QbD Supported Optimization of the Alginate-Chitosan Nanoparticles of Simvastatin in Enhancing the Anti-Proliferative Activity against Tongue Carcinoma
Abstract
:1. Introduction
2. Results
2.1. FTIR (the Fourier Transform Infrared Radiation) Studies
2.2. Formulation Optimization
2.3. Zeta Potential and Polydispersity Index (PDI)
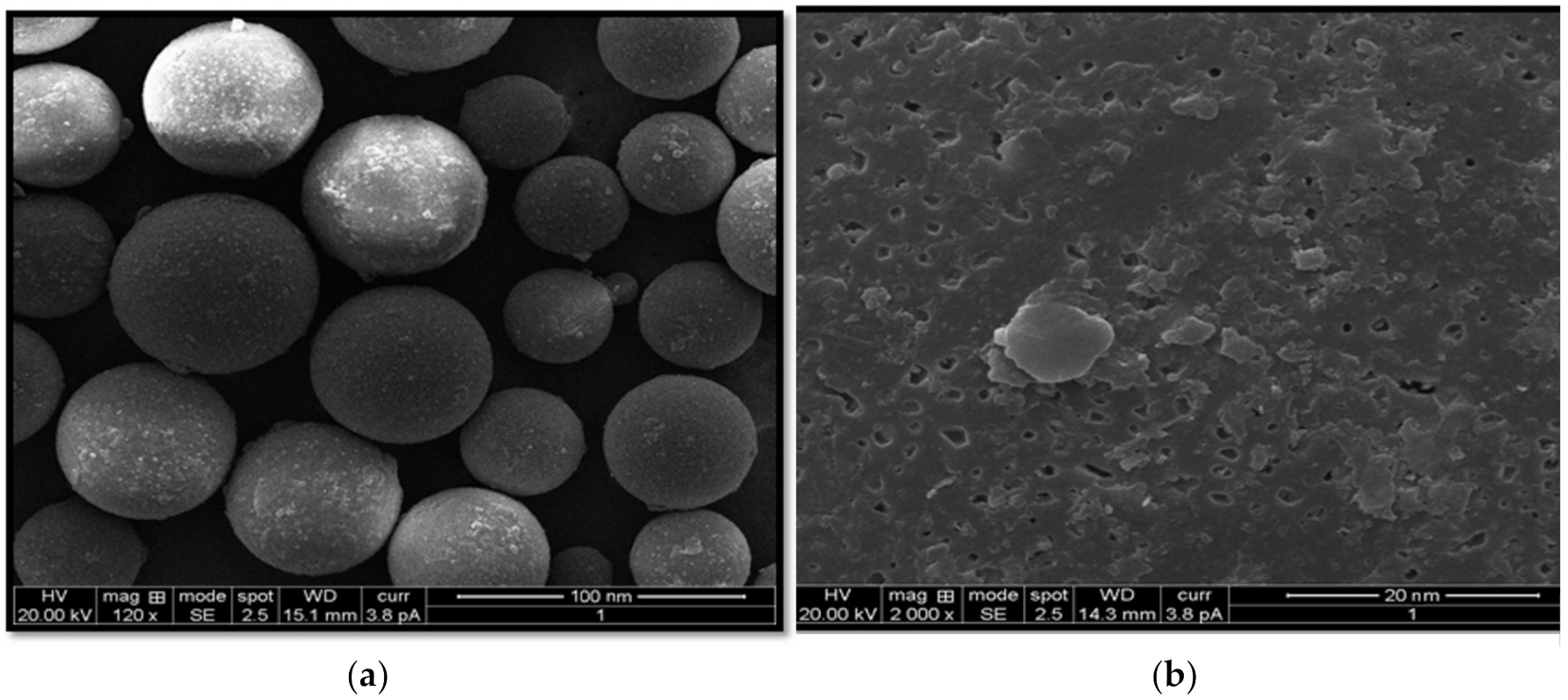
2.4. Scanning Electron Microscopic (SEM) Studies
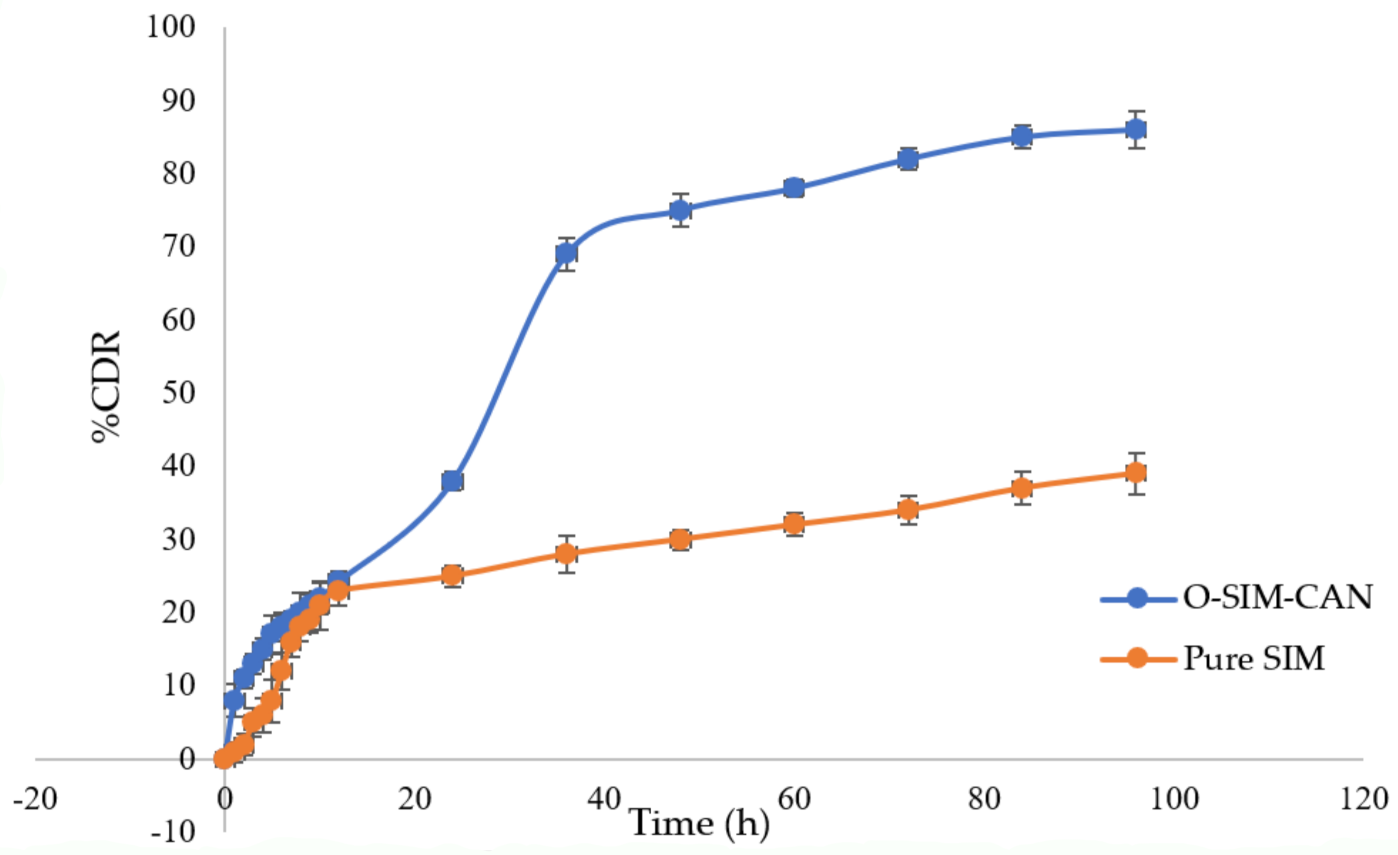
2.5. Drug Release Studies
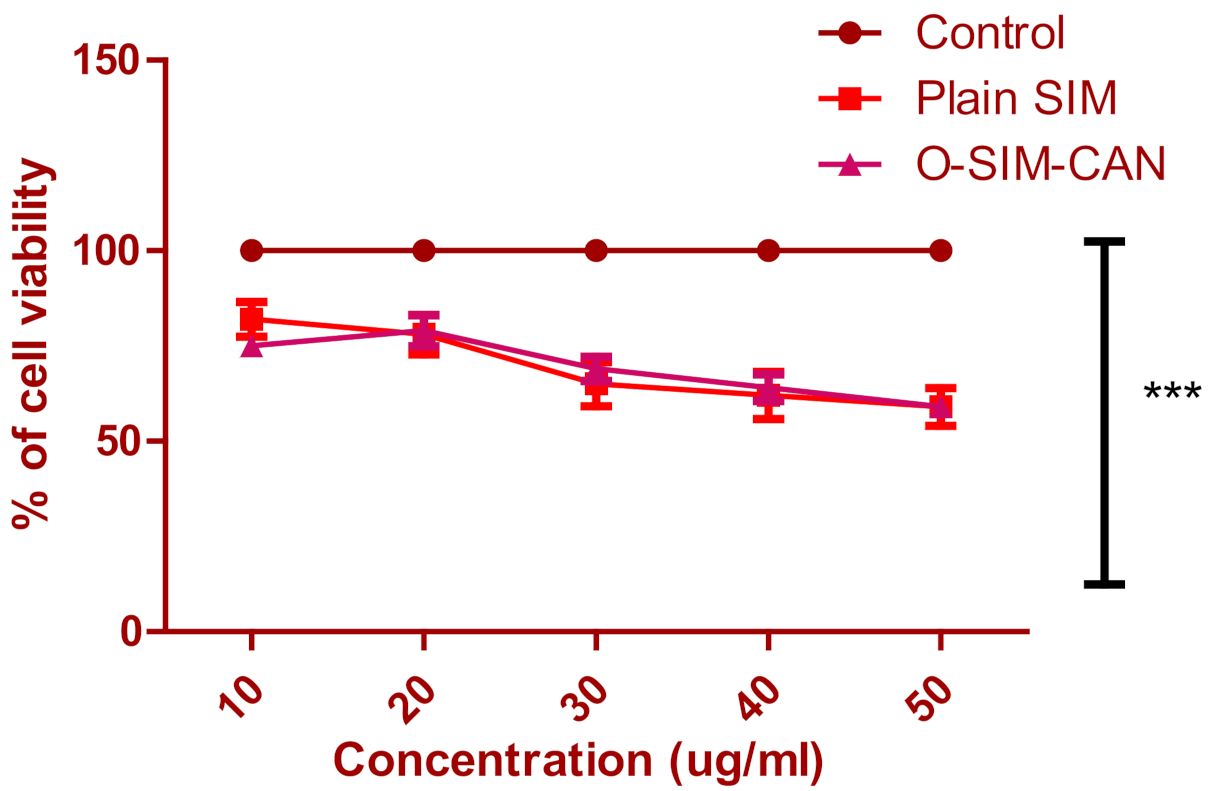
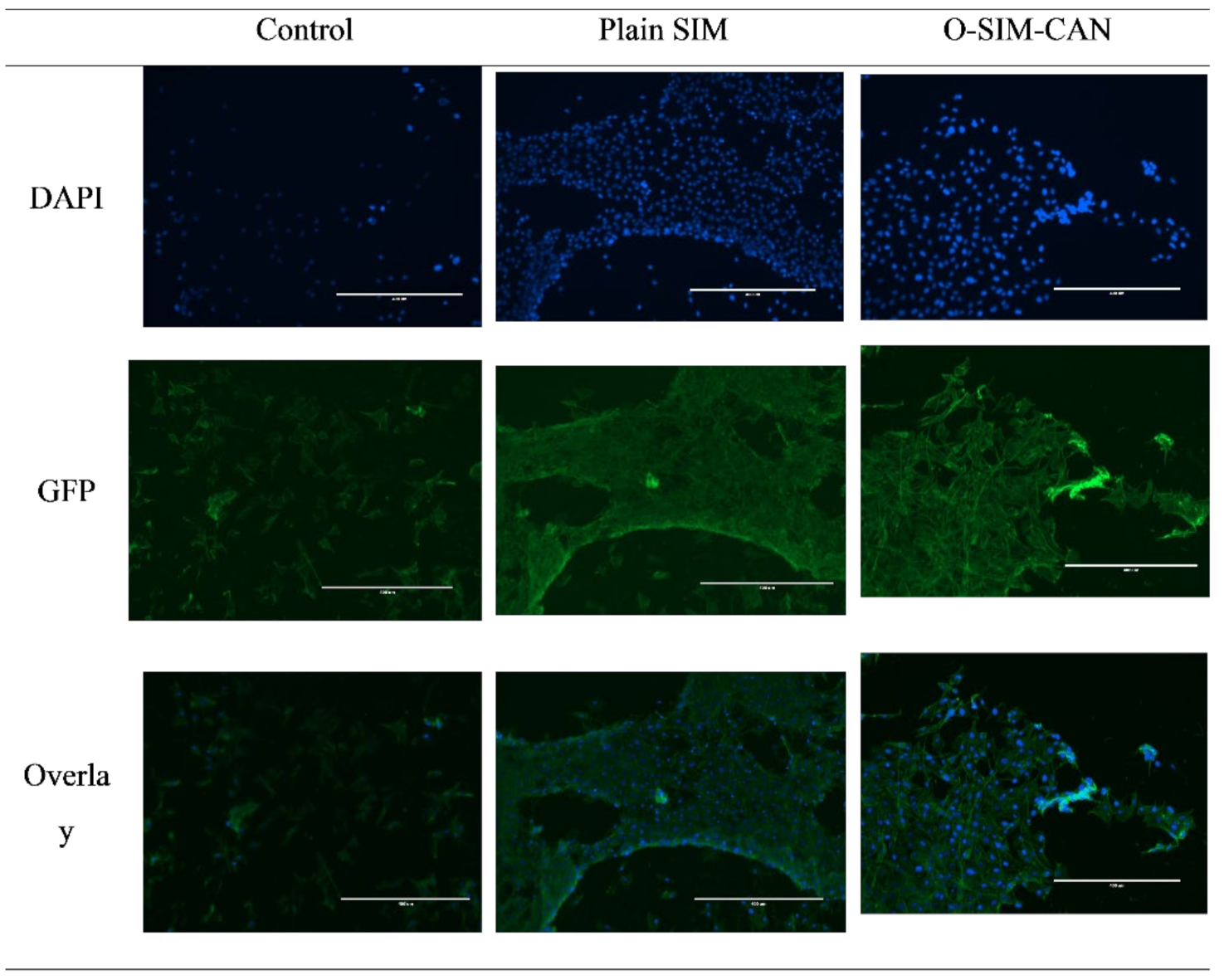
2.6. Cell Viability Studies
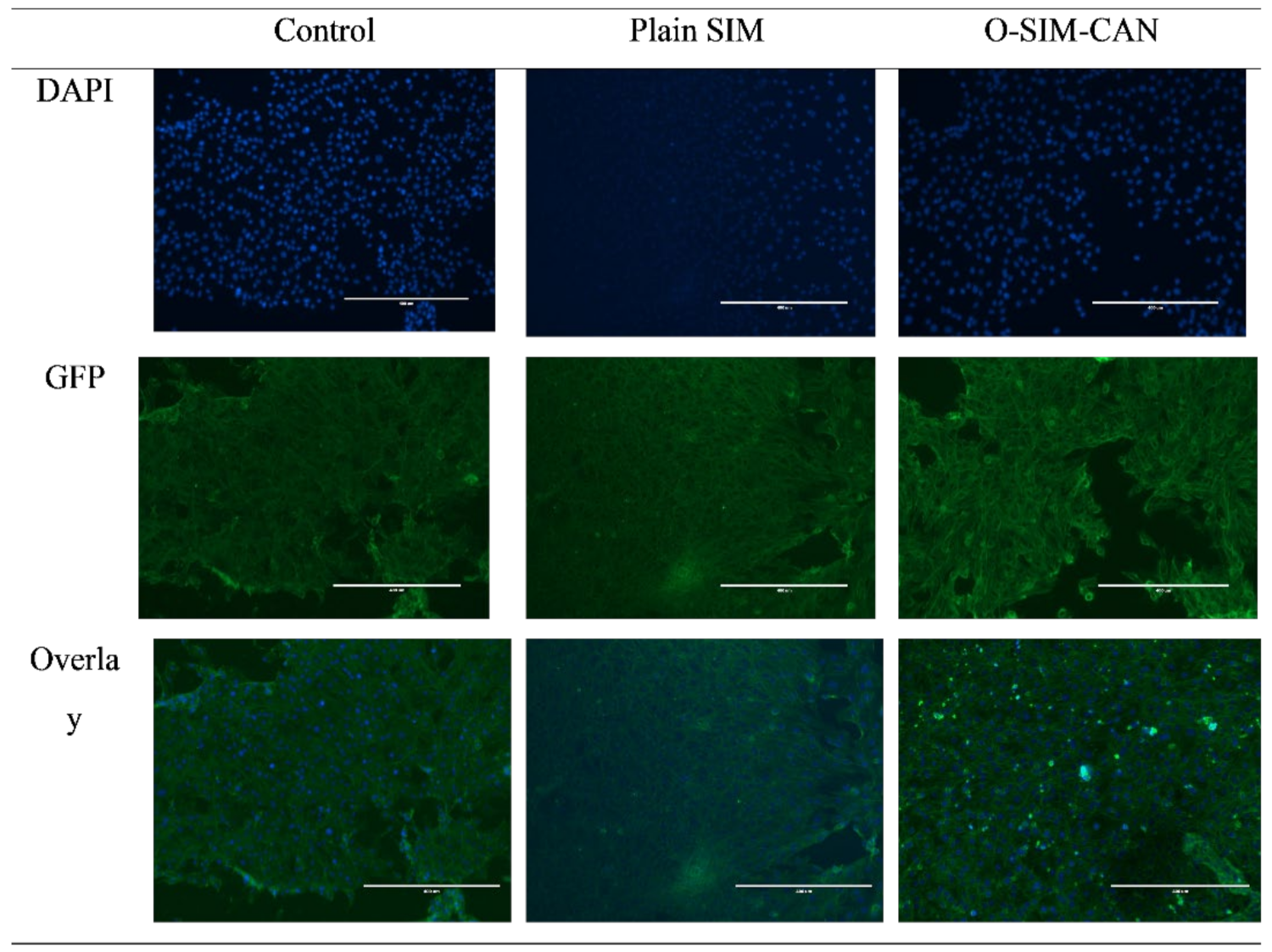
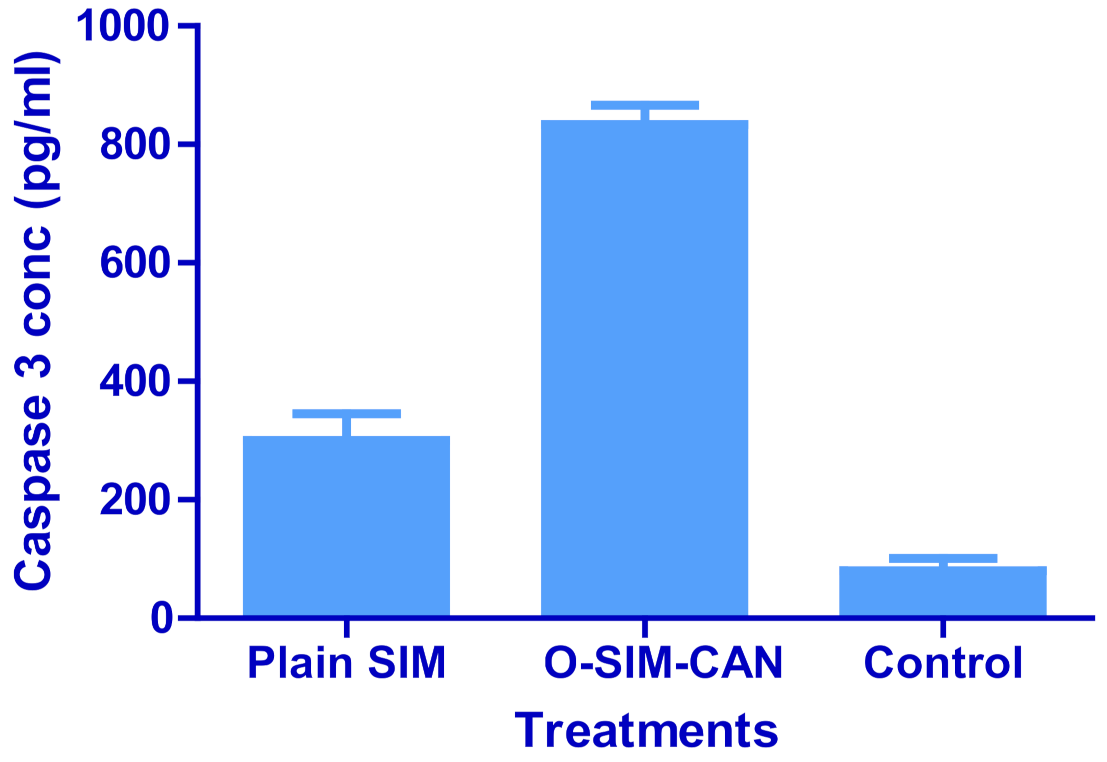
2.7. Caspase-3 Enzyme Assay
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. FTIR Studies
4.2.2. Preparation of SIM Loaded CA-NP
4.2.3. Optimization of SIM-CA-NP Preparation
4.2.4. Characterization
Estimation of PS and Polydispersity Index (PDI)
EE
4.2.5. Standardization and Validation of Optimization Outcome
4.2.6. The Rationale of Experimental Design
4.2.7. PDI and Zeta Potential Determination
4.2.8. SEM
4.2.9. Drug Release Studies
4.2.10. In Vitro Cell Viability Assay
4.2.11. Caspase-3 Enzyme Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jawahar, N.; Meyyanathan, S.N. Polymeric nanoparticles for drug delivery and targeting: A comprehensive review. Int. J. Health Allied Sci. 2012, 1, 217. [Google Scholar] [CrossRef]
- El-Say, K.M.; El-Sawy, H.S. Polymeric nanoparticles: Promising platform for drug delivery. Int. J. Pharm. 2017, 528, 675–691. [Google Scholar] [CrossRef]
- Witzigmann, D.; Kulkarni, J.A.; Leung, J.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev. 2020, 159, 344–363. [Google Scholar] [CrossRef] [PubMed]
