Leveraging Deep Learning and Generative AI for Predicting Rheological Properties and Material Compositions of 3D Printed Polyacrylamide Hydrogels
Abstract
1. Introduction
2. Results and Discussion
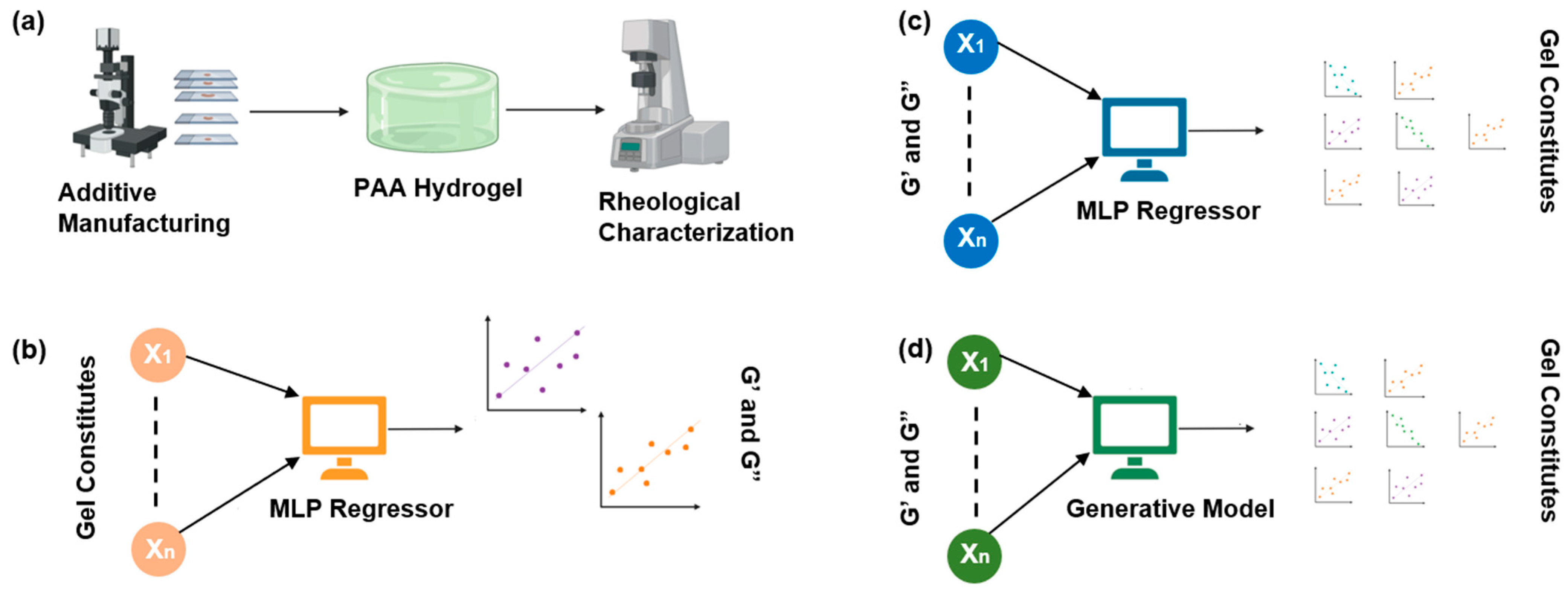
2.1. The Overall Workflow Is Divided into Four Stages
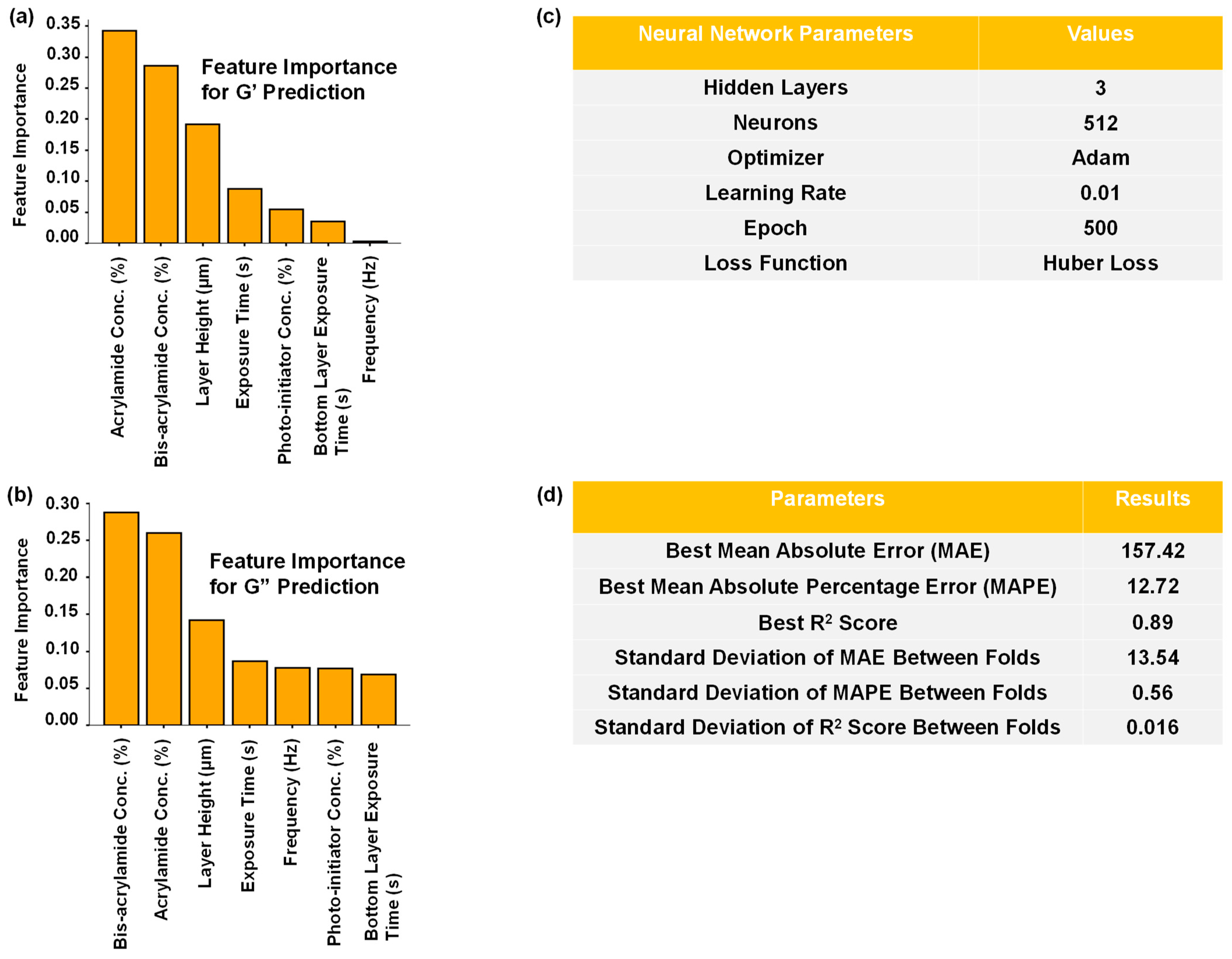
2.2. Determining the Contribution of Each Parameter for Predicting G’ and G”
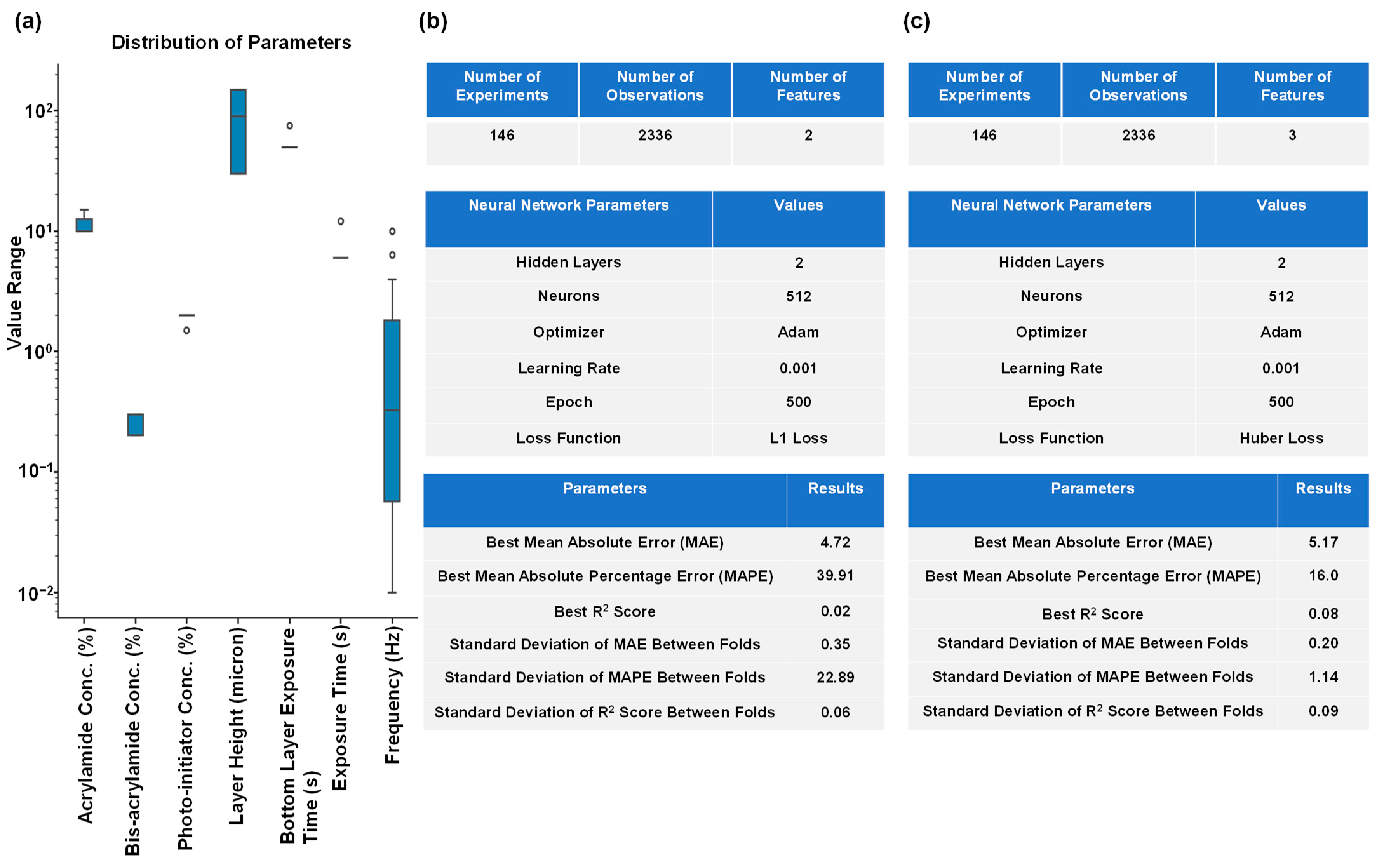
2.3. Predicting the Hydrogel Constituents Using an MLP for a Specific Pair of G’ and G”
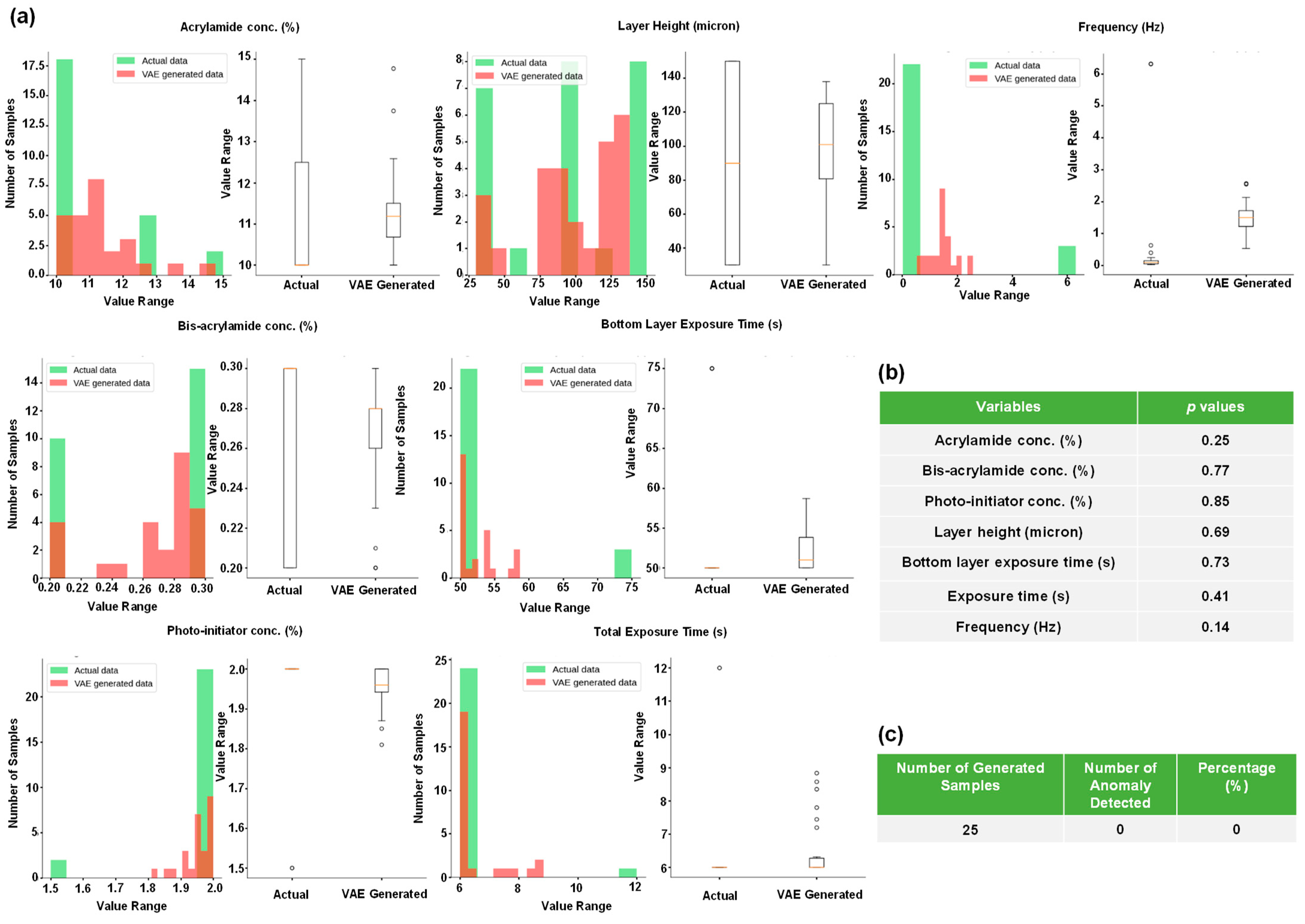
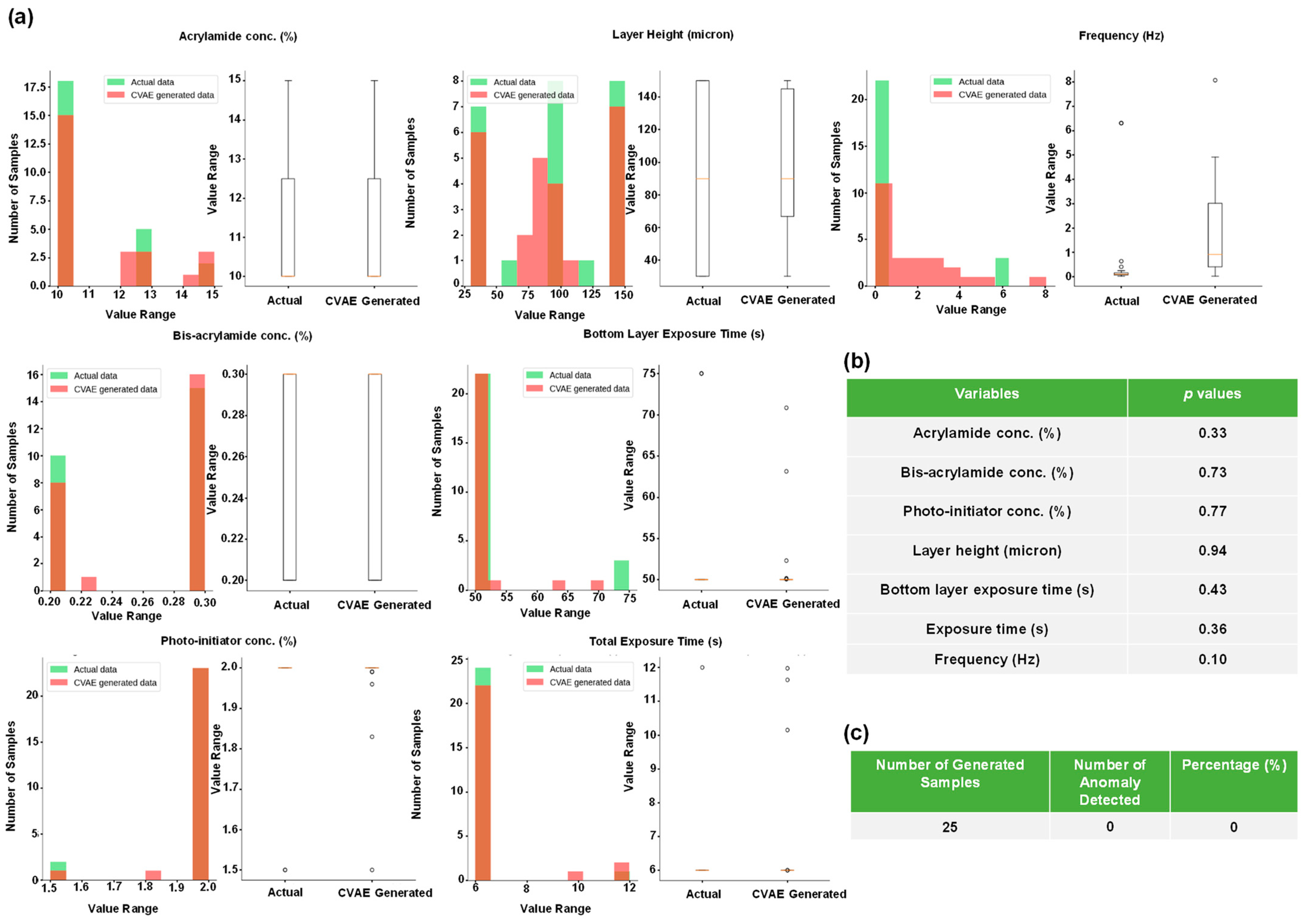
2.4. Using Generative Models to Produce Multiple Gel Constituent Compositions for Paired G’ and G” Values
3. Conclusions
4. Materials and Methods
4.1. Fabrication of PAA Hydrogels by 3D Printing
4.1.1. Measuring Rheological Properties of 3D-Printed PAA Hydrogels
4.1.2. Rheology Data Collection and Processing
4.2. DL Experiments for Predicting Material Properties and Printing Parameters
