Tiny but Mighty: Small RNAs—The Micromanagers of Bacterial Survival, Virulence, and Host–Pathogen Interactions
Abstract
1. Introduction
2. sRNA as Orchestrators of Bacterial Pathogenesis
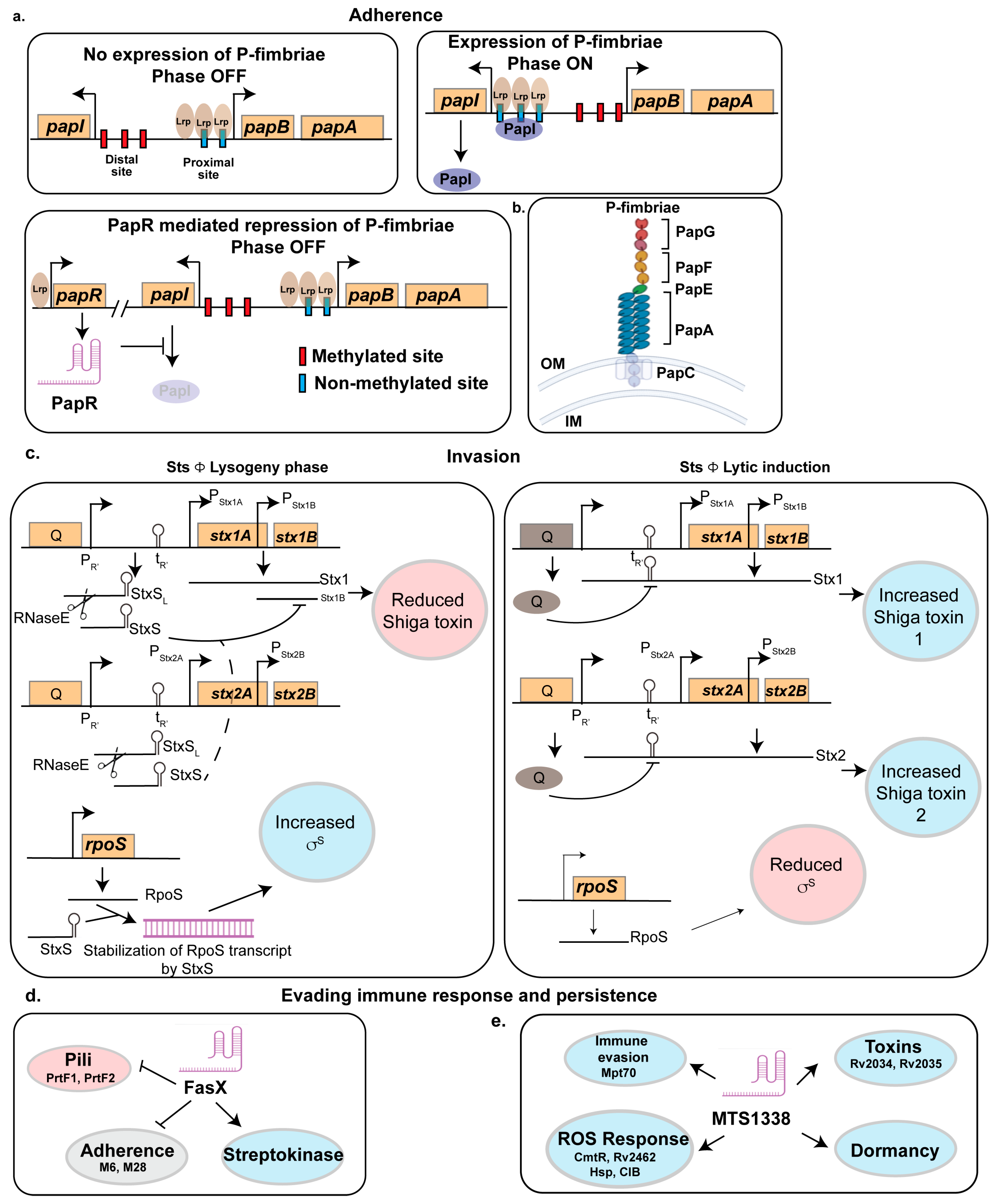
2.1. Adherence to the Host
2.1.1. PapR
2.1.2. RyfA
2.1.3. LhrC
2.1.4. STnc640
2.2. Invasion of the Host
2.2.1. StxS
2.2.2. RNA III
2.2.3. BtsR1
2.3. Evading Immune Response
2.3.1. FasX
2.3.2. MTS1338
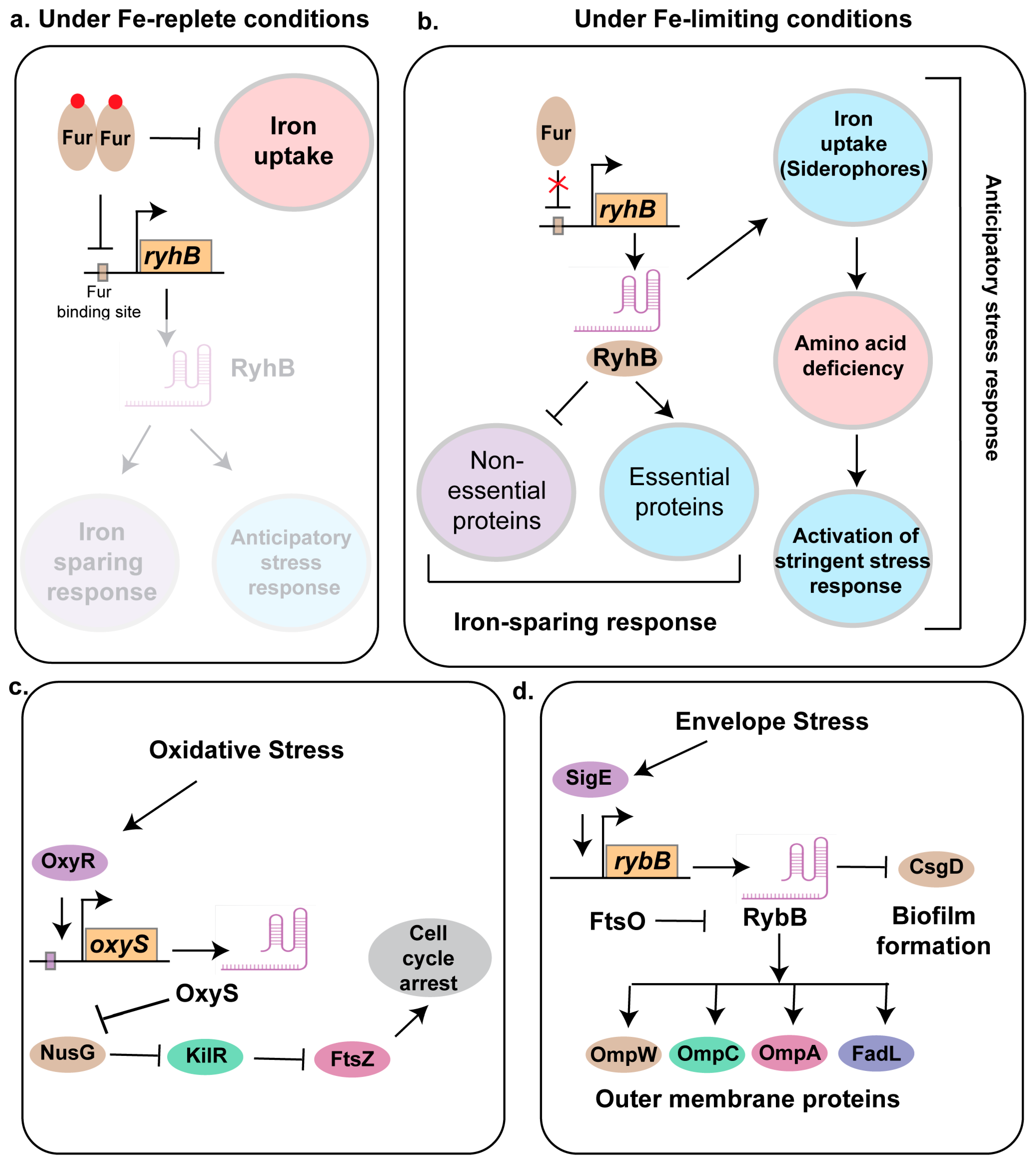
3. Role of Small RNA in Stress Response
3.1. RyhB
3.2. MsrI and Mcr11
3.3. OxyS
3.4. DsrA
3.5. RybB
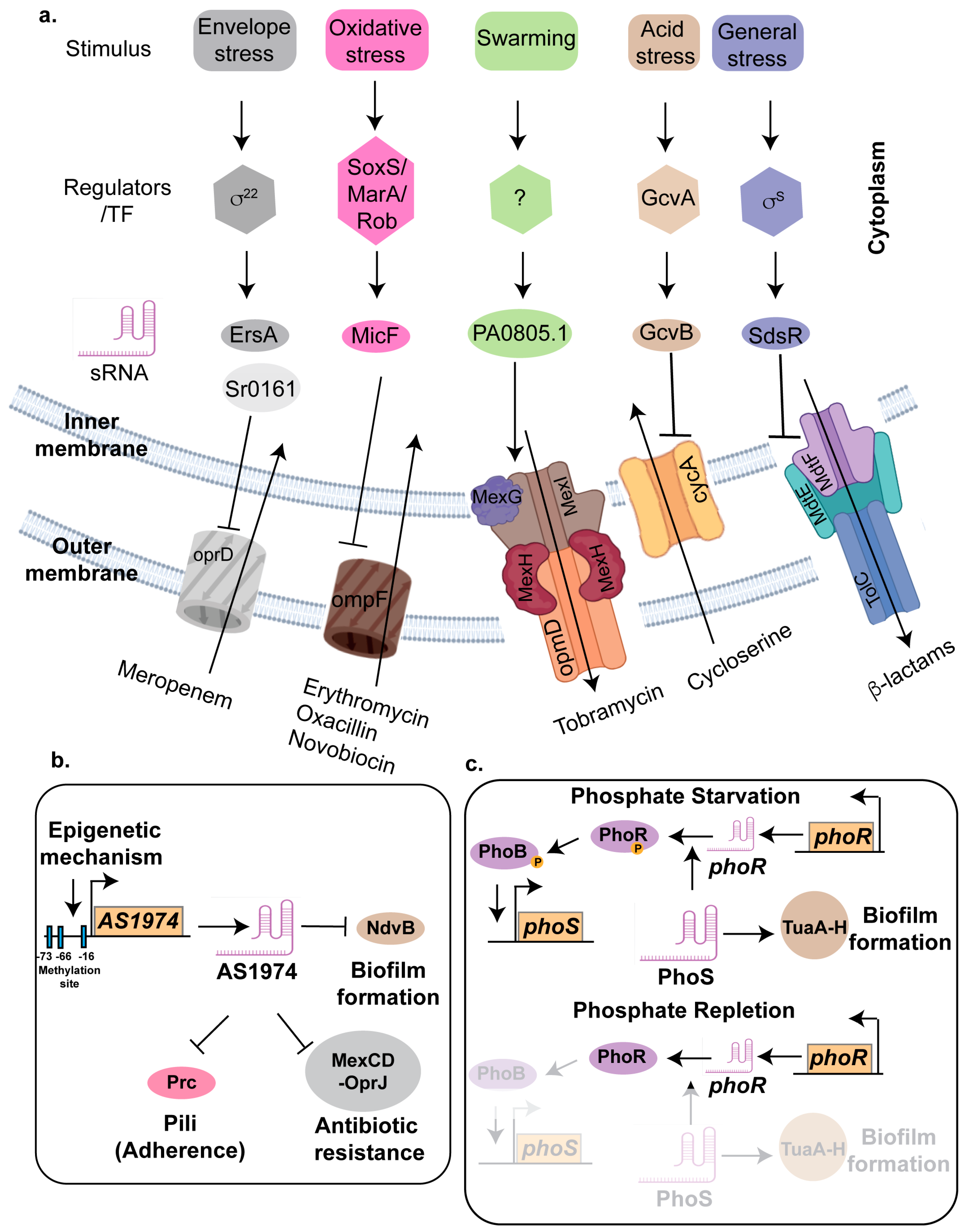
4. Role of sRNA in Antibiotic Resistance
4.1. Inhibiting Drug Uptake Systems
4.1.1. GcvB
4.1.2. MicF
4.1.3. EsrA and Sr0161
4.2. Modification of Cell Envelope
4.2.1. SprX
4.2.2. Sr006
4.3. Activation of Efflux Pumps
4.3.1. CsiR
4.3.2. PA0805.1
4.3.3. SdsR and RyeA
4.4. Biofilm-Mediated Increased Drug Resistance in Bacterial Pathogens
4.4.1. AS1974
4.4.2. PhoS
4.4.3. SrbA
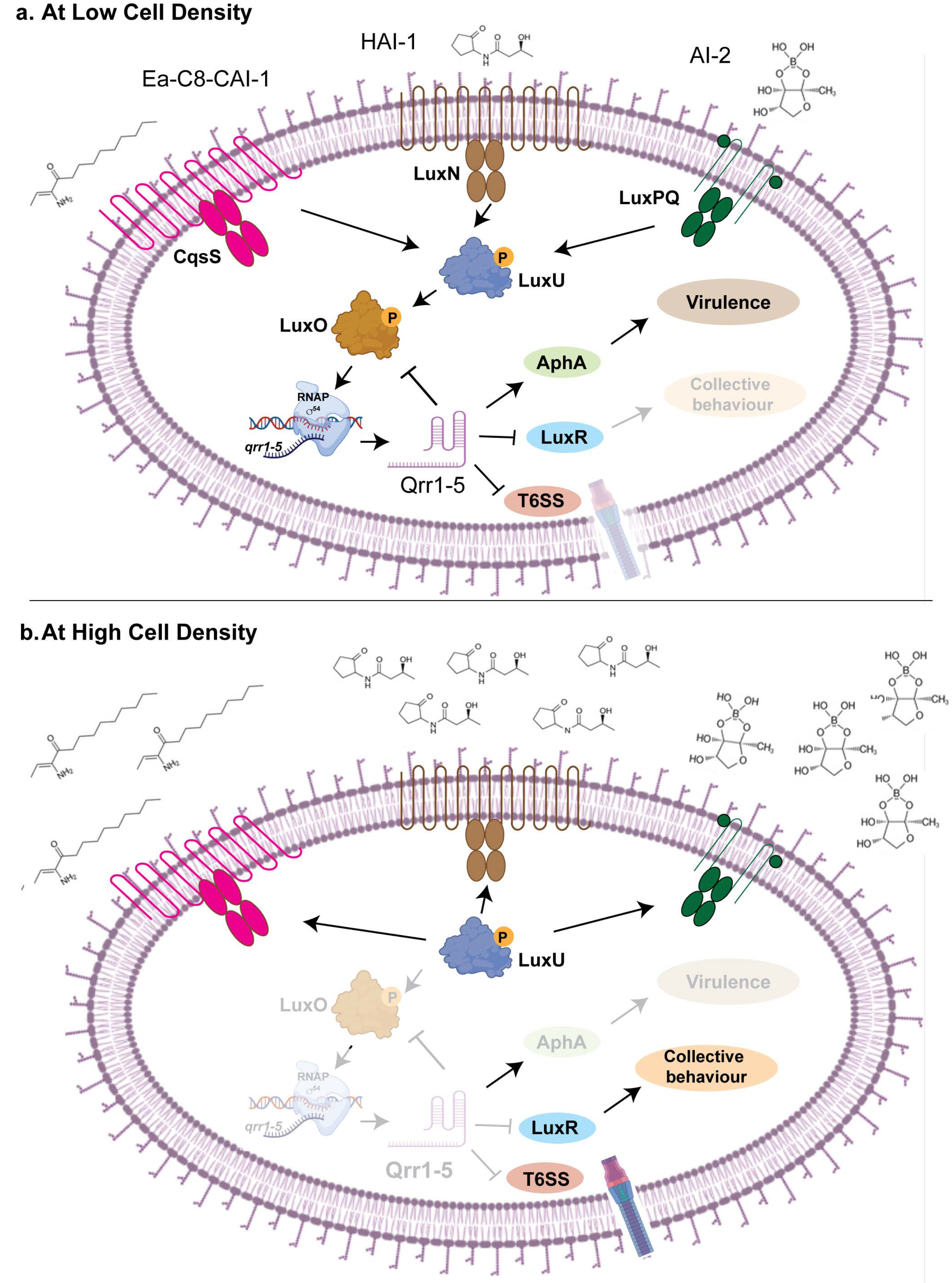
5. Role of sRNA in Quorum Sensing
5.1. Qrr 1-5
5.2. PhrS and ReaL
5.3. AmiL and PqsS
5.4. SprD
6. Discussion
Supplementary Materials
Funding
Data Availability Statement
Conflicts of Interest
References
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, S.; Kim, K.-S. Host immunomodulation strategies to combat pandemic-associated antimicrobial-resistant secondary bacterial infections. Int. J. Antimicrob. Agents 2024, 64, 107308. [Google Scholar] [CrossRef] [PubMed]
- Afshar, M.; Gallo, R.L. Innate immune defense system of the skin. Vet. Dermatol. 2013, 24, 32-e9. [Google Scholar] [CrossRef]
- Xuan, J.; Feng, W.; Wang, J.; Wang, R.; Zhang, B.; Bo, L.; Chen, Z.-S.; Yang, H.; Sun, L. Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resist. Updates 2023, 68, 100954. [Google Scholar] [CrossRef] [PubMed]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef]
- Dlozi, P.N.; Gladchuk, A.; Crutchley, R.D.; Keuler, N.; Coetzee, R.; Dube, A. Cathelicidins and defensins antimicrobial host defense peptides in the treatment of TB and HIV: Pharmacogenomic and nanomedicine approaches towards improved therapeutic outcomes. Biomed. Pharmacother. 2022, 151, 113189. [Google Scholar] [CrossRef]
- Lewis, L.A.; Ram, S. Meningococcal disease and the complement system. Virulence 2014, 5, 98–126. [Google Scholar] [CrossRef]
- Subashchandrabose, S.; Mobley, H.L.T. Virulence and Fitness Determinants of Uropathogenic Escherichia coli. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Murdoch, C.C.; Skaar, E.P. Nutritional immunity: The battle for nutrient metals at the host-pathogen interface. Nat. Rev. Microbiol. 2022, 20, 657–670. [Google Scholar] [CrossRef]
- Bengoechea, J.A.; Sa Pessoa, J. Klebsiella pneumoniae infection biology: Living to counteract host defences. FEMS Microbiol. Rev. 2018, 43, 123–144. [Google Scholar] [CrossRef]
- Cooper, A.M. Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 2009, 27, 393–422. [Google Scholar] [CrossRef]
- Hornef, M.W.; Wick, M.J.; Rhen, M.; Normark, S. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 2002, 3, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Ribet, D.; Cossart, P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015, 17, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.B.; McFadden, G. Anti-Immunology: Evasion of the Host Immune System by Bacterial and Viral Pathogens. Cell 2006, 124, 767–782. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Sun, J.; Deng, Z.; Yan, A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, VMBF-0016-2015. [Google Scholar] [CrossRef]
- Raina, M.; King, A.; Bianco, C.; Vanderpool, C.K. Dual-Function RNAs. Microbiol. Spectr. 2018, 6, rwr-0032-2018. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011, 3, a003798. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Svenningsen, S.L. Small RNA-Based Regulation of Bacterial Quorum Sensing and Biofilm Formation. Microbiol. Spectr. 2018, 6, rwr-0017-2018. [Google Scholar] [CrossRef]
- Lenz, D.H.; Mok, K.C.; Lilley, B.N.; Kulkarni, R.V.; Wingreen, N.S.; Bassler, B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 2004, 118, 69–82. [Google Scholar] [CrossRef]
- Sy, B.M.; Tree, J.J. Small RNA Regulation of Virulence in Pathogenic Escherichia coli. Front. Cell Infect. Microbiol. 2020, 10, 622202. [Google Scholar] [CrossRef]
- Li, W.; Ying, X.; Lu, Q.; Chen, L. Predicting sRNAs and Their Targets in Bacteria. Genom. Proteom. Bioinform. 2012, 10, 276–284. [Google Scholar] [CrossRef]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef]
- Grešová, K.; Alexiou, P.; Giassa, I.C. Small RNA Targets: Advances in Prediction Tools and High-Throughput Profiling. Biology 2022, 11, 1798. [Google Scholar] [CrossRef]
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vázquez-Torres, A. Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20, 133–143. [Google Scholar] [CrossRef]
- Everest, P. Stress and bacteria: Microbial endocrinology. Gut 2007, 56, 1037–1038. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 2019, 294, 11685–11700. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, S.; Banerjee, S.K.; Banerjee, R.; Mukhopadhyay, J.; Kundu, M. Polyphosphate kinase 1, a central node in the stress response network of Mycobacterium tuberculosis, connects the two-component systems MprAB and SenX3-RegX3 and the extracytoplasmic function sigma factor, sigma E. Microbiology 2013, 159, 2074–2086. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Chatterjee, A.; Gupta, S.; Banerjee, R.; Mandal, S.; Mukhopadhyay, J.; Basu, J.; Kundu, M. MtrA, an essential response regulator of the MtrAB two-component system, regulates the transcription of resuscitation-promoting factor B of Mycobacterium tuberculosis. Microbiology 2015, 161, 1271–1281. [Google Scholar] [CrossRef]
- Durand, S.; Storz, G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 2010, 75, 1215–1231. [Google Scholar] [CrossRef]
- Papenfort, K.; Storz, G. Insights into bacterial metabolism from small RNAs. Cell Chem. Biol. 2024, 31, 1571–1577. [Google Scholar] [CrossRef]
- Singh, S.; Nirban, R.; Dutta, T. MTS1338 in Mycobacterium tuberculosis promotes detoxification of reactive oxygen species under oxidative stress. Tuberculosis 2021, 131, 102142. [Google Scholar] [CrossRef]
- Villa, J.K.; Han, R.; Tsai, C.-H.; Chen, A.; Sweet, P.; Franco, G.; Vaezian, R.; Tkavc, R.; Daly, M.J.; Contreras, L.M. A small RNA regulates pprM, a modulator of pleiotropic proteins promoting DNA repair, in Deinococcus radiodurans under ionizing radiation. Sci. Rep. 2021, 11, 12949. [Google Scholar] [CrossRef]
- Chambers, J.R.; Sauer, K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013, 21, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Bak, G.; Lee, J.; Suk, S.; Kim, D.; Young Lee, J.; Kim, K.-S.; Choi, B.-S.; Lee, Y. Identification of novel sRNAs involved in biofilm formation, motility and fimbriae formation in Escherichia coli. Sci. Rep. 2015, 5, 15287. [Google Scholar] [CrossRef]
