Autophagy Is a Crucial Path in Chondrogenesis of Adipose-Derived Mesenchymal Stromal Cells Laden in Hydrogel
Abstract
1. Introduction
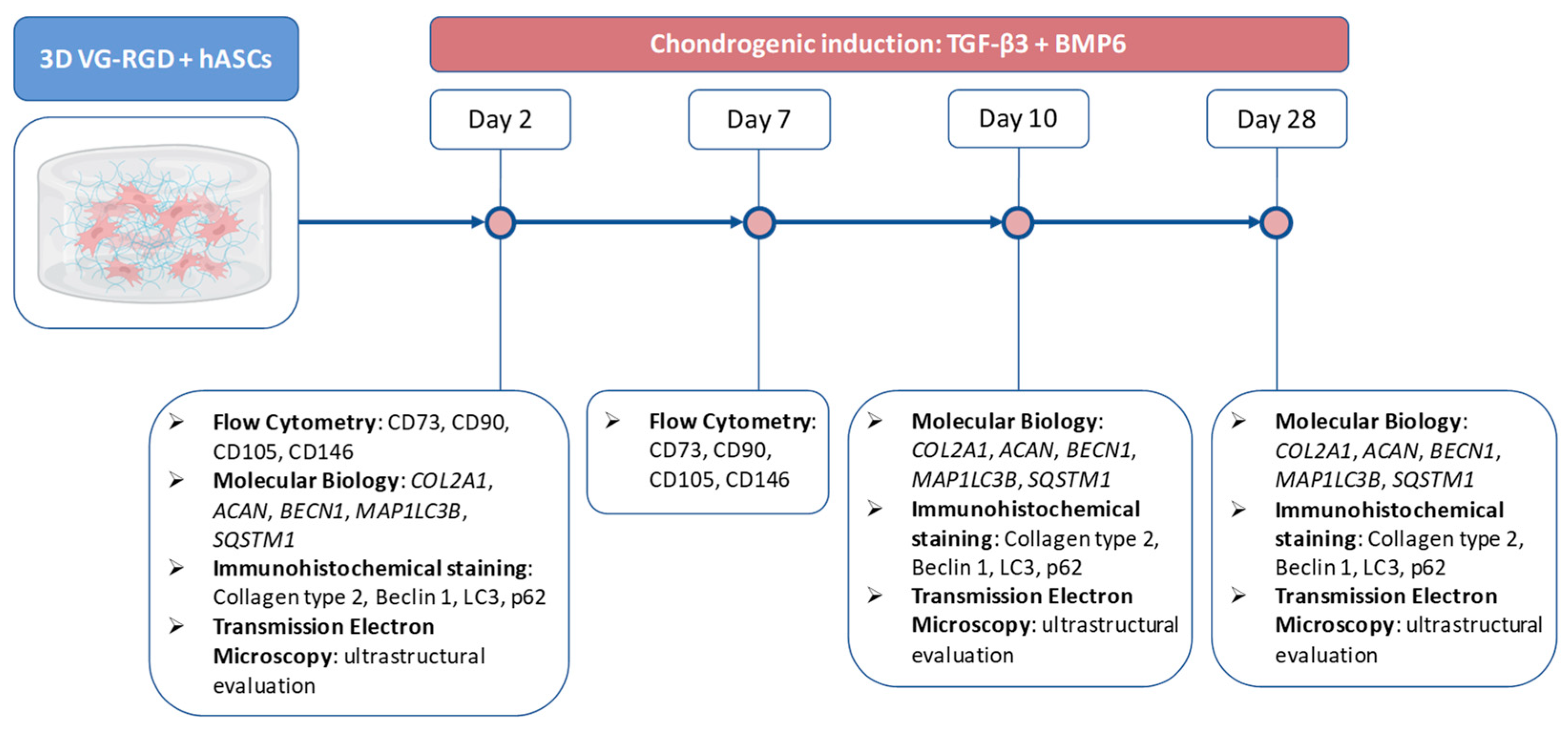
2. Results and Discussion
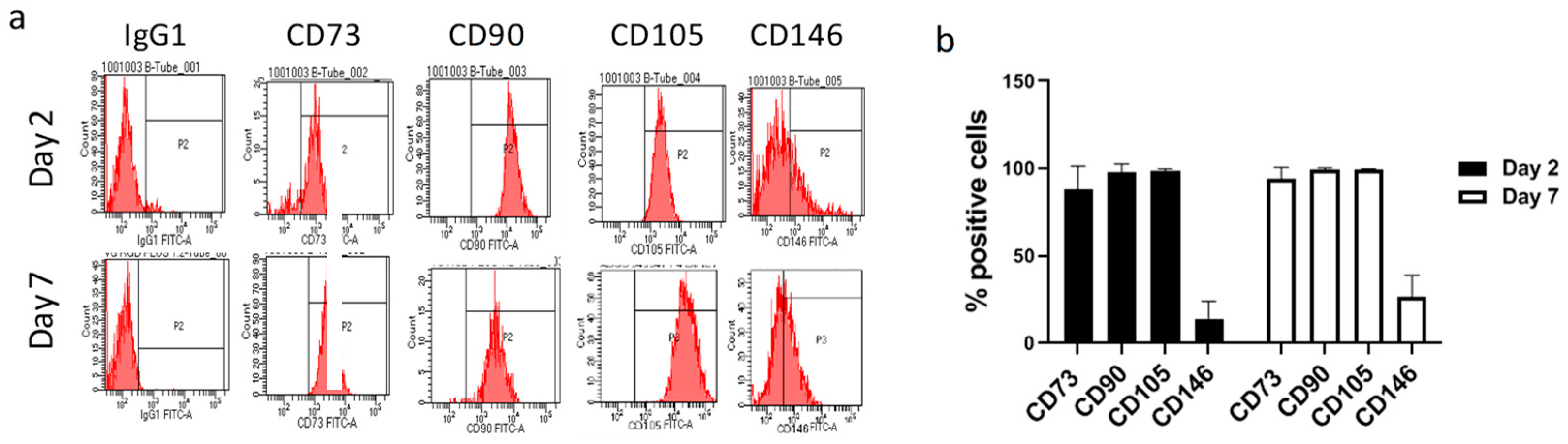
2.1. VG-RGD Hydrogel Retains hASCs Typical Markers Well
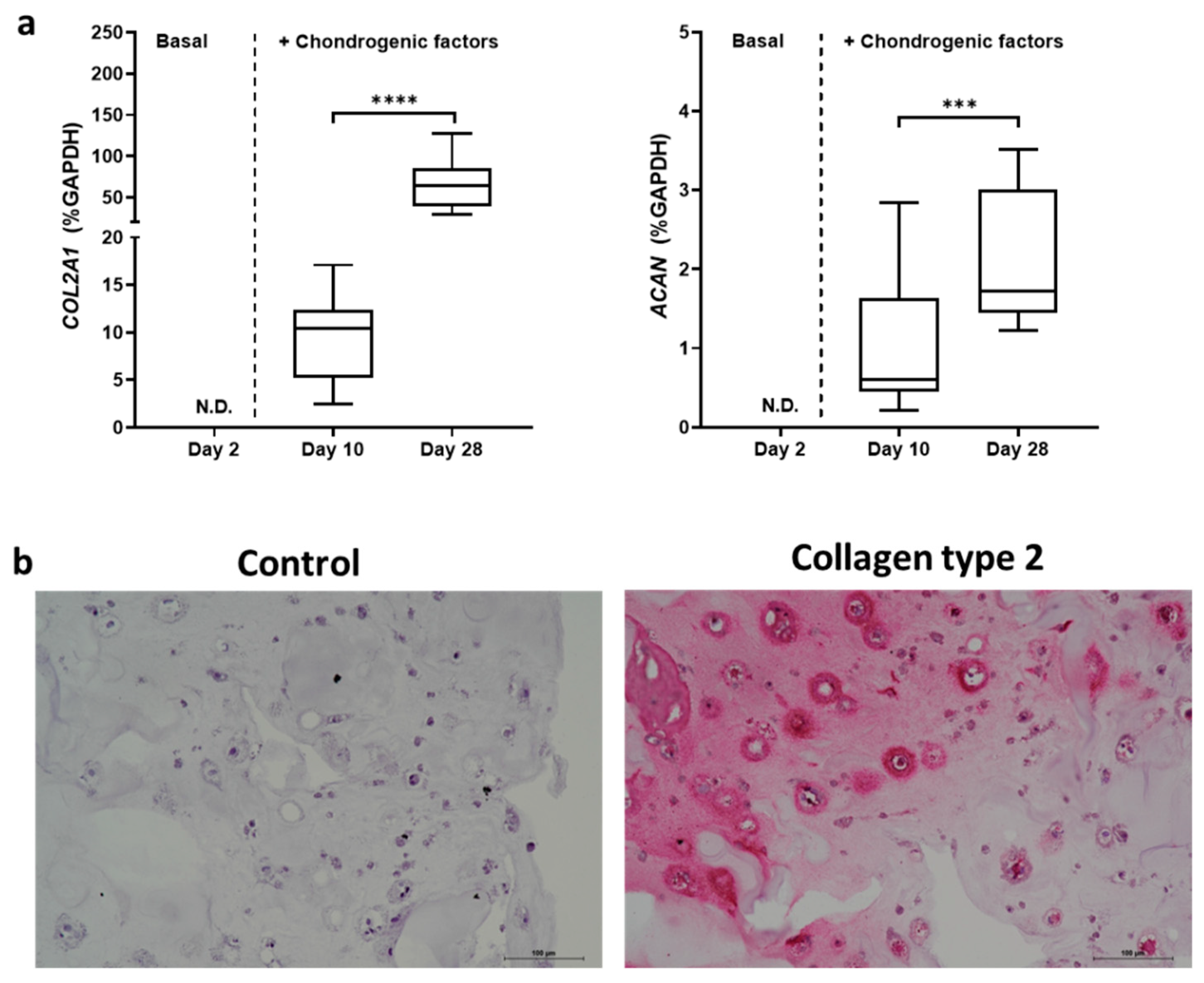
2.2. TGFβ3 Combined with BMP6 Support the Chondrogenic Processes Well
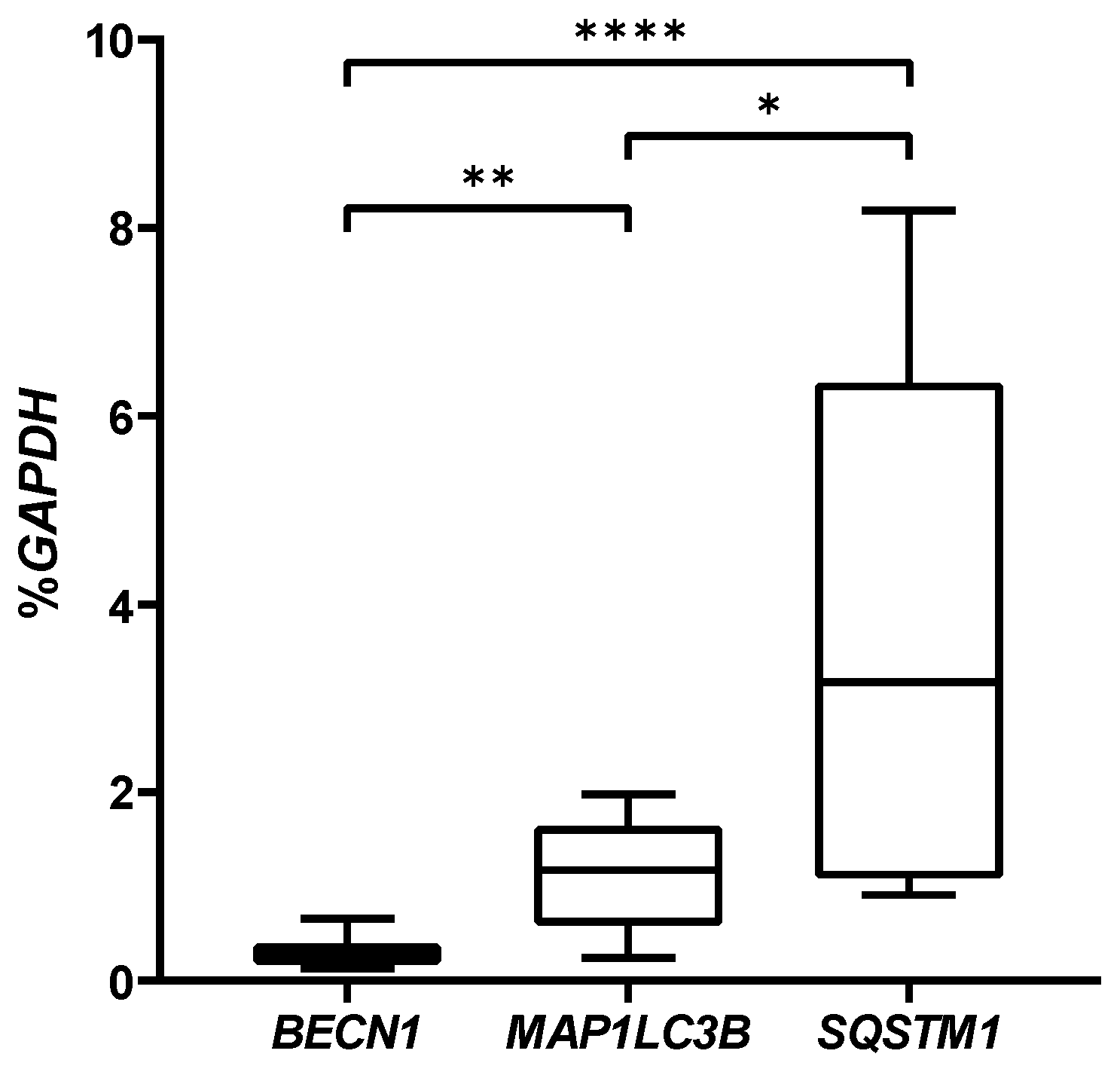
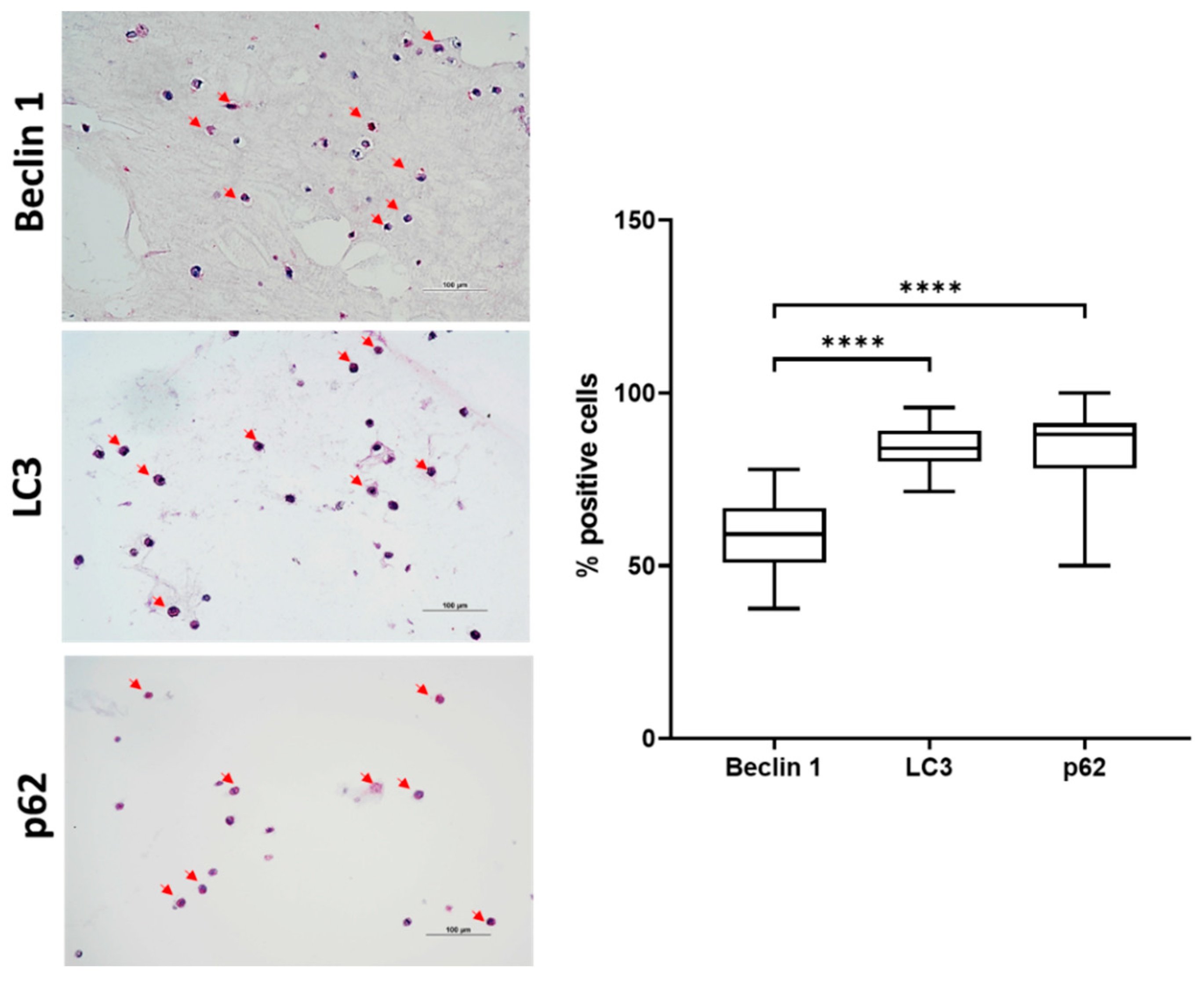
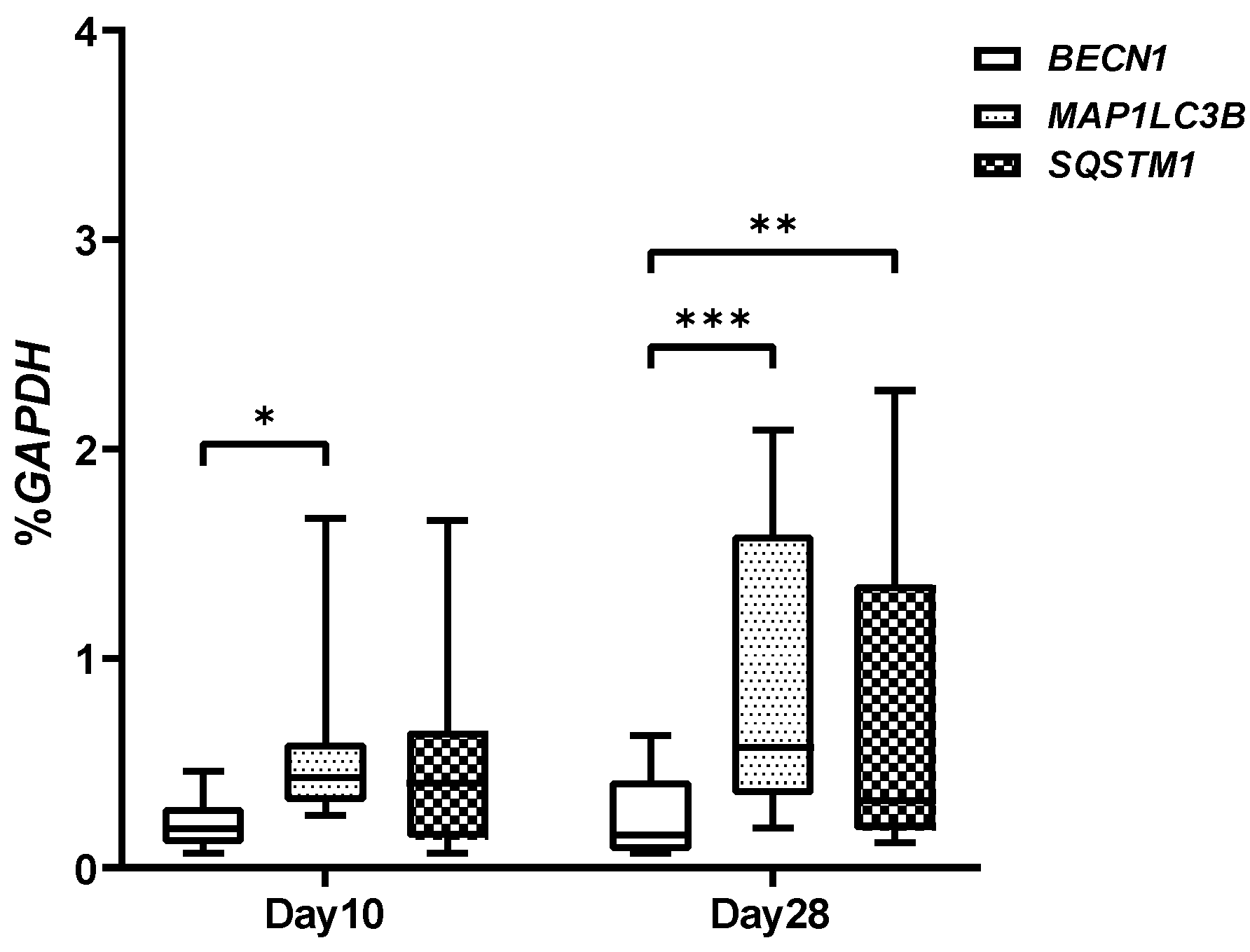
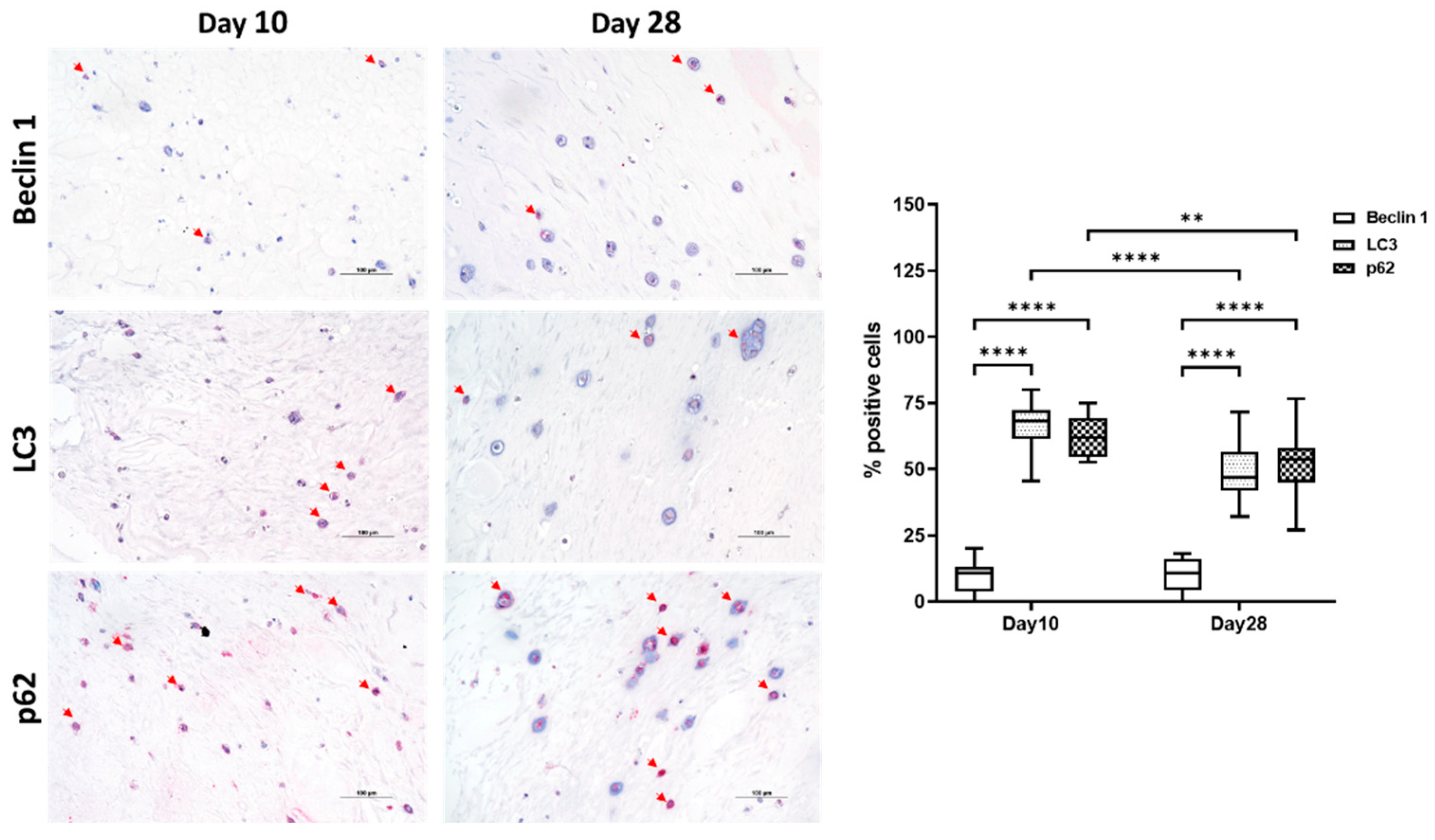
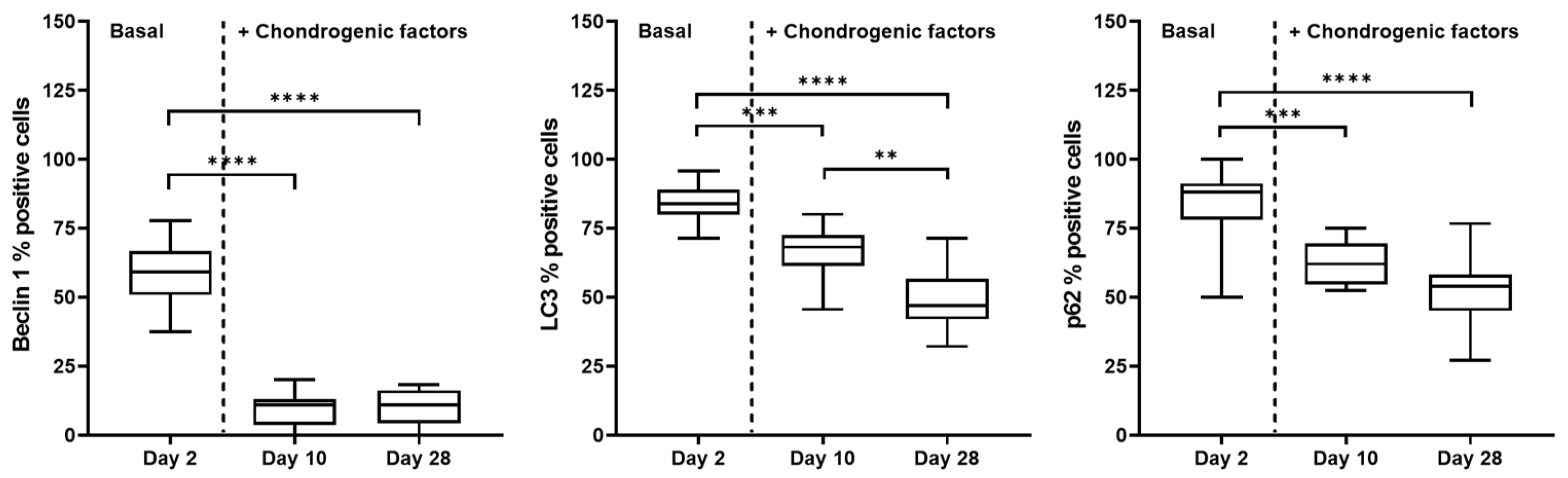
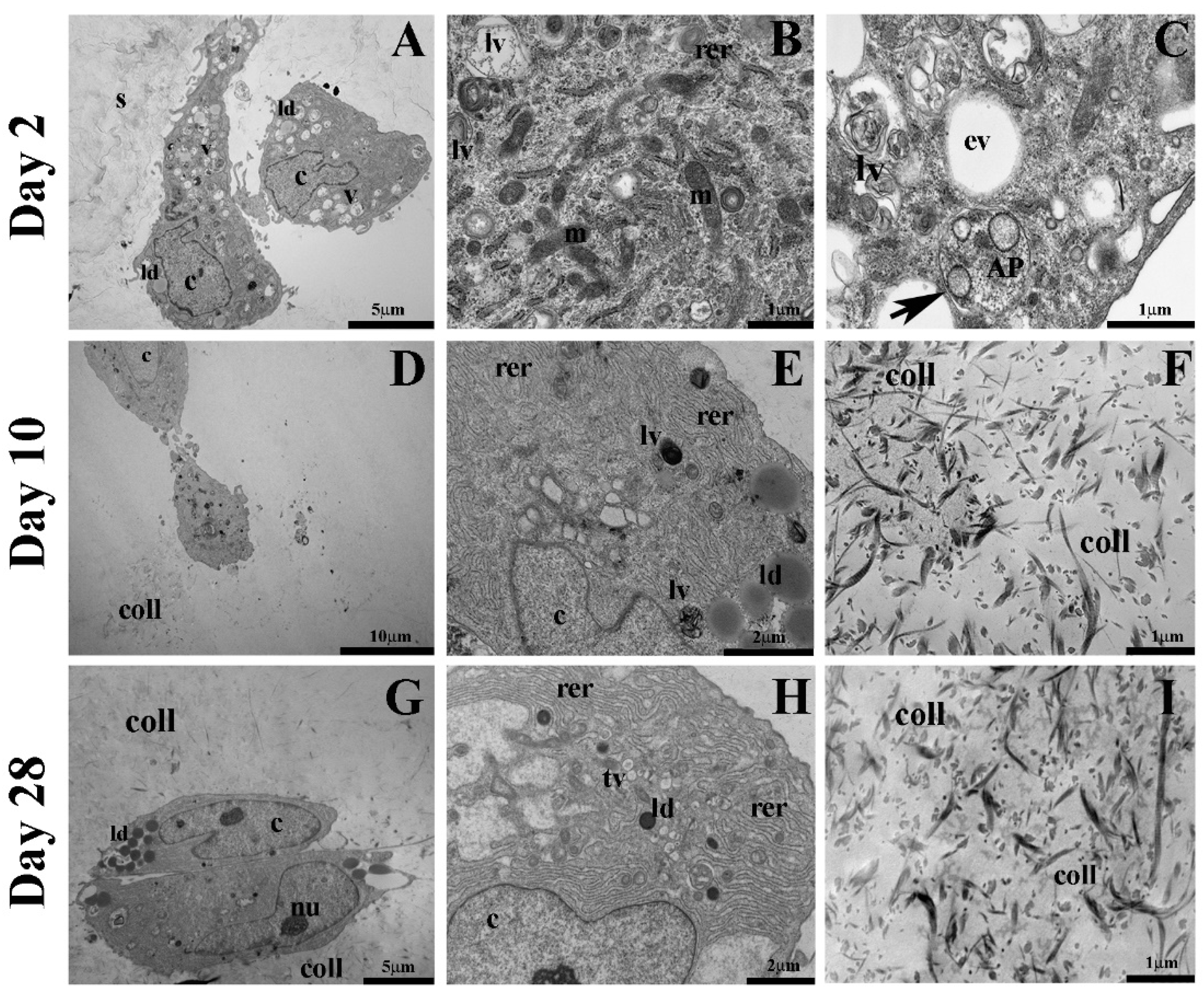
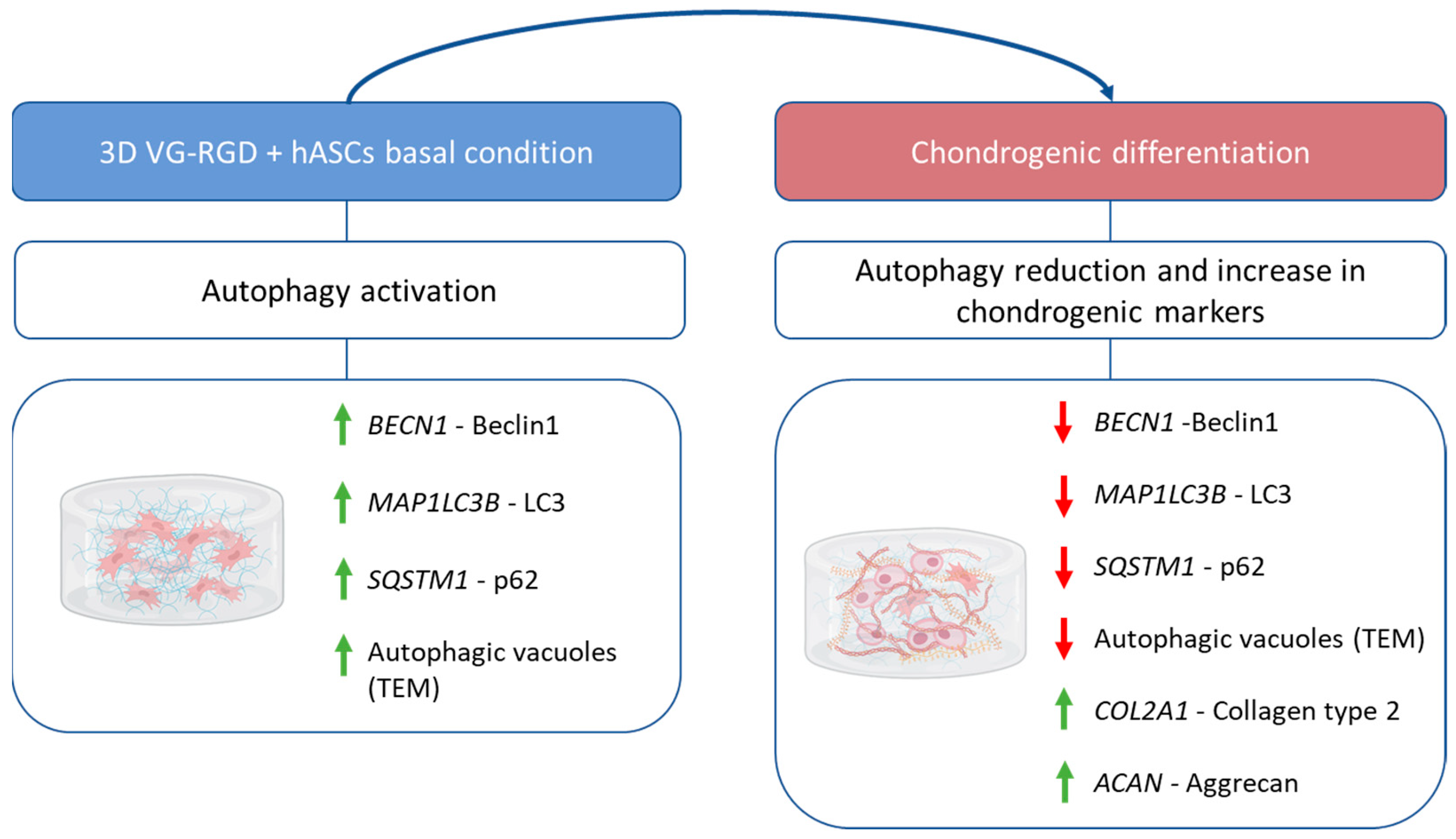
2.3. Autophagic Markers Modulation Associated with Chondrogenic Differentiation
3. Conclusions
4. Materials and Methods
4.1. Human ASC Culture
4.2. hASCs Embedding in Hydrogel
4.3. Molecular Biology
4.4. Immunohistochemical Staining
4.5. Transmission Electron Microscopy
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gomez-Virgilio, L.; Silva-Lucero, M.D.; Flores-Morelos, D.S.; Gallardo-Nieto, J.; Lopez-Toledo, G.; Abarca-Fernandez, A.M.; Zacapala-Gomez, A.E.; Luna-Munoz, J.; Montiel-Sosa, F.; Soto-Rojas, L.O.; et al. Autophagy: A Key Regulator of Homeostasis and Disease: An Overview of Molecular Mechanisms and Modulators. Cells 2022, 11, 2262. [Google Scholar] [CrossRef]
- Ugland, H.; Naderi, S.; Brech, A.; Collas, P.; Blomhoff, H.K. cAMP induces autophagy via a novel pathway involving ERK, cyclin E and Beclin 1. Autophagy 2011, 7, 1199–1211. [Google Scholar] [CrossRef]