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Charrier, E.E.; Pogoda, K.; Wells, R.G.; Janmey, P.A. Control of cell morphology and differentiation by substrates with independently tunable elasticity and viscous dissipation. Nat. Commun. 2018, 9, 449. [Google Scholar] [CrossRef] [PubMed]
- Protick, F.K.; Amit, S.K.; Amar, K.; Nath, S.D.; Akand, R.; Davis, V.A.; Nilufar, S.; Chowdhury, F. Additive Manufacturing of Viscoelastic Polyacrylamide Substrates for Mechanosensing Studies. ACS Omega 2022, 7, 24384–24395. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Sohn, K.; Lee, H.; Yan, X. Learning Structured Output Representation using Deep Conditional Generative Models. In Proceedings of the NIPS, Montreal, QC, Canada, 7–12 December 2015. [Google Scholar]
- Ballard, D.H. Modular learning in neural networks. In Proceedings of the Sixth National Conference on Artificial Intelligence—Volume 1, Seattle, WA, USA, 13–17 July 1987; pp. 279–284. [Google Scholar]
- Mohammad, S.; Roy, A.; Karatzas, A.; Sarver, S.L.; Anagnostopoulos, I.; Chowdhury, F. Deep Learning Powered Identification of Differentiated Early Mesoderm Cells from Pluripotent Stem Cells. Cells 2024, 13, 534. [Google Scholar] [CrossRef] [PubMed]
