Unusual Novel SnoRNA-Like RNAs in Drosophila melanogaster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Computational Analysis
2.2. RNA Extraction and Analysis
3. Results
| Name | Class | Length | Genomic Location | Accession | Host Gene | Opposite to | ||
|---|---|---|---|---|---|---|---|---|
| snoRNA:Or-ACA6 | ACA | 137 | chr2R | 5,968,693–5,968,830 | - | KJ808674 | Uhg46E3 | egr1 |
| snoRNA:Or-ACA7 | ACA | 81 | chr2R | 5,968,118–5,968,199 | - | KJ808675 | Uhg46E3 | egr1 |
| snoRNA:Or-CD13 | CD | 69 | chr2R | 5,969,758–5,969,827 | - | KJ808676 | Uhg46E3 | egr2 |
| snoRNA:Or-CD15 | CD | 96 | chr3L | 1,667,151–1,667,247 | - | KJ808681 | RpL23A3* 4 | |
| snoRNA:Me28S-A2629 | CD | 140 | chr3L | 10,530,718–10,530,858 | + | KJ808678 | A2bp12 | |
| snoRNA:Or-ACA8 | ACA | 93 | chr3L | 11,882,942–11,883,035 | + | KJ808677 | CG58971 | |
| snoRNA:Or-CD14 | CD | 126 | chr2L | 2,014,449–2,014,575 | - | KJ808682 | CG42381 | |
| snoRNA:Me28S-C2789 | CD | 148 | chr2L | 21,034,775–21,034,923 | + | KJ808683 | CG422381 | |
| snoRNA:Me18S-G1506 | CD | 147 | chr2L | 16,561,050–16,561,197 | - | KJ808679 | CG423891 | |
| scaRNA:MeU1:95C-A24 | CD | 106 | chr2L | 901,976–902,082 | - | KJ808680 | ||
| snoRNA:Or-ACA9 | ACA | 600 | chr3R | 11,297,723–11,298,370 †‡ | - | KJ808684 | AdamTS-A1 | |
| snoRNA:Or-CD16 | CD | 400 | chr2L | 3,504,032–3,504,182 ‡ | - | Suppl. data | tim2 | |
| scaRNA:MeU4:25F-C137 | CD | 600 | chr3R | 1,410,224–1,410,366 ‡ | - | Suppl. data | CG29263 | |
| snoRNA:Me28S-C993 | CD | 400,1200 | chr3L | 10,189,262–10,189,354 ‡ | - | Suppl. data | ||
| snoRNA:Or-CD17 | CD | 500 | chr2R | 12,958,091–12,958,193 ‡ | + | Suppl. data | ||
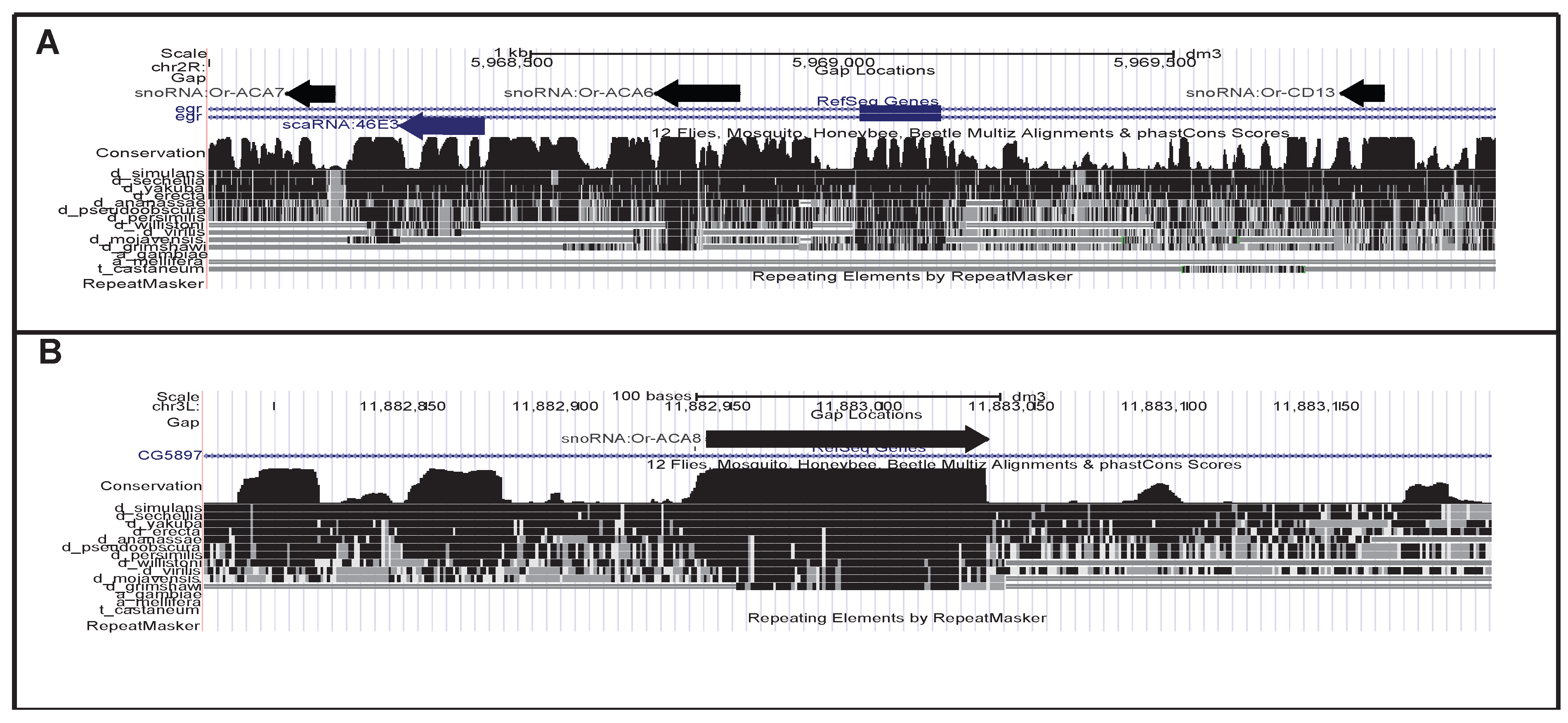
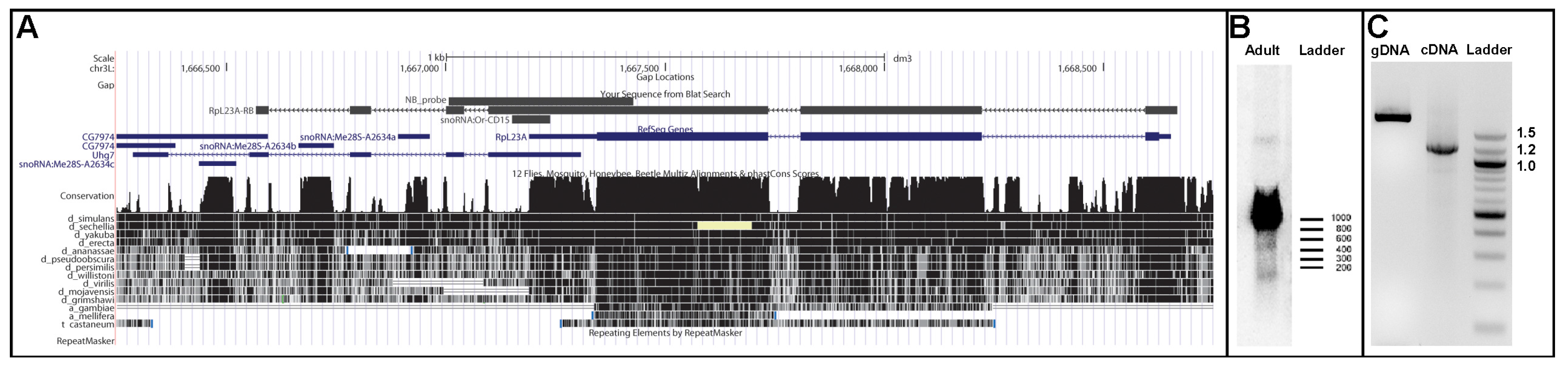
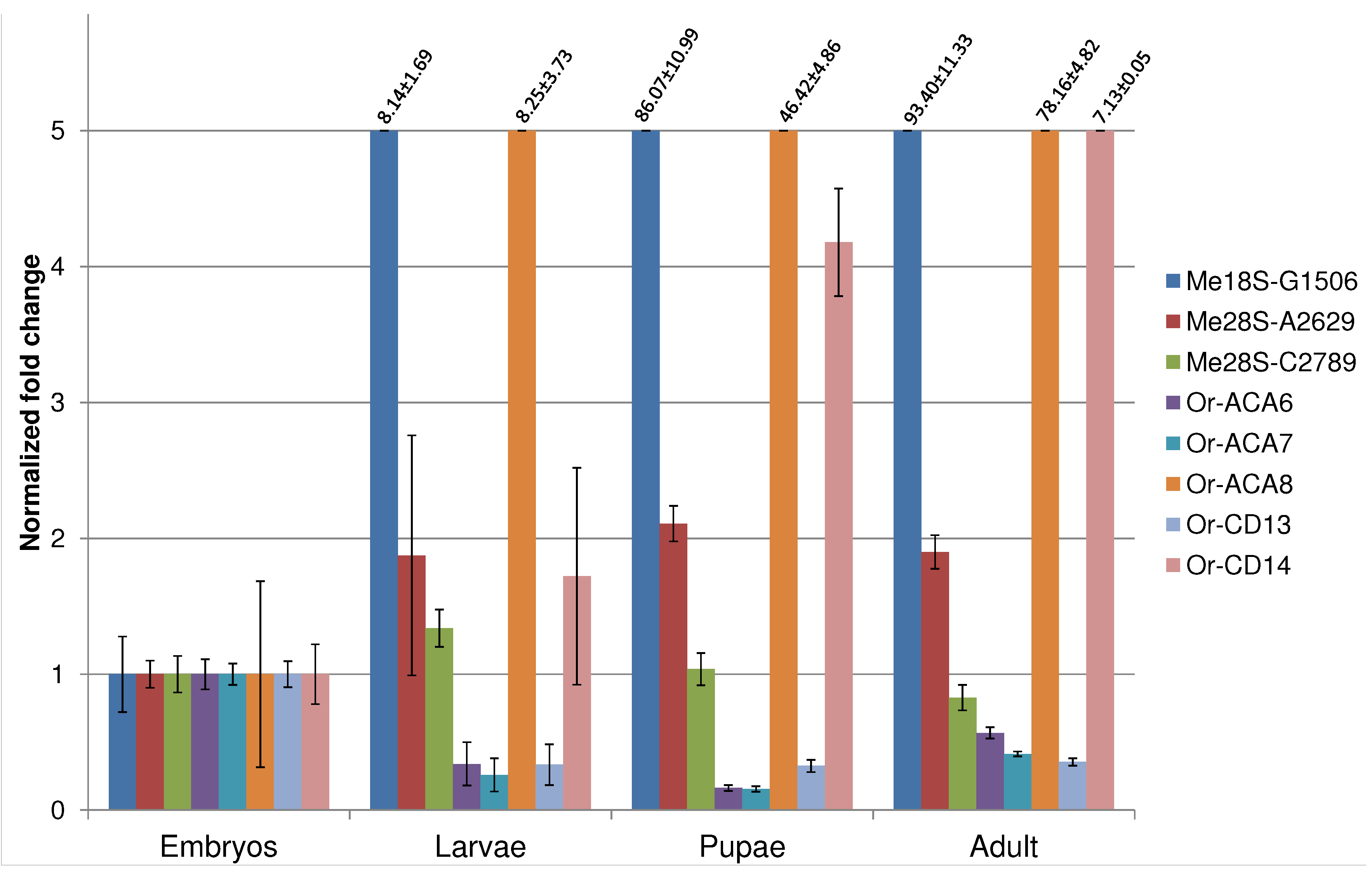