- Poovi, G.; Damodharan, N. Lipid nanoparticles: A challenging approach for oral delivery of BCS Class-II drugs. Future J. Pharm. Sci. 2018, 4, 191–205. [Google Scholar] [CrossRef]
- Wu, K.; Su, D.; Liu, J.; Saha, R.; Wang, J.-P. Magnetic nanoparticles in nanomedicine: A review of recent advances. Nanotechnology 2019, 30, 502003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pansieri, J.; Gerstenmayer, M.; Lux, F.; Mériaux, S.; Tillement, O.; Forge, V.; Larrat, B.; Marquette, C. Magnetic Nanoparticles Applications for Amyloidosis Study and Detection: A Review. Nanomaterials 2018, 8, 740. [Google Scholar] [CrossRef] [Green Version]
- Panahi, Y.; Farshbaf, M.; Mohammadhosseini, M.; Mirahadi, M.; Khalilov, R.; Saghfi, S.; Akbarzadeh, A. Recent advances on liposomal nanoparticles: Synthesis, characterization and biomedical applications. Artif. Cells Nanomed. Biotechnol. 2017, 45, 788–799. [Google Scholar] [CrossRef] [Green Version]
- Rao, J.P.; Geckeler, K.E. Polymer nanoparticles: Preparation techniques and size-control parameters. Prog. Polym. Sci. 2011, 36, 887–913. [Google Scholar] [CrossRef]
- Liechty, W.B.; Kryscio, D.R.; Slaughter, B.V.; Peppas, N.A. Polymers for Drug Delivery Systems. Annu. Rev. Chem. Biomol. Eng. 2010, 1, 149–173. [Google Scholar] [CrossRef] [Green Version]
- Salleh, S.N.; Fairus, A.A.H.; Zahary, M.N.; Raj, N.B.; Jalil, A.M.M. Unravelling the Effects of Soluble Dietary Fibre Supplementation on Energy Intake and Perceived Satiety in Healthy Adults: Evidence from Systematic Review and Meta-Analysis of Randomised-Controlled Trials. Foods 2019, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Pawar, S.N.; Edgar, K.J. Alginate derivatization: A review of chemistry, properties and applications. Biomaterials 2012, 33, 3279–3305. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.; Sun, A.M. Microencapsulated Islets as Bioartificial Endocrine Pancreas. Science 1980, 210, 908–910. [Google Scholar] [CrossRef] [PubMed]