- Ichimiya, T.; Yamakawa, T.; Hirano, T.; Yokoyama, Y.; Hayashi, Y.; Hirayama, D.; Wagatsuma, K.; Itoi, T.; Nakase, H. Autophagy and Autophagy-Related Diseases: A Review. Int. J. Mol. Sci. 2020, 21, 8974. [Google Scholar] [CrossRef]
- Nakamura, S.; Yoshimori, T. New insights into autophagosome-lysosome fusion. J. Cell Sci. 2017, 130, 1209–1216. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef]
- Levine, B.; Deretic, V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007, 7, 767–777. [Google Scholar] [CrossRef]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef]
- Stolz, A.; Ernst, A.; Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell. Biol. 2014, 16, 495–501. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef]
- Jung, M.; Choi, H.; Mun, J.Y. The autophagy research in electron microscopy. Appl. Microsc. 2019, 49, 11. [Google Scholar] [CrossRef]
- Cecconi, F.; Levine, B. The role of autophagy in mammalian development: Cell makeover rather than cell death. Dev. Cell 2008, 15, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Moss, J.J.; Wirth, M.; Tooze, S.A.; Lane, J.D.; Hammond, C.L. Autophagy coordinates chondrocyte development and early joint formation in zebrafish. FASEB J. 2021, 35, e22002. [Google Scholar] [CrossRef] [PubMed]
- Ramesova, A.; Svandova, E.; Vesela, B.; Vacek, L.; Lesot, H.; Matalova, E. Autophagy-related proteases accompany the transition of pre-chondrogenic cells into chondroblasts. Ann. Anat. 2022, 239, 151781. [Google Scholar] [CrossRef]
- Horigome, Y.; Ida-Yonemochi, H.; Waguri, S.; Shibata, S.; Endo, N.; Komatsu, M. Loss of autophagy in chondrocytes causes severe growth retardation. Autophagy 2020, 16, 501–511. [Google Scholar] [CrossRef]