- Sy, B.M.; Lan, R.; Tree, J.J. Early termination of the Shiga toxin transcript generates a regulatory small RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 25055–25065. [Google Scholar] [CrossRef]
- Djapgne, L.; Oglesby, A.G. Impacts of Small RNAs and Their Chaperones on Bacterial Pathogenicity. Front. Cell Infect. Microbiol. 2021, 11, 604511. [Google Scholar] [CrossRef]
- Danger, J.L.; Makthal, N.; Kumaraswami, M.; Sumby, P. The FasX Small Regulatory RNA Negatively Regulates the Expression of Two Fibronectin-Binding Proteins in Group A Streptococcus. J. Bacteriol. 2015, 197, 3720–3730. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Bacterial Priority Pathogens List. 2024. Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 17 May 2024).
- Lopatkin, A.J.; Sysoeva, T.A.; You, L. Dissecting the effects of antibiotics on horizontal gene transfer: Analysis suggests a critical role of selection dynamics. Bioessays 2016, 38, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef]
- Mediati, D.G.; Wu, S.; Wu, W.; Tree, J.J. Networks of Resistance: Small RNA Control of Antibiotic Resistance. Trends Genet. 2021, 37, 35–45. [Google Scholar] [CrossRef]
- Dersch, P.; Khan, M.A.; Mühlen, S.; Görke, B. Roles of Regulatory RNAs for Antibiotic Resistance in Bacteria and Their Potential Value as Novel Drug Targets. Front. Microbiol. 2017, 8, 803. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef]
- Bejerano-Sagie, M.; Xavier, K.B. The role of small RNAs in quorum sensing. Curr. Opin. Microbiol. 2007, 10, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, P.; Wang, R. Small RNA-mediated switch-like regulation in bacterial quorum sensing. IET Syst. Biol. 2013, 7, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Elena, R.C.; Marquez, P.H. The roles of small RNAs: Insights from bacterial quorum sensing. ExRNA 2019, 1, 32. [Google Scholar] [CrossRef]
- Federle, M.J.; Bassler, B.L. Interspecies communication in bacteria. J. Clin. Investig. 2003, 112, 1291–1299. [Google Scholar] [CrossRef]
- Arnold, B.J.; Huang, I.T.; Hanage, W.P. Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Updegrove, T.B.; Shabalina, S.A.; Storz, G. How do base-pairing small RNAs evolve? FEMS Microbiol. Rev. 2015, 39, 379–391. [Google Scholar] [CrossRef]
- Raad, N.; Tandon, D.; Hapfelmeier, S.; Polacek, N. The stationary phase-specific sRNA FimR2 is a multifunctional regulator of bacterial motility, biofilm formation and virulence. Nucleic Acids Res. 2022, 50, 11858–11875. [Google Scholar] [CrossRef]
- Meng, X.; He, M.; Xia, P.; Wang, J.; Wang, H.; Zhu, G. Functions of Small Non-Coding RNAs in Salmonella-Host Interactions. Biology 2022, 11, 1283. [Google Scholar] [CrossRef]
- Coburn, B.; Sekirov, I.; Finlay, B.B. Type III secretion systems and disease. Clin. Microbiol. Rev. 2007, 20, 535–549. [Google Scholar] [CrossRef]
- Cakar, F.; Golubeva, Y.A.; Vanderpool, C.K.; Slauch, J.M. The Small RNA MicC Downregulates hilD Translation To Control the Salmonella Pathogenicity Island 1 Type III Secretion System in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2022, 204, e0037821. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, S.Z.; Kim, K.; Azam, M.S.; Golubeva, Y.A.; Cakar, F.; Slauch, J.M.; Vanderpool, C.K. Small RNAs Activate Salmonella Pathogenicity Island 1 by Modulating mRNA Stability through the hilD mRNA 3′ Untranslated Region. J. Bacteriol. 2023, 205, e0033322. [Google Scholar] [CrossRef]
- Ostrik, A.A.; Azhikina, T.L.; Salina, E.G. Small Noncoding RNAs and Their Role in the Pathogenesis of Mycobacterium tuberculosis Infection. Biochemistry 2021, 86, S109–S119. [Google Scholar] [CrossRef] [PubMed]
- Peñaloza, D.; Acuña, L.G.; Barros, M.J.; Núñez, P.; Montt, F.; Gil, F.; Fuentes, J.A.; Calderón, I.L. The Small RNA RyhB Homologs from Salmonella Typhimurium Restrain the Intracellular Growth and Modulate the SPI-1 Gene Expression within RAW264.7 Macrophages. Microorganisms 2021, 9, 635. [Google Scholar] [CrossRef]
- Melican, K.; Sandoval, R.M.; Kader, A.; Josefsson, L.; Tanner, G.A.; Molitoris, B.A.; Richter-Dahlfors, A. Uropathogenic Escherichia coli P and Type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 2011, 7, e1001298. [Google Scholar] [CrossRef]
- Snyder, J.A.; Haugen, B.J.; Lockatell, C.V.; Maroncle, N.; Hagan, E.C.; Johnson, D.E.; Welch, R.A.; Mobley, H.L.T. Coordinate Expression of Fimbriae in Uropathogenic Escherichia coli. Infect. Immun. 2005, 73, 7588–7596. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.M.; Aberg, A.; Straseviçiene, J.; Emody, L.; Uhlin, B.E.; Balsalobre, C. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 2009, 5, e1000303. [Google Scholar] [CrossRef]
- Miyazaki, J.; Ba-Thein, W.; Kumao, T.; Obata Yasuoka, M.; Akaza, H.; Hayshi, H. Type 1, P and S fimbriae, and afimbrial adhesin I are not essential for uropathogenic Escherichia coli to adhere to and invade bladder epithelial cells. FEMS Immunol. Med. Microbiol. 2002, 33, 23–26. [Google Scholar] [CrossRef]
- Bryan, A.; Roesch, P.; Davis, L.; Moritz, R.; Pellett, S.; Welch, R.A. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 2006, 74, 1072–1083. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Wiles, T.J.; Kulesus, R.R.; Mulvey, M.A. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 2008, 85, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, Z.; Zheng, L.; Gong, Z.; Li, Y.; Jin, Y.; Huang, Y.; Chi, M. Urinary Tract Infections Caused by Uropathogenic Escherichia coli: Mechanisms of Infection and Treatment Options. Int. J. Mol. Sci. 2023, 24, 10537. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.J.; Hultgren, S.J. Sticky fibers and uropathogenesis: Bacterial adhesins in the urinary tract. Future Microbiol. 2006, 1, 75–87. [Google Scholar] [CrossRef]
- Khandige, S.; Kronborg, T.; Uhlin, B.E.; Møller-Jensen, J. sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic Escherichia coli. PLoS Pathog. 2015, 11, e1005109. [Google Scholar] [CrossRef] [PubMed]
- Biggel, M.; Xavier, B.B.; Johnson, J.R.; Nielsen, K.L.; Frimodt-Møller, N.; Matheeussen, V.; Goossens, H.; Moons, P.; Van Puyvelde, S. Horizontally acquired papGII-containing pathogenicity islands underlie the emergence of invasive uropathogenic Escherichia coli lineages. Nat. Commun. 2020, 11, 5968. [Google Scholar] [CrossRef]
- Bien, J.; Sokolova, O.; Bozko, P. Role of Uropathogenic Escherichia coli Virulence Factors in Development of Urinary Tract Infection and Kidney Damage. Int. J. Nephrol. 2012, 2012, 681473. [Google Scholar] [CrossRef]
- Holden, N.J.; Gally, D.L. Switches, cross-talk and memory in Escherichia coli adherence. J. Med. Microbiol. 2004, 53, 585–593. [Google Scholar] [CrossRef]