- Paques, J.P.; van der Linden, E.; van Rijn, C.J.; Sagis, L.M. Preparation methods of alginate nanoparticles. Adv. Colloid Interface Sci. 2014, 209, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Ching, S.H.; Bansal, N.; Bhandari, B. Alginate gel particles–A review of production techniques and physical properties. Crit. Rev. Food Sci. Nutr. 2015, 57, 1133–1152. [Google Scholar] [CrossRef]
- Kurakula, M.; Naveen, N.R. Electrospraying: A facile technology unfolding the chitosan based drug delivery and biomedical applications. Eur. Polym. J. 2021, 147, 110326. [Google Scholar] [CrossRef]
- Kurakula, M.; Naveen, N.R. Prospection of recent chitosan biomedical trends: Evidence from patent analysis (2009–2020). Int. J. Biol. Macromol. 2020, 165, 1924–1938. [Google Scholar] [CrossRef]
- De, S. Polymer relationships during preparation of chitosan–alginate and poly-l-lysine–alginate nanospheres. J. Control. Release 2003, 89, 101–112. [Google Scholar] [CrossRef]
- Motwani, S.K.; Chopra, S.; Talegaonkar, S.; Kohli, K.; Ahmad, F.; Khar, R.K. Chitosan–sodium alginate nanoparticles as submicroscopic reservoirs for ocular delivery: Formulation, optimisation and in vitro characterisation. Eur. J. Pharm. Biopharm. 2008, 68, 513–525. [Google Scholar] [CrossRef]
- Mi, F.-L.; Sung, H.-W.; Shyu, S.-S. Drug release from chitosan–alginate complex beads reinforced by a naturally occurring cross-linking agent. Carbohydr. Polym. 2002, 48, 61–72. [Google Scholar] [CrossRef]
- Balata, G.F.; Zidan, A.S.; Abourehab, M.A.; Essa, E.A. Rapid disintegrating tablets of simvastatin dispersions in polyoxyethylene–polypropylene block copolymer for maximized disintegration and dissolution. Drug Des. Dev. Ther. 2016, ume 10, 3211–3223. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Arnold, R.S.; Sun, C.Q.; Petros, J.A. A mitochondrial DNA mutation influences the apoptotic effect of statins on prostate cancer. Prostate 2015, 75, 1916–1925. [Google Scholar] [CrossRef]