4. Discussion and Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bachellerie, J.P.; Cavaillé, J.; Hüttenhofer, A. The expanding snoRNA world. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef]
- Henras, A.K.; Dez, C.; Henry, Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 2004, 14, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Bratkovič, T.; Rogelj, B. The many faces of small nucleolar RNAs. Biochim. Biophys. Acta 2014, 1839, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Makarova, J.A.; Ivanova, S.M.; Tonevitsky, A.G.; Grigoriev, A.I. New functions of small nucleolar RNAs. Biochemistry (Mosc.) 2013, 78, 638–650. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Stamm, S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays 2013, 35, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Herter, E.K.; Stauch, M.; Gallant, M.; Wolf, E.; Raabe, T.; Gallant, P. snoRNAs are a novel class of biologically relevant Myc targets. BMC Biol. 2015, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Todoerti, K.; Tuana, G.; Agnelli, L.; Mosca, L.; Lionetti, M.; Fabris, S.; Colapietro, P.; Miozzo, M.; Ferrarini, M.; et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J 2012, 2, e96. [Google Scholar] [CrossRef] [PubMed]
- Angrisani, A.; Tafer, H.; Stadler, P.F.; Furia, M. Developmentally regulated expression and expression strategies of Drosophila snoRNAs. Insect Biochem. Mol. Biol. 2015, 61, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Chen, Y.; Ardal, B.R.; Lilje, B.; Waage, J.; Sandelin, A.; Jensen, T.H. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014, 28, 2498–2517. [Google Scholar] [CrossRef] [PubMed]