- Bessaiah, H.; Pokharel, P.; Loucif, H.; Kulbay, M.; Sasseville, C.; Habouria, H.; Houle, S.; Bernier, J.; Massé, É.; Van Grevenynghe, J.; et al. The RyfA small RNA regulates oxidative and osmotic stress responses and virulence in uropathogenic Escherichia coli. PLoS Pathog. 2021, 17, e1009617. [Google Scholar] [CrossRef]
- Fris, M.E.; Broach, W.H.; Klim, S.E.; Coschigano, P.W.; Carroll, R.K.; Caswell, C.C.; Murphy, E.R. Sibling sRNA RyfA1 Influences Shigella dysenteriae Pathogenesis. Genes 2017, 8, 50. [Google Scholar] [CrossRef]
- Sievers, S.; Sternkopf Lillebæk, E.M.; Jacobsen, K.; Lund, A.; Mollerup, M.S.; Nielsen, P.K.; Kallipolitis, B.H. A multicopy sRNA of Listeria monocytogenes regulates expression of the virulence adhesin LapB. Nucleic Acids Res. 2014, 42, 9383–9398. [Google Scholar] [CrossRef]
- Sievers, S.; Lund, A.; Menendez-Gil, P.; Nielsen, A.; Storm Mollerup, M.; Lambert Nielsen, S.; Buch Larsson, P.; Borch-Jensen, J.; Johansson, J.; Kallipolitis, B.H. The multicopy sRNA LhrC controls expression of the oligopeptide-binding protein OppA in Listeria monocytogenes. RNA Biol. 2015, 12, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Meng, X.; Wang, J.; Wang, H.; Zhu, C.; Ni, J.; Zhu, G. Small non-coding RNA STnc640 regulates expression of fimA fimbrial gene and virulence of Salmonella enterica serovar Enteritidis. BMC Vet. Res. 2019, 15, 319. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, B.K.; Mulvey, M.A. The UPEC pore-forming toxin α-hemolysin triggers proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways. Cell Host Microbe 2012, 11, 58–69. [Google Scholar] [CrossRef]
- Ristow, L.C.; Welch, R.A. Hemolysin of uropathogenic Escherichia coli: A cloak or a dagger? Biochim. Biophys. Acta 2016, 1858, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.I.; Lee, A.; Reddy, B.; Muir, A.; Soong, G.; Pitt, A.; Cheung, A.; Prince, A. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 2004, 10, 842–848. [Google Scholar] [CrossRef]
- Carbonetti, N.H. Pertussis toxin and adenylate cyclase toxin: Key virulence factors of Bordetella pertussis and cell biology tools. Future Microbiol. 2010, 5, 455–469. [Google Scholar] [CrossRef]
- Michalska, M.; Wolf, P. Pseudomonas Exotoxin A: Optimized by evolution for effective killing. Front. Microbiol. 2015, 6, 963. [Google Scholar] [CrossRef]
- Di Bella, S.; Ascenzi, P.; Siarakas, S.; Petrosillo, N.; di Masi, A. Clostridium difficile Toxins A and B: Insights into Pathogenic Properties and Extraintestinal Effects. Toxins 2016, 8, 134. [Google Scholar] [CrossRef]
- Melton-Celsa, A.R. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol. Spectr. 2014, 2, Ehec-0024-2013. [Google Scholar] [CrossRef]
- Ruggenenti, P.; Noris, M.; Remuzzi, G. Thrombotic microangiopathy, hemolytic uremic syndrome, and thrombotic thrombocytopenic purpura. Kidney Int. 2001, 60, 831–846. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Haarmann, N.; Schwidder, M.; Muniesa, M.; Schmidt, H. Bacteriophages of Shiga Toxin-Producing Escherichia coli and Their Contribution to Pathogenicity. Pathogens 2021, 10, 404. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.L.; Neely, M.N.; Zhang, X.; Acheson, D.W.; Waldor, M.K.; Friedman, D.I. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 2001, 183, 2081–2085. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.B.; Mekalanos, J.J. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 1987, 169, 4759–4764. [Google Scholar] [CrossRef]
- Melson, E.M.; Kendall, M.M. The sRNA DicF integrates oxygen sensing to enhance enterohemorrhagic Escherichia coli virulence via distinctive RNA control mechanisms. Proc. Natl. Acad. Sci. USA 2019, 116, 14210–14215. [Google Scholar] [CrossRef]
- Wilke, G.A.; Wardenburg, J.B. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus α-hemolysin-mediated cellular injury. Proc. Natl. Acad. Sci. USA 2010, 107, 13473–13478. [Google Scholar] [CrossRef]
- Tam, K.; Torres, V.J. Staphylococcus aureus Secreted Toxins and Extracellular Enzymes. Microbiol. Spectr. 2019, 7, gpp3-0039-2018. [Google Scholar] [CrossRef]
- Le, K.Y.; Otto, M. Quorum-sensing regulation in staphylococci—An overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007, 21, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Luong, T.T.; Lee, C.Y. RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. Proc. Natl. Acad. Sci. USA 2015, 112, 14036–14041. [Google Scholar] [CrossRef]
- Le Huyen, K.B.; Gonzalez, C.D.; Pascreau, G.; Bordeau, V.; Cattoir, V.; Liu, W.; Bouloc, P.; Felden, B.; Chabelskaya, S. A small regulatory RNA alters Staphylococcus aureus virulence by titrating RNAIII activity. Nucleic Acids Res. 2021, 49, 10644–10656. [Google Scholar] [CrossRef]
- Morales-Filloy, H.G.; Zhang, Y.; Nübel, G.; George, S.E.; Korn, N.; Wolz, C.; Jäschke, A. The 5’ NAD Cap of RNAIII Modulates Toxin Production in Staphylococcus aureus Isolates. J. Bacteriol. 2020, 202, JB.00591-19. [Google Scholar] [CrossRef] [PubMed]
- Wiedermannová, J.; Babu, R.; Yuzenkova, Y. Stochastic nature and physiological implications of 5′-NAD RNA cap in bacteria. Nucleic Acids Res. 2024, 52, 11838–11852. [Google Scholar] [CrossRef]
- Peng, D.; Luo, X.; Zhang, N.; Guo, S.; Zheng, J.; Chen, L.; Sun, M. Small RNA-mediated Cry toxin silencing allows Bacillus thuringiensis to evade Caenorhabditis elegans avoidance behavioral defenses. Nucleic Acids Res. 2018, 46, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Porcheron, G.; Habib, R.; Houle, S.; Caza, M.; Lépine, F.; Daigle, F.; Massé, E.; Dozois, C.M. The small RNA RyhB contributes to siderophore production and virulence of uropathogenic Escherichia coli. Infect. Immun. 2014, 82, 5056–5068. [Google Scholar] [CrossRef]
- Meng, X.; Chen, Y.; Wang, P.; He, M.; Shi, Y.; Lai, Y.; Zhu, G.; Wang, H. RyhB in Avian Pathogenic Escherichia coli Regulates the Expression of Virulence-Related Genes and Contributes to Meningitis Development in a Mouse Model. Int. J. Mol. Sci. 2022, 23, 15532. [Google Scholar] [CrossRef]
- Baussier, C.; Oriol, C.; Durand, S.; Py, B.; Mandin, P. Small RNA OxyS induces resistance to aminoglycosides during oxidative stress by controlling Fe-S cluster biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2024, 121, e2317858121. [Google Scholar] [CrossRef] [PubMed]
- Mey, A.R.; Craig, S.A.; Payne, S.M. Characterization of Vibrio cholerae RyhB: The RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 2005, 73, 5706–5719. [Google Scholar] [CrossRef]
- Matsuura, M. Structural Modifications of Bacterial Lipopolysaccharide that Facilitate Gram-Negative Bacteria Evasion of Host Innate Immunity. Front. Immunol. 2013, 4, 109. [Google Scholar] [CrossRef]
- Sansonetti, P.J.; Di Santo, J.P. Debugging how Bacteria Manipulate the Immune Response. Immunity 2007, 26, 149–161. [Google Scholar] [CrossRef]