- Popescu, M.-C.; Balas, V.E.; Perescu-Popescu, L.; Mastorakis, N. Multilayer perceptron and neural networks. WSEAS Trans. Cir. Sys. 2009, 8, 579–588. [Google Scholar]
- Feurer, M.; Hutter, F. Hyperparameter Optimization. In Automated Machine Learning: Methods, Systems, Challenges; Hutter, F., Kotthoff, L., Vanschoren, J., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 3–33. [Google Scholar] [CrossRef]
- Kirch, W. (Ed.) Pearson’s Correlation Coefficient. In Encyclopedia of Public Health; Springer: Dordrecht, The Netherlands, 2008; pp. 1090–1091. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Hsu, C.W.; Chang, C.C.; Lin, C.J. A Practical Guide to Support Vector Classification; Technical Report; Department of Computer Science and Information Engineering, National Taiwan University: Taipei, Taiwan, 2003; pp. 1–12. [Google Scholar]
- Chicco, D. Ten quick tips for machine learning in computational biology. BioData Min. 2017, 10, 35. [Google Scholar] [CrossRef]
- Huber, P.J. Robust Estimation of a Location Parameter. Ann. Math. Stat. 1964, 35, 492–518. [Google Scholar] [CrossRef]
- Pascanu, R.; Mikolov, T.; Bengio, Y. On the difficulty of training recurrent neural networks. In Proceedings of the International Conference on Machine Learning, Edinburgh, UK, 26 June–1 July 2012. [Google Scholar]
- Ramchandran, S.; Tikhonov, G.; Lönnroth, O.; Tiikkainen, P.; Lähdesmäki, H. Learning conditional variational autoencoders with missing covariates. Pattern Recognit. 2024, 147, 110113. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.C.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Fathallah, M.; Sakr, M.; Eletriby, S. Stabilizing and Improving Training of Generative Adversarial Networks Through Identity Blocks and Modified Loss Function. IEEE Access 2023, 11, 43276–43285. [Google Scholar] [CrossRef]
- Nie, W.; Patel, A.B. JR-GAN: Jacobian Regularization for Generative Adversarial Networks. arXiv 2018, arXiv:1806.09235. [Google Scholar]
- Verheyen, C.A.; Uzel, S.G.M.; Kurum, A.; Roche, E.T.; Lewis, J.A. Integrated data-driven modeling and experimental optimization of granular hydrogel matrices. Matter 2023, 6, 1015–1036. [Google Scholar] [CrossRef]