- Yanae, M.; Tsubaki, M.; Satou, T.; Itoh, T.; Imano, M.; Yamazoe, Y.; Nishida, S. Statin-induced apoptosis via the suppression of ERK1/2 and Akt activation by inhibition of the geranylgeranyl-pyrophosphate biosynthesis in glioblastoma. J. Exp. Clin. Cancer Res. 2011, 30, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Politis, S.N.; Colombo, P.; Colombo, G.; Rekkas, D.M. Design of experiments (DoE) in pharmaceutical development. Drug Dev. Ind. Pharm. 2017, 43, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Dai, Y.-N.; Zhang, J.-P.; Wang, A.-Q.; Wei, Q. Chitosan-Alginate Nanoparticles as a Novel Drug Delivery System for Nifedipine. Int. J. Biomed. Sci. 2008, 4, 221–228. [Google Scholar] [PubMed]
- Kurakula, M.; Naveen, N.R. In Situ Gel Loaded with Chitosan-Coated Simvastatin Nanoparticles: Promising Delivery for Effective Anti-Proliferative Activity against Tongue Carcinoma. Mar. Drugs 2020, 18, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarathchandiran, I.; Koumaravelou, K.; Selvasudha, N. Interaction pattern and in vitro, in vivo release behavior of simvastatin-loaded chitosan nanoformulation. Drug Dev. Ind. Pharm. 2019, 45, 1725–1739. [Google Scholar] [CrossRef]
- Zhang, H.; Choi, J.P.; Moon, S.K.; Ngo, T.H. A hybrid multi-objective optimization of aerosol jet printing process via response surface methodology. Addit. Manuf. 2020, 33, 101096. [Google Scholar] [CrossRef]
- Ahmed, O.A.A.; Kurakula, M.; Banjar, Z.M.; Afouna, M.I.; Zidan, A.S. Quality by Design Coupled with Near Infrared in Formulation of Transdermal Glimepiride Liposomal Films. J. Pharm. Sci. 2015, 104, 2062–2075. [Google Scholar] [CrossRef]