- Wallden, K.; Rivera-Calzada, A.; Waksman, G. Type IV secretion systems: Versatility and diversity in function. Cell Microbiol. 2010, 12, 1203–1212. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Boumart, Z.; Velge, P.; Wiedemann, A. Multiple invasion mechanisms and different intracellular Behaviors: A new vision of Salmonella–host cell interaction. FEMS Microbiol. Lett. 2014, 361, 1–7. [Google Scholar] [CrossRef]
- Robbins, J.R.; Barth, A.I.; Marquis, H.; de Hostos, E.L.; Nelson, W.J.; Theriot, J.A. Listeria monocytogenes exploits normal host cell processes to spread from cell to cell. J. Cell Biol. 1999, 146, 1333–1350. [Google Scholar] [CrossRef]
- Lalaouna, D.; Baude, J.; Wu, Z.; Tomasini, A.; Chicher, J.; Marzi, S.; Vandenesch, F.; Romby, P.; Caldelari, I.; Moreau, K. RsaC sRNA modulates the oxidative stress response of Staphylococcus aureus during manganese starvation. Nucleic Acids Res. 2019, 47, 9871–9887. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.W. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 2000, 13, 470–511. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.W. Pathogenesis of group A streptococcal infections and their sequelae. Adv. Exp. Med. Biol. 2008, 609, 29–42. [Google Scholar] [CrossRef]
- Danger, J.L.; Cao, T.N.; Cao, T.H.; Sarkar, P.; Treviño, J.; Pflughoeft, K.J.; Sumby, P. The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets. Mol. Microbiol. 2015, 96, 249–262. [Google Scholar] [CrossRef]
- Ramirez-Peña, E.; Treviño, J.; Liu, Z.; Perez, N.; Sumby, P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol. Microbiol. 2010, 78, 1332–1347. [Google Scholar] [CrossRef]
- Martini, B.A.; Grigorov, A.S.; Skvortsova, Y.V.; Bychenko, O.S.; Salina, E.G.; Azhikina, T.L. Small RNA MTS1338 Configures a Stress Resistance Signature in Mycobacterium tuberculosis. Int. J. Mol. Sci. 2023, 24, 7928. [Google Scholar] [CrossRef]
- Arnvig, K.; Young, D. Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol. 2012, 9, 427–436. [Google Scholar] [CrossRef]
- Moores, A.; Riesco, A.B.; Schwenk, S.; Arnvig, K.B. Expression, maturation and turnover of DrrS, an unusually stable, DosR regulated small RNA in Mycobacterium tuberculosis. PLoS ONE 2017, 12, e0174079. [Google Scholar] [CrossRef] [PubMed]
- Salina, E.G.; Grigorov, A.; Skvortsova, Y.; Majorov, K.; Bychenko, O.; Ostrik, A.; Logunova, N.; Ignatov, D.; Kaprelyants, A.; Apt, A.; et al. MTS1338, A Small Mycobacterium tuberculosis RNA, Regulates Transcriptional Shifts Consistent With Bacterial Adaptation for Entering Into Dormancy and Survival Within Host Macrophages. Front. Cell Infect. Microbiol. 2019, 9, 405. [Google Scholar] [CrossRef]
- Banerjee, R.; Rudra, P.; Prajapati, R.K.; Sengupta, S.; Mukhopadhyay, J. Optimization of recombinant Mycobacterium tuberculosis RNA polymerase expression and purification. Tuberculosis 2014, 94, 397–404. [Google Scholar] [CrossRef]
- Banerjee, R.; Rudra, P.; Saha, A.; Mukhopadhyay, J. Recombinant Reporter Assay Using Transcriptional Machinery of Mycobacterium tuberculosis. J. Bacteriol. 2015, 197, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kumar, R.; Jain, A.; Kumar, M.; Gauttam, R.; Banerjee, R.; Mukhopadhyay, J.; Tyagi, J.S. Functional insights into Mycobacterium tuberculosis DevR-dependent transcriptional machinery utilizing Escherichia coli. Biochem. J. 2021, 478, 3079–3098. [Google Scholar] [CrossRef]
- Kumar, K.; Dutta, T. Transcriptional activation of the Mycobacterium tuberculosis virulence-associated small RNA MTS1338 by the response regulators DosR and PhoP. FEBS Lett. 2024, 598, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.; Schaible, U.E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015, 264, 182–203. [Google Scholar] [CrossRef]
- Horrocks, V.; King, O.G.; Yip, A.Y.G.; Marques, I.M.; McDonald, J.A.K. Role of the gut microbiota in nutrient competition and protection against intestinal pathogen colonization. Microbiology 2023, 169, 001377. [Google Scholar] [CrossRef]
- Nguyen, J.; Lara-Gutiérrez, J.; Stocker, R. Environmental fluctuations and their effects on microbial communities, populations and individuals. FEMS Microbiol. Rev. 2021, 45, fuaa068. [Google Scholar] [CrossRef]
- Cox, G.M.; Harrison, T.S.; McDade, H.C.; Taborda, C.P.; Heinrich, G.; Casadevall, A.; Perfect, J.R. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 2003, 71, 173–180. [Google Scholar] [CrossRef]
- Carlioz, A.; Touati, D. Isolation of superoxide dismutase mutants in Escherichia coli: Is superoxide dismutase necessary for aerobic life? EMBO J. 1986, 5, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Yan, L.J.; Jana, C.K.; Das, N. Role of Catalase in Oxidative Stress- and Age-Associated Degenerative Diseases. Oxidative Med. Cell. Longev. 2019, 2019, 9613090. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.R.; Smith, C.J. Role of the Alkyl Hydroperoxide Reductase (ahpCF) Gene in Oxidative Stress Defense of the Obligate Anaerobe Bacteroides fragilis. J. Bacteriol. 1999, 181, 5701–5710. [Google Scholar] [CrossRef]
- Lund, P.A.; De Biase, D.; Liran, O.; Scheler, O.; Mira, N.P.; Cetecioglu, Z.; Fernández, E.N.; Bover-Cid, S.; Hall, R.; Sauer, M.; et al. Understanding How Microorganisms Respond to Acid pH Is Central to Their Control and Successful Exploitation. Front. Microbiol. 2020, 11, 556140. [Google Scholar] [CrossRef]
- Konieczna, I.; Zarnowiec, P.; Kwinkowski, M.; Kolesinska, B.; Fraczyk, J.; Kaminski, Z.; Kaca, W. Bacterial urease and its role in long-lasting human diseases. Curr. Protein Pept. Sci. 2012, 13, 789–806. [Google Scholar] [CrossRef]
- Alteri, C.J.; Mobley, H.L. Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 2012, 15, 3–9. [Google Scholar] [CrossRef]
- Massé, E.; Vanderpool, C.K.; Gottesman, S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005, 187, 6962–6971. [Google Scholar] [CrossRef]
- Calderón, P.F.; Morales, E.H.; Acuña, L.G.; Fuentes, D.N.; Gil, F.; Porwollik, S.; McClelland, M.; Saavedra, C.P.; Calderón, I.L. The small RNA RyhB homologs from Salmonella typhimurium participate in the response to S-nitrosoglutathione-induced stress. Biochem. Biophys. Res. Commun. 2014, 450, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Weisenhorn, E.; Schwartz, K.J.; Myers, K.S.; Glasner, J.D.; Perna, N.T.; Coon, J.J.; Welch, R.A.; Kiley, P.J. Tailoring a Global Iron Regulon to a Uropathogen. mBio 2020, 11, e00351-20. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, S.; Wu, N.; Yuan, Y.; Zhang, W.; Zhang, Y. Small Non-coding RNA RyhB Mediates Persistence to Multiple Antibiotics and Stresses in Uropathogenic Escherichia coli by Reducing Cellular Metabolism. Front. Microbiol. 2018, 9, 136. [Google Scholar] [CrossRef]