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-King, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D.; et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Chowdhury, F.; Li, Y.; Poh, Y.C.; Yokohama-Tamaki, T.; Wang, N.; Tanaka, T.S. Soft substrates promote homogeneous self-renewal of embryonic stem cells via downregulating cell-matrix tractions. PLoS ONE 2010, 5, e15655. [Google Scholar] [CrossRef] [PubMed]
- Keung, A.J.; de Juan-Pardo, E.M.; Schaffer, D.V.; Kumar, S. Rho GTPases mediate the mechanosensitive lineage commitment of neural stem cells. Stem Cells 2011, 29, 1886–1897. [Google Scholar] [CrossRef]
- Wang, N.; Ostuni, E.; Whitesides, G.M.; Ingber, D.E. Micropatterning tractional forces in living cells. Cell Motil. 2002, 52, 97–106. [Google Scholar] [CrossRef]
- Leight, J.L.; Wozniak, M.A.; Chen, S.; Lynch, M.L.; Chen, C.S. Matrix rigidity regulates a switch between TGF-β1-induced apoptosis and epithelial-mesenchymal transition. Mol. Biol. Cell 2012, 23, 781–791. [Google Scholar] [CrossRef]
- Winer, J.P.; Janmey, P.A.; McCormick, M.E.; Funaki, M. Bone marrow-derived human mesenchymal stem cells become quiescent on soft substrates but remain responsive to chemical or mechanical stimuli. Tissue Eng. Part A 2009, 15, 147–154. [Google Scholar] [CrossRef]
- Wang, Y.-l.; Li, D. Creating Complex Polyacrylamide Hydrogel Structures Using 3D Printing with Applications to Mechanobiology. Macromol. Biosci. 2020, 20, 2000082. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Huang, A.H.; Cai, Y.; Tanase, M.; Sheetz, M.P. Rigidity sensing at the leading edge through alphavbeta3 integrins and RPTPalpha. Biophys. J. 2006, 90, 1804–1809. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Joyce, J.M. Kullback-Leibler Divergence. In International Encyclopedia of Statistical Science; Lovric, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 720–722. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- McKinney, W. Pandas: A Foundational Python Library for Data Analysis and Statistics. Python High Perform. Sci. Comput. 2011, 14, 1–9. