Optimized Selective Media Enhance the Isolation and Characterization of Gut-Derived Probiotic Yeasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibiotic Susceptibility Testing and Growth Assessment of Yeast Isolates
2.2. Fecal Suspension and Preparation
2.3. Optimization of Growth Condition Based on Media, Temperature, Antibiotic Dose, and Incubation Atmosphere
2.4. Isolation Gut Yeast from Human Fecal Samples
2.5. Identification of Gut Yeast from Human Fecal Samples
2.6. Probiotic Characterization
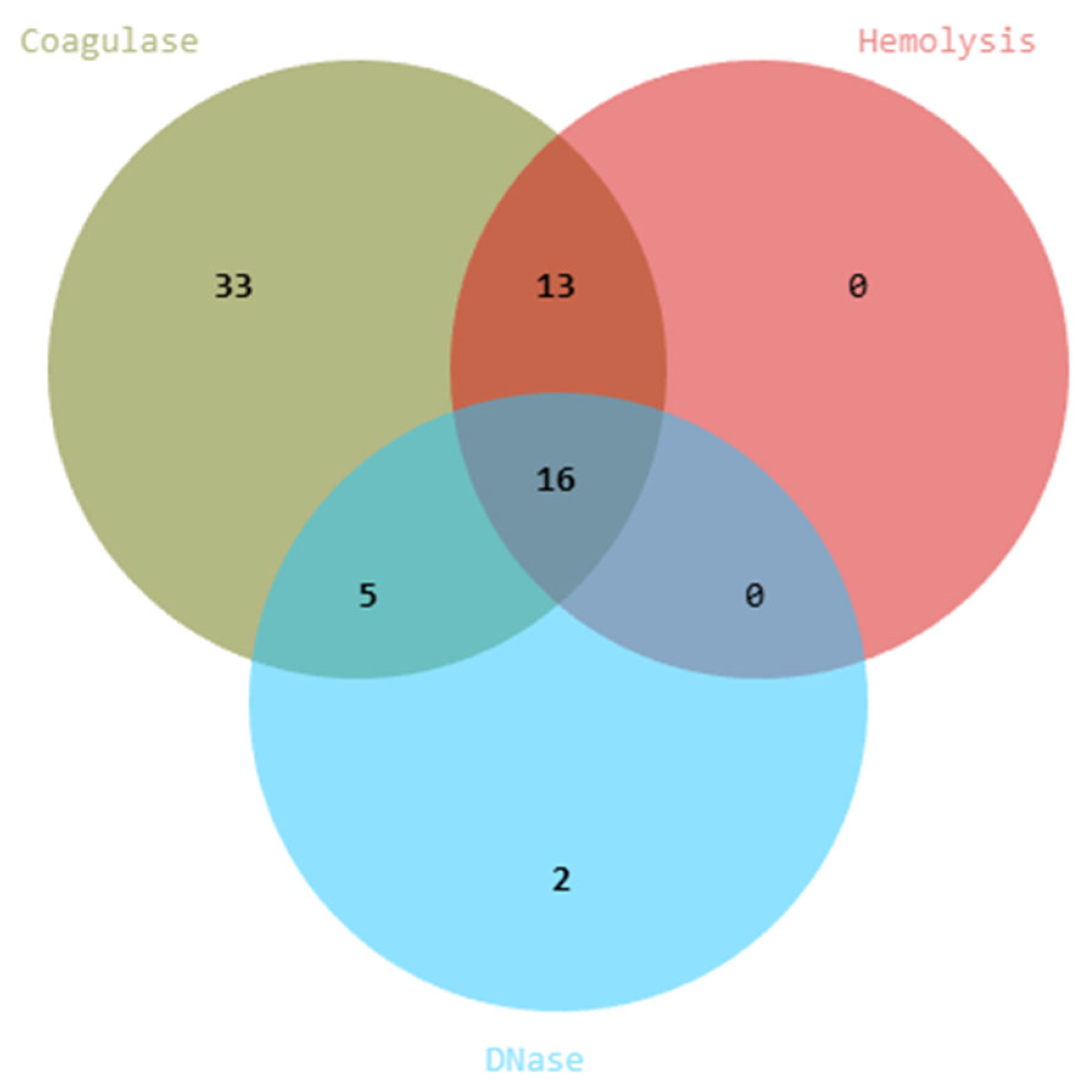
2.6.1. Assessment of Safety Profiles in Yeast Isolates