- Acuña, L.G.; Barros, M.J.; Montt, F.; Peñaloza, D.; Núñez, P.; Valdés, I.; Gil, F.; Fuentes, J.A.; Calderón, I.L. Participation of two sRNA RyhB homologs from the fish pathogen Yersinia ruckeri in bacterial physiology. Microbiol. Res. 2021, 242, 126629. [Google Scholar] [CrossRef] [PubMed]
- Coronel-Tellez, R.H.; Pospiech, M.; Barrault, M.; Liu, W.; Bordeau, V.; Vasnier, C.; Felden, B.; Sargueil, B.; Bouloc, P. sRNA-controlled iron sparing response in Staphylococci. Nucleic Acids Res. 2022, 50, 8529–8546. [Google Scholar] [CrossRef] [PubMed]
- Gerrick, E.R.; Barbier, T.; Chase, M.R.; Xu, R.; François, J.; Lin, V.H.; Szucs, M.J.; Rock, J.M.; Ahmad, R.; Tjaden, B.; et al. Small RNA profiling in Mycobacterium tuberculosis identifies MrsI as necessary for an anticipatory iron sparing response. Proc. Natl. Acad. Sci. USA 2018, 115, 6464–6469. [Google Scholar] [CrossRef]
- Girardin, R.C.; McDonough, K.A. Small RNA Mcr11 requires the transcription factor AbmR for stable expression and regulates genes involved in the central metabolism of Mycobacterium tuberculosis. Mol. Microbiol. 2020, 113, 504–520. [Google Scholar] [CrossRef] [PubMed]
- Rudra, P.; Prajapati, R.K.; Banerjee, R.; Sengupta, S.; Mukhopadhyay, J. Novel mechanism of gene regulation: The protein Rv1222 of Mycobacterium tuberculosis inhibits transcription by anchoring the RNA polymerase onto DNA. Nucleic Acids Res. 2015, 43, 5855–5867. [Google Scholar] [CrossRef][Green Version]
- Altuvia, S.; Weinstein-Fischer, D.; Zhang, A.; Postow, L.; Storz, G. A Small, Stable RNA Induced by Oxidative Stress: Role as a Pleiotropic Regulator and Antimutator. Cell 1997, 90, 43–53. [Google Scholar] [CrossRef]
- Åslund, F.; Zheng, M.; Beckwith, J.; Storz, G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol—Disulfide status. Proc. Natl. Acad. Sci. USA 1999, 96, 6161–6165. [Google Scholar] [CrossRef]
- Štih, V.; Amenitsch, H.; Plavec, J.; Podbevšek, P. Spatial arrangement of functional domains in OxyS stress response sRNA. RNA 2023, 29, 1520–1534. [Google Scholar] [CrossRef]
- Soper, T.; Mandin, P.; Majdalani, N.; Gottesman, S.; Woodson, S.A. Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. USA 2010, 107, 9602–9607. [Google Scholar] [CrossRef]
- Barshishat, S.; Elgrably-Weiss, M.; Edelstein, J.; Georg, J.; Govindarajan, S.; Haviv, M.; Wright, P.R.; Hess, W.R.; Altuvia, S. OxyS small RNA induces cell cycle arrest to allow DNA damage repair. EMBO J. 2018, 37, 413–426. [Google Scholar] [CrossRef]
- Cho, H.; Kim, K.-S. Escherichia coli OxyS RNA triggers cephalothin resistance by modulating the expression of CRP-associated genes. Biochem. Biophys. Res. Commun. 2018, 506, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Sledjeski, D.D.; Gupta, A.; Gottesman, S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 1996, 15, 3993–4000. [Google Scholar] [CrossRef] [PubMed]
- Lease, R.A.; Smith, D.; McDonough, K.; Belfort, M. The small noncoding DsrA RNA is an acid resistance regulator in Escherichia coli. J. Bacteriol. 2004, 186, 6179–6185. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Liang, Y.; He, S.; Cui, Y.; Shi, C.; He, Y.; Shi, X. DsrA Modulates Central Carbon Metabolism and Redox Balance by Directly Repressing pflB Expression in Salmonella typhimurium. Microbiol. Spectr. 2022, 10, e0152221. [Google Scholar] [CrossRef]
- Sledjeski, D.; Gottesman, S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 2003–2007. [Google Scholar] [CrossRef]
- Rozanov, D.V.; D’Ari, R.; Sineoky, S.P. RecA-independent pathways of lambdoid prophage induction in Escherichia coli. J. Bacteriol. 1998, 180, 6306–6315. [Google Scholar] [CrossRef]
- Zhu, W.; Xi, L.; Qiao, J.; Du, D.; Wang, Y.; Morigen. Involvement of OxyR and Dps in the repression of replication initiation by DsrA small RNA in Escherichia coli. Gene 2023, 882, 147659. [Google Scholar] [CrossRef]
- Ades, S.E. Regulation by destruction: Design of the sigmaE envelope stress response. Curr. Opin. Microbiol. 2008, 11, 535–540. [Google Scholar] [CrossRef]
- Thompson, K.M.; Rhodius, V.A.; Gottesman, S. SigmaE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 2007, 189, 4243–4256. [Google Scholar] [CrossRef]
- Papenfort, K.; Bouvier, M.; Mika, F.; Sharma, C.M.; Vogel, J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 20435–20440. [Google Scholar] [CrossRef]
- Adams, P.P.; Baniulyte, G.; Esnault, C.; Chegireddy, K.; Singh, N.; Monge, M.; Dale, R.K.; Storz, G.; Wade, J.T. Regulatory roles of Escherichia coli 5′ UTR and ORF-internal RNAs detected by 3′ end mapping. eLife 2021, 10, e62438. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef] [PubMed]
- Kounatidis, D.; Dalamaga, M.; Grivakou, E.; Karampela, I.; Koufopoulos, P.; Dalopoulos, V.; Adamidis, N.; Mylona, E.; Kaziani, A.; Vallianou, N.G. Third-Generation Tetracyclines: Current Knowledge and Therapeutic Potential. Biomolecules 2024, 14, 783. [Google Scholar] [CrossRef] [PubMed]
- Bosnar, M.; Kelnerić, Z.; Munić, V.; Eraković, V.; Parnham, M.J. Cellular uptake and efflux of azithromycin, erythromycin, clarithromycin, telithromycin, and cethromycin. Antimicrob. Agents Chemother. 2005, 49, 2372–2377. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Topoisomerase Inhibitors: Fluoroquinolone Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025320. [Google Scholar] [CrossRef]
- Ovung, A.; Bhattacharyya, J. Sulfonamide drugs: Structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 2021, 13, 259–272. [Google Scholar] [CrossRef]
- Zeng, D.; Debabov, D.; Hartsell, T.L.; Cano, R.J.; Adams, S.; Schuyler, J.A.; McMillan, R.; Pace, J.L. Approved Glycopeptide Antibacterial Drugs: Mechanism of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a026989. [Google Scholar] [CrossRef]
- Howden, B.P.; Beaume, M.; Harrison, P.F.; Hernandez, D.; Schrenzel, J.; Seemann, T.; Francois, P.; Stinear, T.P. Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure. Antimicrob. Agents Chemother. 2013, 57, 3864–3874. [Google Scholar] [CrossRef]
- Molina-Santiago, C.; Daddaoua, A.; Gómez-Lozano, M.; Udaondo, Z.; Molin, S.; Ramos, J.L. Differential transcriptional response to antibiotics by Pseudomonas putida DOT-T1E. Environ. Microbiol. 2015, 17, 3251–3262. [Google Scholar] [CrossRef]
- Jin, Y.; Watt, R.M.; Danchin, A.; Huang, J.-D. Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics 2009, 10, 165. [Google Scholar] [CrossRef]
- Muto, A.; Goto, S.; Kurita, D.; Ushida, C.; Himeno, H. Involvement of GcvB small RNA in intrinsic resistance to multiple aminoglycoside antibiotics in Escherichia coli. J. Biochem. 2020, 169, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fang, J.; Chen, H.; Sun, Y.; Yang, S.; Gao, Q.; Zhang, Y.; Chen, C. GcvB Regulon Revealed by Transcriptomic and Proteomic Analysis in Vibrio alginolyticus. Int. J. Mol. Sci. 2022, 23, 9399. [Google Scholar] [CrossRef] [PubMed]