- Tycowski, K.T.; Steitz, J.A. Non-coding snoRNA host genes in Drosophila: expression strategies for modification guide snoRNAs. Eur. J. Cell Biol. 2001, 80, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Hansen, M.A.; Makunin, I.V.; Korbie, D.J.; Mattick, J.S. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC Genomics 2010, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Renalier, M.H.; Nicoloso, M.; Qu, L.H.; Bachellerie, J.P. SnoRNA U21 is also intron-encoded in Drosophila melanogaster but in a different host-gene as compared to warm-blooded vertebrates. FEBS Lett. 1996, 379, 212–216. [Google Scholar] [CrossRef]
- Enerly, E.; Mikkelsen, O.L.; Lyamouri, M.; Lambertsson, A. Evolutionary profiling of the U49 snoRNA gene. Hereditas 2003, 138, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Accardo, M.C.; Giordano, E.; Riccardo, S.; Digilio, F.A.; Iazzetti, G.; Calogero, R.A.; Furia, M. A computational search for box C/D snoRNA genes in the Drosophila melanogaster genome. Bioinformatics 2004, 20, 3293–3301. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.P.; Zhou, H.; Liang, D.; Qu, L.H. Different expression strategy: multiple intronic gene clusters of box H/ACA snoRNA in Drosophila melanogaster. J. Mol. Biol. 2004, 341, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.P.; Zhou, H.; He, H.L.; Chen, C.L.; Liang, D.; Qu, L.H. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA 2005, 11, 1303–1316. [Google Scholar] [CrossRef] [PubMed]
- Riccardo, S.; Tortoriello, G.; Giordano, E.; Turano, M.; Furia, M. The coding/non-coding overlapping architecture of the gene encoding the Drosophila pseudouridine synthase. BMC Mol. Biol. 2007, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Tortoriello, G.; Accardo, M.C.; Scialò, F.; Angrisani, A.; Turano, M.; Furia, M. A novel Drosophila antisense scaRNA with a predicted guide function. Gene 2009, 436, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. A computational screen for methylation guide snoRNAs in yeast. Science 1999, 283, 1168–1171. [Google Scholar] [CrossRef] [PubMed]