- Singh, B.; Kapil, R.; Nandi, M.; Ahuja, N. Developing oral drug delivery systems using formulation by design: Vital precepts, retrospect and prospects. Expert Opin. Drug Deliv. 2011, 8, 1341–1360. [Google Scholar] [CrossRef]
- Adetunji, A.I.; Olaniran, A.O. Statistical modelling and optimization of protease production by an autochthonous Bacillus aryabhattai Ab15-ES: A response surface methodology approach. Biocatal. Agric. Biotechnol. 2020, 24, 101528. [Google Scholar] [CrossRef]
- Alhakamy, N.; Ahmed, O.; Kurakula, M.; Caruso, G.; Caraci, F.; Asfour, H.; Alfarsi, A.; Eid, B.; Mohamed, A.; Alruwaili, N.; et al. Chitosan-Based Microparticles Enhance Ellagic Acid’s Colon Targeting and Proapoptotic Activity. Pharmaceutics 2020, 12, 652. [Google Scholar] [CrossRef] [PubMed]
- Kurakula, M.; Naveen, N.R.; Patel, B.; Manne, R.; Patel, D.B. Preparation, Optimization and Evaluation of Chitosan-Based Avanafil Nanocomplex Utilizing Antioxidants for Enhanced Neuroprotective Effect on PC12 Cells. Gels 2021, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Katuwavila, N.P.; Perera, A.D.L.C.; Samarakoon, S.R.; Soysa, P.; Karunaratne, V.; Amaratunga, G.A.J.; Karunaratne, D.N. Chitosan-Alginate Nanoparticle System Efficiently Delivers Doxorubicin to MCF-7 Cells. J. Nanomater. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Naveen, N.R.; Gopinath, C.; Rao, D.S. Design expert supported mathematical optimization of repaglinide gastroretentive floating tablets: In vitro and in vivo evaluation. Future J. Pharm. Sci. 2017, 3, 140–147. [Google Scholar] [CrossRef]
- Naveen, N.R.; Kurakula, M.; Gowthami, B. Process optimization by response surface methodology for preparation and evaluation of methotrexate loaded chitosan nanoparticles. Mater. Today Proc. 2020, 33, 2716–2724. [Google Scholar] [CrossRef]