- Liu, W.; Luo, H.; Wang, R.; Kang, Y.; Liao, W.; Sun, Y.; Chen, G.; Shao, L. Rapamycin-Induced Autophagy Promotes the Chondrogenic Differentiation of Synovium-Derived Mesenchymal Stem Cells in the Temporomandibular Joint in Response to IL-1beta. Biomed. Res. Int. 2020, 2020, 4035306. [Google Scholar] [CrossRef] [PubMed]
- Vuppalapati, K.K.; Bouderlique, T.; Newton, P.T.; Kaminskyy, V.O.; Wehtje, H.; Ohlsson, C.; Zhivotovsky, B.; Chagin, A.S. Targeted Deletion of Autophagy Genes Atg5 or Atg7 in the Chondrocytes Promotes Caspase-Dependent Cell Death and Leads to Mild Growth Retardation. J. Bone Miner. Res. 2015, 30, 2249–2261. [Google Scholar] [CrossRef] [PubMed]
- Bouderlique, T.; Vuppalapati, K.K.; Newton, P.T.; Li, L.; Barenius, B.; Chagin, A.S. Targeted deletion of Atg5 in chondrocytes promotes age-related osteoarthritis. Ann. Rheum. Dis. 2016, 75, 627–631. [Google Scholar] [CrossRef]
- Kao, W.C.; Chen, J.C.; Liu, P.C.; Lu, C.C.; Lin, S.Y.; Chuang, S.C.; Wu, S.C.; Chang, L.H.; Lee, M.J.; Yang, C.D.; et al. The Role of Autophagy in Osteoarthritic Cartilage. Biomolecules 2022, 12, 1357. [Google Scholar] [CrossRef]
- Valenti, M.T.; Dalle Carbonare, L.; Zipeto, D.; Mottes, M. Control of the Autophagy Pathway in Osteoarthritis: Key Regulators, Therapeutic Targets and Therapeutic Strategies. Int. J. Mol. Sci. 2021, 22, 2700. [Google Scholar] [CrossRef] [PubMed]
- Jakovljevic, J.; Harrell, C.R.; Fellabaum, C.; Arsenijevic, A.; Jovicic, N.; Volarevic, V. Modulation of autophagy as new approach in mesenchymal stem cell-based therapy. Biomed. Pharmacother. 2018, 104, 404–410. [Google Scholar] [CrossRef]