2.6.2. Coagulase Activity
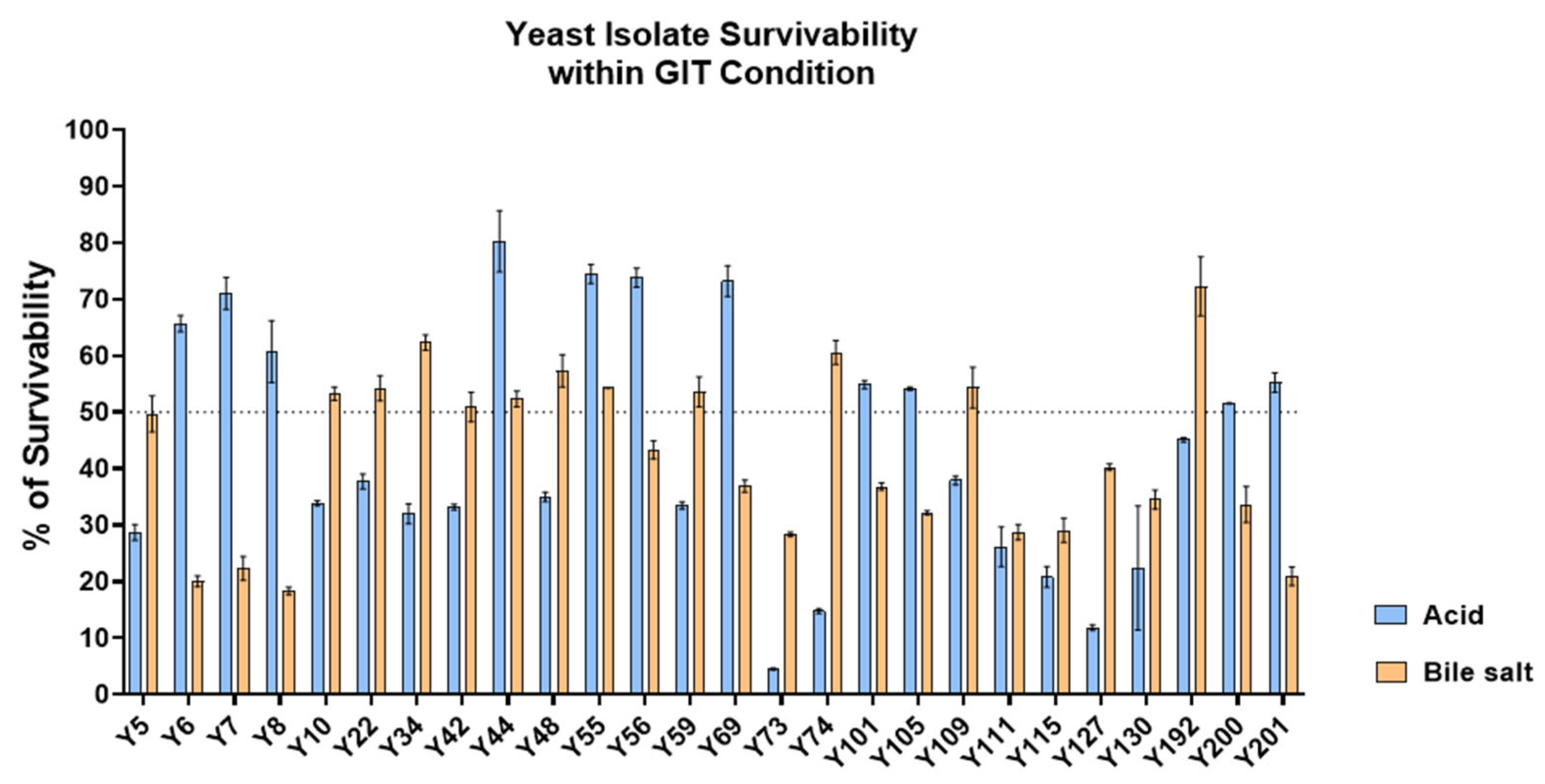
2.6.3. Acid and Bile Salt Tolerance Assay of Yeast Isolates
2.6.4. Assessment of Anti-Pathogenic Activity Against Enteropathogenic Bacteria
2.6.5. Assessment of Cholesterol-Lowering Potential of Yeast Based on BSH Activity
2.7. Statistical Analysis
3. Results
3.1. Optimization of Selective Medium and Conditions
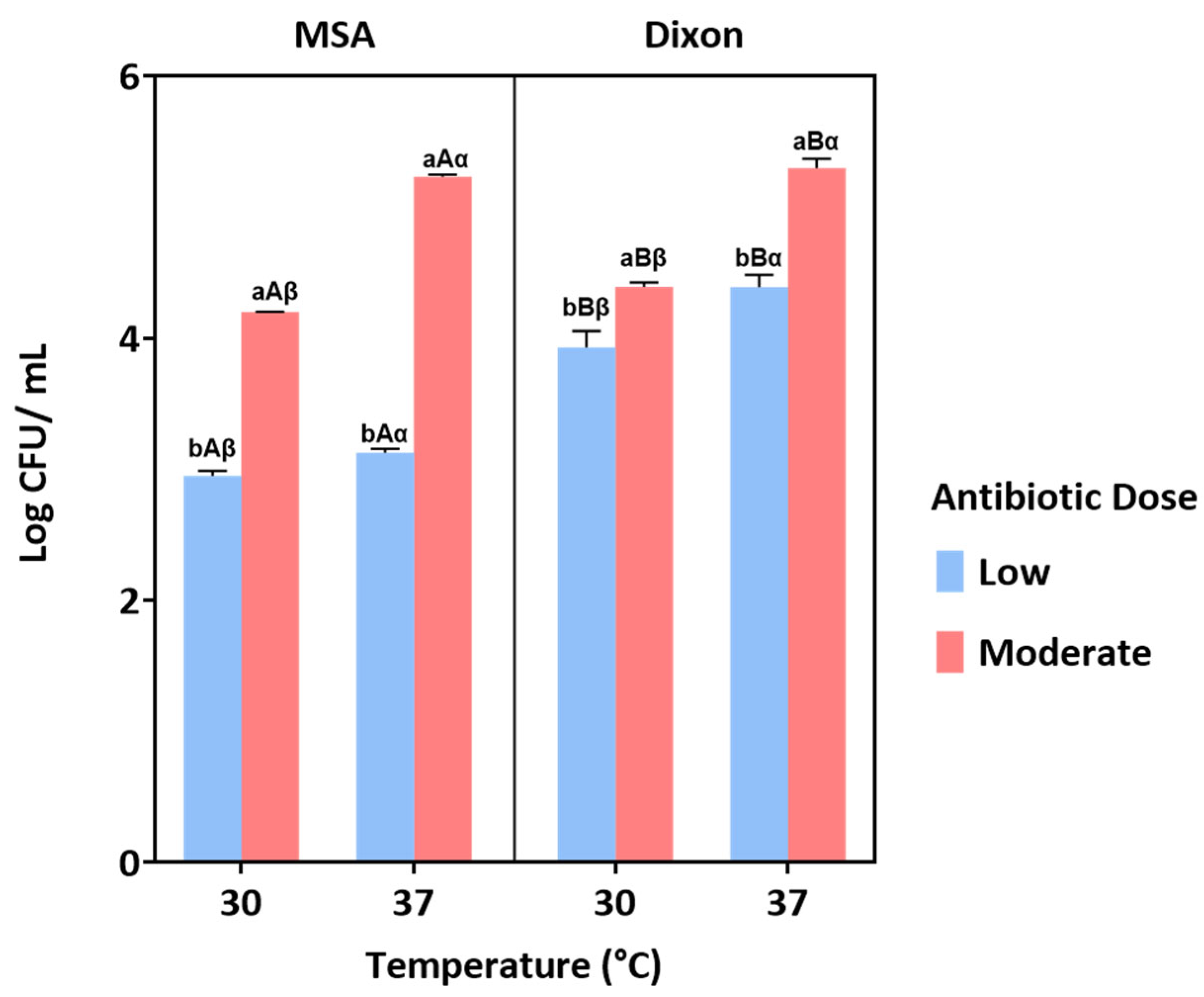
3.1.1. Antibiotic Concentration Screening
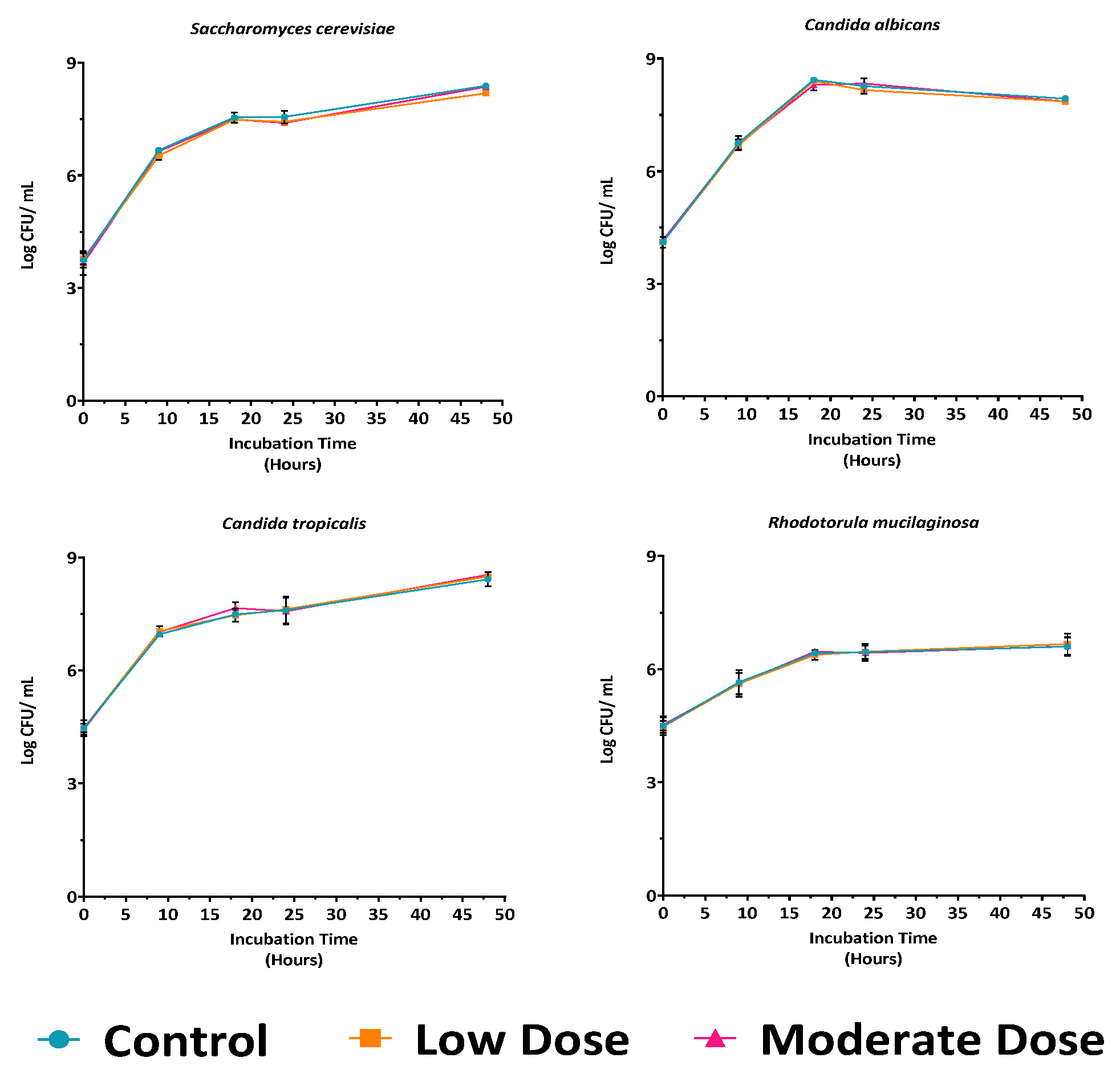
3.1.2. Influence of Medium, Temperature, and Antibiotic Dose Under Aerobic Condition
3.1.3. Effect of Oxygen Availability