- Hertel, J.; Hofacker, I.L.; Stadler, P.F. snoReport: Computational identification of snoRNAs with unknown targets. Bioinformatics 2008, 24, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Bernhart, S.H.; Hofacker, I.L.; Will, S.; Gruber, A.R.; Stadler, P.F. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics 2008, 9, 474. [Google Scholar] [CrossRef] [PubMed]
- Tafer, H.; Kehr, S.; Hertel, J.; Stadler, P.F. RNAsnoop: Efficient target prediction for box H/ACA snoRNAs. Bioinformatics 2010, 26, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Kehr, S.; Bartschat, S.; Stadler, P.F.; Tafer, H. PLEXY: Efficient Target Prediction for Box C/D snoRNAs. Bioinformatics 2011, 27, 279–280. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Untergrasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3=–new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005, 15. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long noncoding RNAs with snoRNA ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Yin, Q.F.; Wang, H.B.; Zhang, Y.; Chen, T.; Zheng, P.; Lu, X.; Chen, L.L.; Yang, L. Species-specific alternative splicing leads to unique expression of sno-lncRNAs. BMC Genomics 2014, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Igaki, T.; Miura, M. The Drosophila TNF ortholog Eiger: emerging physiological roles and evolution of the TNF system. Semin. Immunol. 2014, 26, 267–274. [Google Scholar] [CrossRef] [PubMed]
- McQuilton, P.; St Pierre, S.E.; Thurmond, J.; FlyBase Consortium. FlyBase 101—The basics of navigating FlyBase. Nucleic Acids Res 2012, 40, D706–D714. [Google Scholar] [CrossRef] [PubMed]