- Wang, L.; Fan, J.; Lin, Y.S.; Guo, Y.S.; Gao, B.; Shi, Q.Y.; Wei, B.Y.; Chen, L.; Yang, L.; Liu, J.; et al. Glucocorticoids induce autophagy in rat bone marrow mesenchymal stem cells. Mol. Med. Rep. 2015, 11, 2711–2716. [Google Scholar] [CrossRef] [PubMed]
- Sin, J.; Andres, A.M.; Taylor, D.J.; Weston, T.; Hiraumi, Y.; Stotland, A.; Kim, B.J.; Huang, C.; Doran, K.S.; Gottlieb, R.A. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 2016, 12, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.P.; Lai, W.Y.; Lin, T.W.; Lin, Y.Y.; Chien, Y.; Tsai, Y.C.; Tai, H.Y.; Wang, C.L.; Liu, Y.Y.; Huang, P.I.; et al. Autophagy reprogramming stem cell pluripotency and multiple-lineage differentiation. J. Chin. Med. Assoc. 2022, 85, 667–671. [Google Scholar] [CrossRef]
- Cao, Z.; Huang, S.; Dou, C.; Xiang, Q.; Dong, S. Cyanidin suppresses autophagic activity regulating chondrocyte hypertrophic differentiation. J. Cell. Physiol. 2018, 233, 2332–2342. [Google Scholar] [CrossRef]
- Wang, X.; Lin, Q.; Zhang, T.; Wang, X.; Cheng, K.; Gao, M.; Xia, P.; Li, X. Low-intensity pulsed ultrasound promotes chondrogenesis of mesenchymal stem cells via regulation of autophagy. Stem Cell Res. Ther. 2019, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, M.; Guo, Q.Y.; Lu, S.B.; Peng, J. Mesenchymal Stem Cells for Treating Articular Cartilage Defects and Osteoarthritis. Cell Transpl. 2015, 24, 1661–1678. [Google Scholar] [CrossRef] [PubMed]