- Kumar, V.D.; Verma, P.R.P.; Singh, S.K. Development and evaluation of biodegradable polymeric nanoparticles for the effective delivery of quercetin using a quality by design approach. LWT 2015, 61, 330–338. [Google Scholar] [CrossRef]
- Bhatia, M.; Ahuja, M. Psyllium arabinoxylan: Carboxymethylation, characterization and evaluation for nanoparticulate drug delivery. Int. J. Biol. Macromol. 2015, 72, 495–501. [Google Scholar] [CrossRef]
- Sarin, N.; Kurakula, M.; Singh, K.; Kumar, A.; Singh, D.; Arora, S. Strontium and selenium doped bioceramics incorporated polyacrylamide-carboxymethylcellulose hydrogel scaffolds: Mimicking key features of bone regeneration. J. Asian Ceram. Soc. 2021, 9, 531–548. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Hsieh, P.-L.; Liao, Y.-W.; Peng, C.-Y.; Lu, M.-Y.; Yang, C.-H.; Yu, C.-C.; Liu, C.-M. Berberine-targeted miR-21 chemosensitizes oral carcinomas stem cells. Oncotarget 2017, 8, 80900–80908. [Google Scholar] [CrossRef] [Green Version]



| S. No | Wavenumber Identified in SIM (cm−1) | Functional Group Assigned | Wavenumber Identified in Formulation (cm−1) |
|---|---|---|---|
| 1. | 3550 | O-H Stretch | 3559 |
| 2. | 2931 | C-H stretch: Methyl/methylene asymmetric | 2942 |
| 3. | 1465 | Methylene symmetric C-H bend | 1472 |
| 4. | 1267 | -C-O-C bend (Lactone) | 1271 |
| 5. | 1165 | -C-O-C bend (Ester) | 1169 |
| 6. | 1072 | C-O Stretch (secondary alcohol) | 1073 |
| 7. | 1566 | N-H stretch | 1569 |
| Factor 1 | Factor 2 | Response 1 | Response 2 | ||
|---|---|---|---|---|---|
| Std | Run | A: Chitosan | B: Sodium Alginate | PS | EE |
| (g) | (g) | (nm) | (%) | ||
| 10 | 1 | 0.25 | 0.3 | 109 | 66 |
| 4 | 2 | 0.4 | 0.5 | 296 | 79 |
| 2 | 3 | 0.4 | 0.1 | 110 | 41 |