- Miyakoshi, M.; Okayama, H.; Lejars, M.; Kanda, T.; Tanaka, Y.; Itaya, K.; Okuno, M.; Itoh, T.; Iwai, N.; Wachi, M. Mining RNA-seq data reveals the massive regulon of GcvB small RNA and its physiological significance in maintaining amino acid homeostasis in Escherichia coli. Mol. Microbiol. 2022, 117, 160–178. [Google Scholar] [CrossRef] [PubMed]
- Miyakoshi, M.; Chao, Y.; Vogel, J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015, 34, 1478–1492. [Google Scholar] [CrossRef]
- Lalaouna, D.; Eyraud, A.; Devinck, A.; Prévost, K.; Massé, E. GcvB small RNA uses two distinct seed regions to regulate an extensive targetome. Mol. Microbiol. 2019, 111, 473–486. [Google Scholar] [CrossRef]
- Stauffer, L.T.; Stauffer, G.V. The Escherichia coli GcvB sRNA Uses Genetic Redundancy to Control cycA Expression. Int. Sch. Res. Not. 2012, 2012, 636273. [Google Scholar] [CrossRef]
- Barreto, B.; Rogers, E.; Xia, J.; Frisch, R.L.; Richters, M.; Fitzgerald, D.M.; Rosenberg, S.M. The Small RNA GcvB Promotes Mutagenic Break Repair by Opposing the Membrane Stress Response. J. Bacteriol. 2016, 198, 3296–3308. [Google Scholar] [CrossRef]
- Delihas, N.; Forst, S. MicF: An antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 2001, 313, 1–12. [Google Scholar] [CrossRef]
- Cowan, S.W.; Garavito, R.M.; Jansonius, J.N.; Jenkins, J.A.; Karlsson, R.; König, N.; Pai, E.F.; Pauptit, R.A.; Rizkallah, P.J.; Rosenbusch, J.P.; et al. The structure of OmpF porin in a tetragonal crystal form. Structure 1995, 3, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Carrier, M.C.; Lalaouna, D.; Massé, E. Hfq protein and GcvB small RNA tailoring of oppA target mRNA to levels allowing translation activation by MicF small RNA in Escherichia coli. RNA Biol. 2023, 20, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Ocampo-Sosa, A.A.; Cabot, G.; Rodríguez, C.; Roman, E.; Tubau, F.; Macia, M.D.; Moya, B.; Zamorano, L.; Suárez, C.; Peña, C.; et al. Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study. Antimicrob. Agents Chemother. 2012, 56, 1703–1713. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Han, K.; Chandler, C.E.; Tjaden, B.; Ernst, R.K.; Lory, S. Probing the sRNA regulatory landscape of P. aeruginosa: Post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol. Microbiol. 2017, 106, 919–937. [Google Scholar] [CrossRef]
- Sonnleitner, E.; Pusic, P.; Wolfinger, M.T.; Bläsi, U. Distinctive Regulation of Carbapenem Susceptibility in Pseudomonas aeruginosa by Hfq. Front. Microbiol. 2020, 11, 1001. [Google Scholar] [CrossRef] [PubMed]
- Schumann, A.; Gaballa, A.; Wiedmann, M. The multifaceted roles of phosphoethanolamine-modified lipopolysaccharides: From stress response and virulence to cationic antimicrobial resistance. Microbiol. Mol. Biol. Rev. 2024, 88, e0019323. [Google Scholar] [CrossRef]
- Wang, J.; Ma, W.; Fang, Y.; Liang, H.; Yang, H.; Wang, Y.; Dong, X.; Zhan, Y.; Wang, X. Core Oligosaccharide Portion of Lipopolysaccharide Plays Important Roles in Multiple Antibiotic Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2021, 65, e0034121. [Google Scholar] [CrossRef]
- Moon, K.; Six, D.A.; Lee, H.-J.; Raetz, C.R.H.; Gottesman, S. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol. Microbiol. 2013, 89, 52–64. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Cui, L.; Kuroda, M.; Ito, T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001, 9, 486–493. [Google Scholar] [CrossRef]
- Bohn, C.; Rigoulay, C.; Chabelskaya, S.; Sharma, C.M.; Marchais, A.; Skorski, P.; Borezée-Durant, E.; Barbet, R.; Jacquet, E.; Jacq, A.; et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010, 38, 6620–6636. [Google Scholar] [CrossRef] [PubMed]
- Kathirvel, M.; Buchad, H.; Nair, M. Enhancement of the pathogenicity of Staphylococcus aureus strain Newman by a small noncoding RNA SprX1. Med. Microbiol. Immunol. 2016, 205, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Eyraud, A.; Tattevin, P.; Chabelskaya, S.; Felden, B. A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res. 2014, 42, 4892–4905. [Google Scholar] [CrossRef] [PubMed]
- Ivain, L.; Bordeau, V.; Eyraud, A.; Hallier, M.; Dreano, S.; Tattevin, P.; Felden, B.; Chabelskaya, S. An in vivo reporter assay for sRNA-directed gene control in Gram-positive bacteria: Identifying a novel sRNA target in Staphylococcus aureus. Nucleic Acids Res. 2017, 45, 4994–5007. [Google Scholar] [CrossRef]
- Buchad, H.; Nair, M. The small RNA SprX regulates the autolysin regulator WalR in Staphylococcus aureus. Microbiol. Res. 2021, 250, 126785. [Google Scholar] [CrossRef]
- Wurtzel, O.; Yoder-Himes, D.R.; Han, K.; Dandekar, A.A.; Edelheit, S.; Greenberg, E.P.; Sorek, R.; Lory, S. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 2012, 8, e1002945. [Google Scholar] [CrossRef]
- Di Maso, A.M.; Ruiz, C. Physiological Effects of TolC-Dependent Multidrug Efflux Pumps in Escherichia coli: Impact on Motility and Growth Under Stress Conditions. MicrobiologyOpen 2024, 13, e70006. [Google Scholar] [CrossRef]
- Du, D.; Wang, Z.; James, N.R.; Voss, J.E.; Klimont, E.; Ohene-Agyei, T.; Venter, H.; Chiu, W.; Luisi, B.F. Structure of the AcrAB-TolC multidrug efflux pump. Nature 2014, 509, 512–515. [Google Scholar] [CrossRef]
- Smith, B.L.; Fernando, S.; King, M.D. Escherichia coli resistance mechanism AcrAB-TolC efflux pump interactions with commonly used antibiotics: A molecular dynamics study. Sci. Rep. 2024, 14, 2742. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.P.; Llabrés, S.; Neuberger, A.; Blaza, J.N.; Bai, X.-C.; Okada, U.; Murakami, S.; van Veen, H.W.; Zachariae, U.; Scheres, S.H.W.; et al. Structure of the MacAB–TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2017, 2, 17070. [Google Scholar] [CrossRef]
- Horiyama, T.; Nishino, K. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS ONE 2014, 9, e108642. [Google Scholar] [CrossRef]
- Yousefian, N.; Ornik-Cha, A.; Poussard, S.; Decossas, M.; Berbon, M.; Daury, L.; Taveau, J.C.; Dupuy, J.W.; Đorđević-Marquardt, S.; Lambert, O.; et al. Structural characterization of the EmrAB-TolC efflux complex from E. coli. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183488. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.F.; Liu, X.; Zhang, S.; Pan, B.; Liu, M.L. Ciprofloxacin derivatives and their antibacterial activities. Eur. J. Med. Chem. 2018, 146, 599–612. [Google Scholar] [CrossRef]
- Zhang, H.; Song, T.; Qin, C.; Xu, H.; Qiao, M. A Novel Non-Coding RNA CsiR Regulates the Ciprofloxacin Resistance in Proteus vulgaris by Interacting with emrB mRNA. Int. J. Mol. Sci. 2021, 22, 10627. [Google Scholar] [CrossRef] [PubMed]
- Coleman, S.R.; Smith, M.L.; Spicer, V.; Lao, Y.; Mookherjee, N.; Hancock, R.E.W. Overexpression of the Small RNA PA0805.1 in Pseudomonas aeruginosa Modulates the Expression of a Large Set of Genes and Proteins, Resulting in Altered Motility, Cytotoxicity, and Tobramycin Resistance. mSystems 2020, 5, e00204-20. [Google Scholar] [CrossRef]