- Somoza, R.A.; Welter, J.F.; Correa, D.; Caplan, A.I. Chondrogenic differentiation of mesenchymal stem cells: Challenges and unfulfilled expectations. Tissue Eng. Part B Rev. 2014, 20, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Estes, B.T.; Diekman, B.O.; Gimble, J.M.; Guilak, F. Isolation of adipose-derived stem cells and their induction to a chondrogenic phenotype. Nat. Protoc. 2010, 5, 1294–1311. [Google Scholar] [CrossRef] [PubMed]
- Puetzer, J.L.; Petitte, J.N.; Loboa, E.G. Comparative review of growth factors for induction of three-dimensional in vitro chondrogenesis in human mesenchymal stem cells isolated from bone marrow and adipose tissue. Tissue Eng. Part B Rev. 2010, 16, 435–444. [Google Scholar] [CrossRef]
- Deng, Y.; Sun, A.X.; Overholt, K.J.; Yu, G.Z.; Fritch, M.R.; Alexander, P.G.; Shen, H.; Tuan, R.S.; Lin, H. Enhancing chondrogenesis and mechanical strength retention in physiologically relevant hydrogels with incorporation of hyaluronic acid and direct loading of TGF-beta. Acta Biomater. 2019, 83, 167–176. [Google Scholar] [CrossRef]
- Levinson, C.; Lee, M.; Applegate, L.A.; Zenobi-Wong, M. An injectable heparin-conjugated hyaluronan scaffold for local delivery of transforming growth factor beta1 promotes successful chondrogenesis. Acta Biomater. 2019, 99, 168–180. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Liu, Y.; Li, D.; Zhang, L.; Wang, Q.; Xiao, Y.; Zhang, X. Influence of hydrogel network microstructures on mesenchymal stem cell chondrogenesis in vitro and in vivo. Acta Biomater. 2019, 91, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nan, L.P.; Zhou, S.F.; Liu, Y.; Wang, Z.Y.; Wang, J.C.; Feng, X.M.; Zhang, L. Injectable Hydrogel Combined with Nucleus Pulposus-Derived Mesenchymal Stem Cells for the Treatment of Degenerative Intervertebral Disc in Rats. Stem Cells Int. 2019, 2019, 8496025. [Google Scholar] [CrossRef] [PubMed]