| 6 | 4 | 0.462132 | 0.3 | 219 | 56 |
| 3 | 5 | 0.1 | 0.5 | 309 | 87 |
| 11 | 6 | 0.25 | 0.3 | 118 | 71 |
| 12 | 7 | 0.25 | 0.3 | 132 | 67 |
| 7 | 8 | 0.25 | 0.0171573 | 206 | 37 |
| 9 | 9 | 0.25 | 0.3 | 127 | 78 |
| 5 | 10 | 0.037868 | 0.3 | 307 | 60 |
| 8 | 11 | 0.25 | 0.582843 | 351 | 91 |
| 1 | 12 | 0.1 | 0.1 | 209 | 46 |
| 13 | 13 | 0.25 | 0.3 | 128 | 70 |
| Responses | Source | Sequential p-Value | Lack of Fit p-Value | Adjusted R² | Predicted R² | |
|---|---|---|---|---|---|---|
| Lag Time | Linear | 0.0811 | 0.0002 | 0.2738 | 0.0095 | |
| 2FI | 0.5958 | 0.0002 | 0.2194 | −0.0863 | ||
| Quadratic | 0.0001 | 0.2114 | 0.9209 | 0.8919 | Suggested | |
| Cubic | 0.5849 | 0.0037 | 0.9106 | −1.1565 | Aliased | |
| T- 95% CDR | Linear | <0.0001 | 0.2628 | 0.8750 | 0.8363 | |
| 2FI | 0.8162 | 0.2116 | 0.8620 | 0.8172 | ||
| Quadratic | 0.0152 | 0.8480 | 0.9463 | 0.9223 | Suggested | |
| Cubic | 0.8315 | 0.5374 | 0.9302 | 0.7696 |
| Parameter | PS | EE |
|---|---|---|
| Std. Dev. | 24.90 | 3.91 |
| Mean | 201.62 | 65.31 |
| C.V. % | 7.35 | 5.98 |
| Adeq Precision | 13.1592 | 20.6875 |
| Lack of Fit F-value | 15.55 | 12.2650 |
| Lack of Fit p-value | 0.0614 | 0.8480 |
| Model F value | 28.92 | 43.33 |
| Model p value | 0.0002 | <0.0001 |
| Intercept | A | B | AB | A² | B² | |
|---|---|---|---|---|---|---|
| PS | 122.8 | −29.5563 | 61.3826 | 21.5 | 60.1625 | 67.9125 |
| p-values | 0.0121 | 0.0002 | 0.1279 | 0.0004 | 0.0002 | |
| EE | 70.4 | −2.33211 | 19.4209 | −0.75 | −5.6375 | −2.6375 |
| p-values | 0.1353 | <0.0001 | 0.7125 | 0.0067 | 0.1183 |
| S. No | Response | Predicted/Theoretical Value | Experimental/Practical Value | Relative Error (%) | Limit for Relative Error (%) |
|---|---|---|---|---|---|
| 1. | PS (nm) | 142.56 | 143.23 | −0.47 | ±5 |
| 2. | EE (%) | 75.18 | 74.72 | 0.61 |
