; Yamasaki, C.; Ishida, Y.; Myoken, Y.; Oofusa, K.; Okada, M.; Tsutsui, N.; Yoshizato, K. Chimeric mice with hepatocyte-humanized liver as an appropriate model to study human peroxisome proliferator-activated receptor-alpha. Toxicol. Pathol. 2015, 43, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Staels, B.; Butruille, L.; Francque, S. Treating NASH by targeting peroxisome proliferator-activated receptors. J. Hepatol. 2023, 79, 1302–1316. [Google Scholar] [CrossRef] [PubMed]
- Belfort, R.; Berria, R.; Cornell, J.; Cusi, K. Fenofibrate reduces systemic inflammation markers independent of its effects on lipid and glucose metabolism in patients with the metabolic syndrome. J. Clin. Endocrinol. Metab. 2010, 95, 829–836. [Google Scholar] [CrossRef]
- Abdelmegeed, M.A.; Yoo, S.H.; Henderson, L.E.; Gonzalez, F.J.; Woodcroft, K.J.; Song, B.J. PPARalpha expression protects male mice from high fat-induced nonalcoholic fatty liver. J. Nutr. 2011, 141, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, A.; Moallem, S.A.; Johnston, T.P.; Sahebkar, A. Liver Protective Effect of Fenofibrate in NASH/NAFLD Animal Models. PPAR Res. 2022, 2022, 5805398. [Google Scholar] [CrossRef]
- Regnier, M.; Polizzi, A.; Smati, S.; Lukowicz, C.; Fougerat, A.; Lippi, Y.; Fouche, E.; Lasserre, F.; Naylies, C.; Betoulieres, C.; et al. Hepatocyte-specific deletion of Pparalpha promotes NAFLD in the context of obesity. Sci. Rep. 2020, 10, 6489. [Google Scholar] [CrossRef]
- Baldini, L.; Labialle, S. Using Native RIP, UV-CLIP or fCLIP to Address Protein-RNA Interactions In Vivo. Methods Mol. Biol. 2021, 2300, 89–98. [Google Scholar] [PubMed]
- Suzuki, K.; Bose, P.; Leong-Quong, R.Y.; Fujita, D.J.; Riabowol, K. REAP: A two minute cell fractionation method. BMC Res. Notes 2010, 3, 294. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integrative Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Zhang, A.Y.; Su, S.; Ng, A.P.; Holik, A.Z.; Asselin-Labat, M.L.; Ritchie, M.E.; Law, C.W. Covering all your bases: Incorporating intron signal from RNA-seq data. NAR Genom. Bioinform. 2020, 2, lqaa073. [Google Scholar] [CrossRef]








| Gene_Name | Description | Log2FC | p-Value |
|---|---|---|---|
| ACAA2 | acetyl-CoA acyltransferase 2 | 0.607 | 1.13 × 10−5 |
| ACADM | acyl-CoA dehydrogenase medium chain | 0.654 | 1.48 × 10−4 |
| ACOT2 | acyl-CoA thioesterase 2 | 0.681 | 4.64 × 10−4 |
| ACSL1 | acyl-CoA synthetase long chain family member 1 | 0.912 | 1.05 × 10−8 |
| ACSL5 | acyl-CoA synthetase long chain family member 5 | 1.113 | 4.77 × 10−8 |