- Manferdini, C.; Trucco, D.; Saleh, Y.; Gabusi, E.; Dolzani, P.; Lenzi, E.; Vannozzi, L.; Ricotti, L.; Lisignoli, G. RGD-Functionalized Hydrogel Supports the Chondrogenic Commitment of Adipose Mesenchymal Stromal Cells. Gels 2022, 8, 382. [Google Scholar] [CrossRef]
- Li, Z.; Cao, B.; Wang, X.; Ye, K.; Li, S.; Ding, J. Effects of RGD nanospacing on chondrogenic differentiation of mesenchymal stem cells. J. Mater. Chem. B 2015, 3, 5197–5209. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Ni, Y.; Liu, B.; Zhou, T.; Yu, C.; Su, Y.; Zhu, X.; Yu, X.; Zhou, Y. Self-crosslinking and injectable hyaluronic acid/RGD-functionalized pectin hydrogel for cartilage tissue engineering. Carbohydr. Polym. 2017, 166, 31–44. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Kollmer, M.; Keskar, V.; Hauk, T.G.; Collins, J.M.; Russell, B.; Gemeinhart, R.A. Stem cell-derived extracellular matrix enables survival and multilineage differentiation within superporous hydrogels. Biomacromolecules 2012, 13, 963–973. [Google Scholar] [CrossRef]
- Rastaldo, R.; Vitale, E.; Giachino, C. Dual Role of Autophagy in Regulation of Mesenchymal Stem Cell Senescence. Front. Cell Dev. Biol. 2020, 8, 276. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hong, Y.; Zhang, H.; Li, X. Mesenchymal Stem Cell Senescence and Rejuvenation: Current Status and Challenges. Front. Cell Dev. Biol. 2020, 8, 364. [Google Scholar] [CrossRef] [PubMed]