- Choi, J.S.; Kim, W.; Suk, S.; Park, H.; Bak, G.; Yoon, J.; Lee, Y. The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol. 2018, 15, 1319–1335. [Google Scholar] [CrossRef]
- Parker, A.; Gottesman, S. Small RNA Regulation of TolC, the Outer Membrane Component of Bacterial Multidrug Transporters. J. Bacteriol. 2016, 198, 1101–1113. [Google Scholar] [CrossRef]
- Gutierrez, A.; Laureti, L.; Crussard, S.; Abida, H.; Rodríguez-Rojas, A.; Blázquez, J.; Baharoglu, Z.; Mazel, D.; Darfeuille, F.; Vogel, J.; et al. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 2013, 4, 1610. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial Biofilm: A Review on Formation, Infection, Antibiotic Resistance, Control Measures, and Innovative Treatment. Microorganisms 2023, 11, 1614. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Waters, E.M.; Rowe, S.E.; O’Gara, J.P.; Conlon, B.P. Convergence of Staphylococcus aureus Persister and Biofilm Research: Can Biofilms Be Defined as Communities of Adherent Persister Cells? PLoS Pathog. 2016, 12, e1006012. [Google Scholar] [CrossRef]
- Sultan, M.; Arya, R.; Kim, K.K. Roles of Two-Component Systems in Pseudomonas aeruginosa Virulence. Int. J. Mol. Sci. 2021, 22, 12152. [Google Scholar] [CrossRef]
- Mukherjee, S.; Moustafa, D.; Smith, C.D.; Goldberg, J.B.; Bassler, B.L. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 2017, 13, e1006504. [Google Scholar] [CrossRef]
- Van Puyvelde, S.; Steenackers, H.P.; Vanderleyden, J. Small RNAs regulating biofilm formation and outer membrane homeostasis. RNA Biol. 2013, 10, 185–191. [Google Scholar] [CrossRef]
- Timmermans, J.; Van Melderen, L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol. Life Sci. 2010, 67, 2897–2908. [Google Scholar] [CrossRef]
- Liu, P.; Yue, C.; Liu, L.; Gao, C.; Lyu, Y.; Deng, S.; Tian, H.; Jia, X. The function of small RNA in Pseudomonas aeruginosa. PeerJ 2022, 10, e13738. [Google Scholar] [CrossRef]
- Law, C.O.K.; Huang, C.; Pan, Q.; Lee, J.; Hao, Q.; Chan, T.F.; Lo, N.W.S.; Ang, I.L.; Koon, A.; Ip, M.; et al. A Small RNA Transforms the Multidrug Resistance of Pseudomonas aeruginosa to Drug Susceptibility. Mol. Ther. Nucleic Acids 2019, 16, 218–228. [Google Scholar] [CrossRef]
- Li, Y.; Cao, X.; Chai, Y.; Chen, R.; Zhao, Y.; Borriss, R.; Ding, X.; Wu, X.; Ye, J.; Hao, D.; et al. A phosphate starvation induced small RNA promotes Bacillus biofilm formation. NPJ Biofilms Microbiomes 2024, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.K.; Van Kessel, A.T.M.; Colavita, A.; Hancock, R.E.W.; Mah, T.F. A novel small RNA is important for biofilm formation and pathogenicity in Pseudomonas aeruginosa. PLoS ONE 2017, 12, e0182582. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. QUORUM SENSING: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Wu, L.; Luo, Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021, 12, 611413. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.S.; Iglewski, B.H. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J. Clin. Investig. 2003, 112, 1460–1465. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 2003, 112, 1620–1625. [Google Scholar] [CrossRef]
- Zhu, J.; Mekalanos, J.J. Quorum Sensing-Dependent Biofilms Enhance Colonization in Vibrio cholerae. Dev. Cell 2003, 5, 647–656. [Google Scholar] [CrossRef]
- Anetzberger, C.; Reiger, M.; Fekete, A.; Schell, U.; Stambrau, N.; Plener, L.; Kopka, J.; Schmitt-Kopplin, P.; Hilbi, H.; Jung, K. Autoinducers act as biological timers in Vibrio harveyi. PLoS ONE 2012, 7, e48310. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Bassler, B.L. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006, 20, 2754–2767. [Google Scholar] [CrossRef]
- Mok, K.C.; Wingreen, N.S.; Bassler, B.L. Vibrio harveyi quorum sensing: A coincidence detector for two autoinducers controls gene expression. EMBO J. 2003, 22, 870–881. [Google Scholar] [CrossRef]
- Papenfort, K.; Silpe, J.E.; Schramma, K.R.; Cong, J.P.; Seyedsayamdost, M.R.; Bassler, B.L. A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat. Chem. Biol. 2017, 13, 551–557. [Google Scholar] [CrossRef]
- Jemielita, M.; Wingreen, N.S.; Bassler, B.L. Quorum sensing controls Vibrio cholerae multicellular aggregate formation. eLife 2018, 7, e42057. [Google Scholar] [CrossRef] [PubMed]
- Tsou, A.M.; Liu, Z.; Cai, T.; Zhu, J. The VarS/VarA two-component system modulates the activity of the Vibrio cholerae quorum-sensing transcriptional regulator HapR. Microbiology 2011, 157, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Bassler, B.L. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol. Microbiol. 2014, 92, 921–930. [Google Scholar] [CrossRef]
- MacIntyre, D.L.; Miyata, S.T.; Kitaoka, M.; Pukatzki, S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. USA 2010, 107, 19520–19524. [Google Scholar] [CrossRef]
- Allsopp, L.P.; Bernal, P. Killing in the name of: T6SS structure and effector diversity. Microbiology 2023, 169, 001367. [Google Scholar] [CrossRef]
- Dong, T.G.; Ho, B.T.; Yoder-Himes, D.R.; Mekalanos, J.J. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2013, 110, 2623–2628. [Google Scholar] [CrossRef]
- Hersch, S.J.; Manera, K.; Dong, T.G. Defending against the Type Six Secretion System: Beyond Immunity Genes. Cell Rep. 2020, 33, 108259. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Lippegaus, A.; Melamed, S.; Siemers, M.; Wucher, B.R.; Hoyos, M.; Nadell, C.; Storz, G.; Papenfort, K. An RNA sponge controls quorum sensing dynamics and biofilm formation in Vibrio cholerae. Nat. Commun. 2022, 13, 7585. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Smith, R.S.; Iglewski, B.H. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 2003, 6, 56–60. [Google Scholar] [CrossRef] [PubMed]
- McKnight, S.L.; Iglewski, B.H.; Pesci, E.C. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 2702–2708. [Google Scholar] [CrossRef]
- Jimenez, P.N.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The Multiple Signaling Systems Regulating Virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef]
- Managò, A.; Becker, K.A.; Carpinteiro, A.; Wilker, B.; Soddemann, M.; Seitz, A.P.; Edwards, M.J.; Grassmé, H.; Szabò, I.; Gulbins, E. Pseudomonas aeruginosa pyocyanin induces neutrophil death via mitochondrial reactive oxygen species and mitochondrial acid sphingomyelinase. Antioxid. Redox Signal 2015, 22, 1097–1110. [Google Scholar] [CrossRef]
- Sonnleitner, E.; Gonzalez, N.; Sorger-Domenigg, T.; Heeb, S.; Richter, A.S.; Backofen, R.; Williams, P.; Hüttenhofer, A.; Haas, D.; Bläsi, U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol. Microbiol. 2011, 80, 868–885. [Google Scholar] [CrossRef]