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammad, S.; Akand, R.; Cook, K.M.; Nilufar, S.; Chowdhury, F. Leveraging Deep Learning and Generative AI for Predicting Rheological Properties and Material Compositions of 3D Printed Polyacrylamide Hydrogels. Gels 2024, 10, 660. https://doi.org/10.3390/gels10100660
Mohammad S, Akand R, Cook KM, Nilufar S, Chowdhury F. Leveraging Deep Learning and Generative AI for Predicting Rheological Properties and Material Compositions of 3D Printed Polyacrylamide Hydrogels. Gels. 2024; 10(10):660. https://doi.org/10.3390/gels10100660
Chicago/Turabian StyleMohammad, Sakib, Rafee Akand, Kaden M. Cook, Sabrina Nilufar, and Farhan Chowdhury. 2024. "Leveraging Deep Learning and Generative AI for Predicting Rheological Properties and Material Compositions of 3D Printed Polyacrylamide Hydrogels" Gels 10, no. 10: 660. https://doi.org/10.3390/gels10100660
APA StyleMohammad, S., Akand, R., Cook, K. M., Nilufar, S., & Chowdhury, F. (2024). Leveraging Deep Learning and Generative AI for Predicting Rheological Properties and Material Compositions of 3D Printed Polyacrylamide Hydrogels. Gels, 10(10), 660. https://doi.org/10.3390/gels10100660