- Marycz, K.; Kornicka, K.; Grzesiak, J.; Smieszek, A.; Szlapka, J. Macroautophagy and Selective Mitophagy Ameliorate Chondrogenic Differentiation Potential in Adipose Stem Cells of Equine Metabolic Syndrome: New Findings in the Field of Progenitor Cells Differentiation. Oxid. Med. Cell. Longev. 2016, 2016, 3718468. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef] [PubMed]
- Heo, D.N.; Kim, H.J.; Lee, D.; Kim, H.; Lee, S.J.; Lee, H.R.; Kwon, I.K.; Do, S.H. Comparison of polysaccharides in articular cartilage regeneration associated with chondrogenic and autophagy-related gene expression. Int. J. Biol. Macromol. 2020, 146, 922–930. [Google Scholar] [CrossRef]
- Oliver, L.; Hue, E.; Priault, M.; Vallette, F.M. Basal autophagy decreased during the differentiation of human adult mesenchymal stem cells. Stem Cells Dev. 2012, 21, 2779–2788. [Google Scholar] [CrossRef]
- Lee, Y.; Jung, J.; Cho, K.J.; Lee, S.K.; Park, J.W.; Oh, I.H.; Kim, G.J. Increased SCF/c-kit by hypoxia promotes autophagy of human placental chorionic plate-derived mesenchymal stem cells via regulating the phosphorylation of mTOR. J. Cell. Biochem. 2013, 114, 79–88. [Google Scholar] [CrossRef]
- Nuschke, A.; Rodrigues, M.; Stolz, D.B.; Chu, C.T.; Griffith, L.; Wells, A. Human mesenchymal stem cells/multipotent stromal cells consume accumulated autophagosomes early in differentiation. Stem Cell Res. Ther. 2014, 5, 140. [Google Scholar] [CrossRef]
- Ceccariglia, S.; Cargnoni, A.; Silini, A.R.; Parolini, O. Autophagy: A potential key contributor to the therapeutic action of mesenchymal stem cells. Autophagy 2020, 16, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Menshikov, M.; Zubkova, E.; Stafeev, I.; Parfyonova, Y. Autophagy, Mesenchymal Stem Cell Differentiation, and Secretion. Biomedicines 2021, 9, 1178. [Google Scholar] [CrossRef]