| HMGCS2 | 3-hydroxy-3-methylglutaryl-CoA synthase 2 | 0.784 | 1.18 × 10−9 |
| PDK4 | pyruvate dehydrogenase kinase 4 | 0.482 | 2.49 × 10−3 |
| SLC27A2 | solute carrier family 27 member 2 | 0.672 | 1.17 × 10−3 |
| SLC27A5 | solute carrier family 27 member 5 | 1.264 | 3.91 × 10−6 |
| Primer Name | Primer Sequence |
|---|---|
| GAPDH-f | ATGGGTGTGAACCATGAGAA |
| GAPDH-r | GTGCTAAGCAGTTGGTGGTG |
| HMGCS2-f | CAGTCCAAGAGGACATCAACTC |
| HMGCS2-r | CAGTGCCTACTTCCAGCCTG |
| PDK4-f | GGAAGCATTGATCCTAACTGTGA |
| PDK4-r | GGTGAGAAGGAACATACACGATG |
| ACADM-f | ACAGGGGTTCAGACTGCTATT |
| ACADM-r | TCCTCCGTTGGTTATCCACAT |
| HuR-f | GCCGTCACCAATGTGAAAGT |
| HuR-r | CCATCGCGGCTTCTTCATAG |
| PPARα-f | ATGGTGGACACGGAAAGCC |
| PPARα-r | CGATGGATTGCGAAATCTCTTGG |
| β-Actin-f | CATGTACGTTGCTATCCAGGC |
| β-Actin-r | CTCCTTAATGTCACGCACGAT |
| HMGCS2_e1i1_f | GTGAAGCGCATTCTGCAACT |
| HMGCS2_e1i1_r | ACAAGGTGCTTCTCAGAAAGTGA |
| HMGCS2_i2e3_f | GTACACAGTAGGCACTCAGTCT |
| HMGCS2_i2e3_r | ACGAGCATTACCACTGGGAT |
| HMGCS2_e4i4_f | CCATCCAGTGCTACTTGCGG |
| HMGCS2_e4i4_r | GGGTACAAACCCTTGGCCTAT |
| HMGCS2_i8e9_f | TTTGTGAGTACTGCCTCTTCTCC |
| HMGCS2_i8e9_r | CGGGCATACTTTCGGCGATG |
| HMGCS2-intron1-f | TGTGTTTGTGAGCAATGGCA |
| HMGCS2-intron1-r | AGGTGGGAATGGTGGTGTAG |
| HMGCS2-intron2-f | GAATTGTCAAGCCTGTGGGA |
| HMGCS2-intron2-r | AAGGCTTTGGCTTCATGCTC |
| HMGCS2-intron9-f | GCAGCCCAGATGTTGTTAGG |
| HMGCS2-intron9-r | GCTAACTGAGCTCGTCCTCT |
| mGapdh-f | AGGTCGGTGTGAACGGATTTG |
| mGapdh-r | GGGGTCGTTGATGGCAACA |
| mHmgcs2_f | AGAGAGCGATGCAGGAAACTT |
| mHmgcs2_r | AAGGATGCCCACATCTTTTGG |
| mPdk4-f1 | TTGGGAGGCTGAAGGGTAAG |
| mPdk4-r1 | CAGGAAAGGCACAGAGCATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takaoka, S.; Jaso-Vera, M.E.; Ruan, X. Human-Specific Suppression of Hepatic Fatty Acid Catabolism by RNA-Binding Protein HuR. Non-Coding RNA 2025, 11, 65. https://doi.org/10.3390/ncrna11050065
Takaoka S, Jaso-Vera ME, Ruan X. Human-Specific Suppression of Hepatic Fatty Acid Catabolism by RNA-Binding Protein HuR. Non-Coding RNA. 2025; 11(5):65. https://doi.org/10.3390/ncrna11050065
Chicago/Turabian StyleTakaoka, Shohei, Marcos E. Jaso-Vera, and Xiangbo Ruan. 2025. "Human-Specific Suppression of Hepatic Fatty Acid Catabolism by RNA-Binding Protein HuR" Non-Coding RNA 11, no. 5: 65. https://doi.org/10.3390/ncrna11050065
APA StyleTakaoka, S., Jaso-Vera, M. E., & Ruan, X. (2025). Human-Specific Suppression of Hepatic Fatty Acid Catabolism by RNA-Binding Protein HuR. Non-Coding RNA, 11(5), 65. https://doi.org/10.3390/ncrna11050065