| Selected Formulation Factors | Levels | Responses/Dependent Variables | Constraints | ||||
|---|---|---|---|---|---|---|---|
| −1.141 | +1 | 0 | +1 | +1.141 | |||
| Chitosan (g)-X1 | 0.037 | 0.1 | 0.25 | 0.4 | 0.462 | Particle size (nm) | Minimum |
| Eudragit RLPO (g)-X2 | 0.017 | 0.1 | 0.3 | 0.5 | 0.582 | EE (%) | Maximum |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rizg, W.Y.; Naveen, N.R.; Kurakula, M.; Bukhary, H.A.; Safhi, A.Y.; Alfayez, E.; Sindi, A.M.; Ali, S.; Murshid, S.S.; Hosny, K.M. QbD Supported Optimization of the Alginate-Chitosan Nanoparticles of Simvastatin in Enhancing the Anti-Proliferative Activity against Tongue Carcinoma. Gels 2022, 8, 103. https://doi.org/10.3390/gels8020103
Rizg WY, Naveen NR, Kurakula M, Bukhary HA, Safhi AY, Alfayez E, Sindi AM, Ali S, Murshid SS, Hosny KM. QbD Supported Optimization of the Alginate-Chitosan Nanoparticles of Simvastatin in Enhancing the Anti-Proliferative Activity against Tongue Carcinoma. Gels. 2022; 8(2):103. https://doi.org/10.3390/gels8020103
Chicago/Turabian StyleRizg, Waleed Y., N. Raghavendra Naveen, Mallesh Kurakula, Haitham A. Bukhary, Awaji Y. Safhi, Eman Alfayez, Amal M. Sindi, Sarah Ali, Samar S. Murshid, and Khaled M. Hosny. 2022. "QbD Supported Optimization of the Alginate-Chitosan Nanoparticles of Simvastatin in Enhancing the Anti-Proliferative Activity against Tongue Carcinoma" Gels 8, no. 2: 103. https://doi.org/10.3390/gels8020103
APA StyleRizg, W. Y., Naveen, N. R., Kurakula, M., Bukhary, H. A., Safhi, A. Y., Alfayez, E., Sindi, A. M., Ali, S., Murshid, S. S., & Hosny, K. M. (2022). QbD Supported Optimization of the Alginate-Chitosan Nanoparticles of Simvastatin in Enhancing the Anti-Proliferative Activity against Tongue Carcinoma. Gels, 8(2), 103. https://doi.org/10.3390/gels8020103