- Manferdini, C.; Gabusi, E.; Sartore, L.; Dey, K.; Agnelli, S.; Almici, C.; Bianchetti, A.; Zini, N.; Russo, D.; Re, F.; et al. Chitosan-based scaffold counteracts hypertrophic and fibrotic markers in chondrogenic differentiated mesenchymal stromal cells. J. Tissue Eng. Regen. Med. 2019, 13, 1896–1911. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabusi, E.; Lenzi, E.; Manferdini, C.; Dolzani, P.; Columbaro, M.; Saleh, Y.; Lisignoli, G. Autophagy Is a Crucial Path in Chondrogenesis of Adipose-Derived Mesenchymal Stromal Cells Laden in Hydrogel. Gels 2022, 8, 766. https://doi.org/10.3390/gels8120766
Gabusi E, Lenzi E, Manferdini C, Dolzani P, Columbaro M, Saleh Y, Lisignoli G. Autophagy Is a Crucial Path in Chondrogenesis of Adipose-Derived Mesenchymal Stromal Cells Laden in Hydrogel. Gels. 2022; 8(12):766. https://doi.org/10.3390/gels8120766
Chicago/Turabian StyleGabusi, Elena, Enrico Lenzi, Cristina Manferdini, Paolo Dolzani, Marta Columbaro, Yasmin Saleh, and Gina Lisignoli. 2022. "Autophagy Is a Crucial Path in Chondrogenesis of Adipose-Derived Mesenchymal Stromal Cells Laden in Hydrogel" Gels 8, no. 12: 766. https://doi.org/10.3390/gels8120766
APA StyleGabusi, E., Lenzi, E., Manferdini, C., Dolzani, P., Columbaro, M., Saleh, Y., & Lisignoli, G. (2022). Autophagy Is a Crucial Path in Chondrogenesis of Adipose-Derived Mesenchymal Stromal Cells Laden in Hydrogel. Gels, 8(12), 766. https://doi.org/10.3390/gels8120766