- Vitali, P.; Basyuk, E.; le Meur, E.; Bertrand, E.; Muscatelli, F.; Cavaillé, J.; Hüttenhofer, A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005, 169, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Raabe, C.A.; Reinhardt, R.; Brosius, J.; Rozhdestvensky, T.S. Alternative processing as evolutionary mechanism for the origin of novel nonprotein coding RNAs. Genome Biol. Evol. 2013, 5, 2061–2071. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.E.; Grant, G.R.; Paquin, C.; Qian, J.; Nitabach, M.N. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012, 22, 1266–1281. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs derived from snoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Langenberger, D.; Çakir, M.V.; Hoffmann, S.; Stadler, P.F. Dicer-Processed Small RNAs: Rules and Exceptions. J. Exp. Zool. Mol. Dev. Evol. 2012, 320, 35–46. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agrisani, A.; Tafer, H.; Stadler, P.F.; Furia, M. Unusual Novel SnoRNA-Like RNAs in Drosophila melanogaster. Non-Coding RNA 2015, 1, 139-150. https://doi.org/10.3390/ncrna1020139
Agrisani A, Tafer H, Stadler PF, Furia M. Unusual Novel SnoRNA-Like RNAs in Drosophila melanogaster. Non-Coding RNA. 2015; 1(2):139-150. https://doi.org/10.3390/ncrna1020139
Chicago/Turabian StyleAgrisani, Alberto, Hakim Tafer, Peter F. Stadler, and Maria Furia. 2015. "Unusual Novel SnoRNA-Like RNAs in Drosophila melanogaster" Non-Coding RNA 1, no. 2: 139-150. https://doi.org/10.3390/ncrna1020139
APA StyleAgrisani, A., Tafer, H., Stadler, P. F., & Furia, M. (2015). Unusual Novel SnoRNA-Like RNAs in Drosophila melanogaster. Non-Coding RNA, 1(2), 139-150. https://doi.org/10.3390/ncrna1020139