- Carloni, S.; Macchi, R.; Sattin, S.; Ferrara, S.; Bertoni, G. The small RNA ReaL: A novel regulatory element embedded in the Pseudomonas aeruginosa quorum sensing networks. Environ. Microbiol. 2017, 19, 4220–4237. [Google Scholar] [CrossRef]
- Pu, J.; Zhang, S.; He, X.; Zeng, J.; Shen, C.; Luo, Y.; Li, H.; Long, Y.; Liu, J.; Xiao, Q.; et al. The Small RNA AmiL Regulates Quorum Sensing-Mediated Virulence in Pseudomonas aeruginosa PAO1. Microbiol. Spectr. 2022, 10, e0221121. [Google Scholar] [CrossRef]
- Clamens, T.; Rosay, T.; Crépin, A.; Grandjean, T.; Kentache, T.; Hardouin, J.; Bortolotti, P.; Neidig, A.; Mooij, M.; Hillion, M.; et al. The aliphatic amidase AmiE is involved in regulation of Pseudomonas aeruginosa virulence. Sci. Rep. 2017, 7, 41178. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, H.; Pu, J.; Xiao, Q.; Zhao, C.; Cai, Y.; Liu, Y.; Wang, L.; Li, Y.; Huang, B.; et al. Identification of a novel RhlI/R-PrrH-LasI/Phzc/PhzD signalling cascade and its implication in P. aeruginosa virulence. Emerg. Microbes Infect. 2019, 8, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Bi, X.; Li, M.; Zhang, C.; Ren, A.; Li, S.; Zhou, T.; Zhang, Y.; Liu, Y.; Liu, X.; et al. Hfq-binding small RNA PqsS regulates Pseudomonas aeruginosa pqs quorum sensing system and virulence. NPJ Biofilms Microbiomes 2024, 10, 82. [Google Scholar] [CrossRef]
- Chabelskaya, S.; Gaillot, O.; Felden, B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010, 6, e1000927. [Google Scholar] [CrossRef]
- Grainger, D.C.; Aiba, H.; Hurd, D.; Browning, D.F.; Busby, S.J. Transcription factor distribution in Escherichia coli: Studies with FNR protein. Nucleic Acids Res. 2007, 35, 269–278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Melamed, S.; Faigenbaum-Romm, R.; Peer, A.; Reiss, N.; Shechter, O.; Bar, A.; Altuvia, Y.; Argaman, L.; Margalit, H. Mapping the small RNA interactome in bacteria using RIL-seq. Nat. Protoc. 2018, 13, 1–33. [Google Scholar] [CrossRef]
- Han, K.; Tjaden, B.; Lory, S. GRIL-seq provides a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation. Nat. Microbiol. 2016, 2, 16239. [Google Scholar] [CrossRef]
- Boudry, P.; Piattelli, E.; Drouineau, E.; Peltier, J.; Boutserin, A.; Lejars, M.; Hajnsdorf, E.; Monot, M.; Dupuy, B.; Martin-Verstraete, I.; et al. Identification of RNAs bound by Hfq reveals widespread RNA partners and a sporulation regulator in the human pathogen Clostridioides difficile. RNA Biol. 2021, 18, 1931–1952. [Google Scholar] [CrossRef]
- Banerjee, R.; Askenasy, I.; Mettert, E.L.; Kiley, P.J. Iron–sulfur Rrf2 transcription factors: An emerging versatile platform for sensing stress. Curr. Opin. Microbiol. 2024, 82, 102543. [Google Scholar] [CrossRef]
- Balderas, D.; Mettert, E.; Lam, H.N.; Banerjee, R.; Gverzdys, T.; Alvarez, P.; Saarunya, G.; Tanner, N.; Zoubedi, A.; Wei, Y.; et al. Genome Scale Analysis Reveals IscR Directly and Indirectly Regulates Virulence Factor Genes in Pathogenic Yersinia. mBio 2021, 12, e0063321. [Google Scholar] [CrossRef]
- Azaldegui, C.A.; Vecchiarelli, A.G.; Biteen, J.S. The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 2021, 120, 1123–1138. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R. Synthetic Biology-Based Approaches to Investigate Host–Pathogen Interactions. SynBio 2025, 3, 4. [Google Scholar] [CrossRef]
- Wang, J.; Lu, X.; Wang, C.; Yue, Y.; Wei, B.; Zhang, H.; Wang, H.; Chen, J. Research Progress on the Combination of Quorum-Sensing Inhibitors and Antibiotics against Bacterial Resistance. Molecules 2024, 29, 1674. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhuang, X.; Dong, W.; Xin, F.; Jia, H.; Wu, X. Advances in the Application of Quorum Sensing to Regulate Electrode Biofilms in Bioelectrochemical Systems. Fermentation 2023, 9, 625. [Google Scholar] [CrossRef]
- Davies, J.; Spiegelman, G.B.; Yim, G. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 2006, 9, 445–453. [Google Scholar] [CrossRef]
- Yu, J.; Schneiders, T. Tigecycline challenge triggers sRNA production in Salmonella enterica serovar Typhimurium. BMC Microbiol. 2012, 12, 195. [Google Scholar] [CrossRef]
- Chen, Y.; Indurthi, D.C.; Jones, S.W.; Papoutsakis, E.T. Small RNAs in the genus Clostridium. mBio 2011, 2, e00340-10. [Google Scholar] [CrossRef] [PubMed]
- Poonawala, H.; Zhang, Y.; Kuchibhotla, S.; Green, A.G.; Cirillo, D.M.; Di Marco, F.; Spitlaeri, A.; Miotto, P.; Farhat, M.R. Transcriptomic responses to antibiotic exposure in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2024, 68, e0118523. [Google Scholar] [CrossRef]
- Adnan, S.N.; Ibrahim, N.; Yaacob, W.A. Transcriptome analysis of methicillin-resistant Staphylococcus aureus in response to stigmasterol and lupeol. J. Glob. Antimicrob. Resist. 2017, 8, 48–54. [Google Scholar] [CrossRef]
- Fernández, M.; Conde, S.; de la Torre, J.; Molina-Santiago, C.; Ramos, J.L.; Duque, E. Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440. Antimicrob. Agents Chemother. 2012, 56, 1001–1009. [Google Scholar] [CrossRef]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef] [PubMed]
- Layton, J.C.; Foster, P.L. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 2003, 50, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Cen, T.; Zhang, X.; Xie, S.; Li, D. Preservatives accelerate the horizontal transfer of plasmid-mediated antimicrobial resistance genes via differential mechanisms. Environ. Int. 2020, 138, 105544. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.; Ho, J.; Liu, X.; Zhang, L.; Wong, S.H.; Chan, M.T.; Wu, W.K. Potential and use of bacterial small RNAs to combat drug resistance: A systematic review. Infect. Drug Resist. 2017, 10, 521–532. [Google Scholar] [CrossRef]
- Qian, W.; Sun, J.; Liu, T.; Yang, Z.; Tsui, S.K. sRNAdeep: A novel tool for bacterial sRNA prediction based on DistilBERT encoding mode and deep learning algorithms. BMC Genom. 2024, 25, 1021. [Google Scholar] [CrossRef]
- Tjaden, B. TargetRNA3: Predicting prokaryotic RNA regulatory targets with machine learning. Genome Biol. 2023, 24, 276. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banerjee, R. Tiny but Mighty: Small RNAs—The Micromanagers of Bacterial Survival, Virulence, and Host–Pathogen Interactions. Non-Coding RNA 2025, 11, 36. https://doi.org/10.3390/ncrna11030036
Banerjee R. Tiny but Mighty: Small RNAs—The Micromanagers of Bacterial Survival, Virulence, and Host–Pathogen Interactions. Non-Coding RNA. 2025; 11(3):36. https://doi.org/10.3390/ncrna11030036
Chicago/Turabian StyleBanerjee, Rajdeep. 2025. "Tiny but Mighty: Small RNAs—The Micromanagers of Bacterial Survival, Virulence, and Host–Pathogen Interactions" Non-Coding RNA 11, no. 3: 36. https://doi.org/10.3390/ncrna11030036
APA StyleBanerjee, R. (2025). Tiny but Mighty: Small RNAs—The Micromanagers of Bacterial Survival, Virulence, and Host–Pathogen Interactions. Non-Coding RNA, 11(3), 36. https://doi.org/10.3390/ncrna11030036