3.2. Isolation and Identification Gut Yeast from Human Fecal Samples
3.3. Probiotic Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huseyin, C.E.; O’Toole, P.W.; Cotter, P.D.; Scanlan, P.D. Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol. Rev. 2017, 41, 479–511. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bonete, M.J.; Rajan, A.; Suriano, F.; Layunta, E. The Underrated Gut Microbiota Helminths, Bacteriophages, Fungi, and Archaea. Life 2023, 13, 1765. [Google Scholar] [CrossRef] [PubMed]
- Tidjani Alou, M.; Naud, S.; Khelaifia, S.; Bonnet, M.; Lagier, J.C.; Raoult, D. State of the Art in the Culture of the Human Microbiota: New Interests and Strategies. Clin. Microbiol. Rev. 2020, 34, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.M.; de Paula, T.O.; Sarmiento, M.R.A.; de Oliveira, M.G.; Pereira, M.L.M.; Toledo, I.V.; Diniz, C.G. Fungal Diversity of Human Gut Microbiota Among Eutrophic, Overweight, and Obese Individuals Based on Aerobic Culture-Dependent Approach. Curr. Microbiol. 2018, 75, 726–735. [Google Scholar] [CrossRef]
- Hallen-Adams, H.E.; Kachman, S.D.; Kim, J.; Legge, R.M.; Martínez, I. Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fungal Ecol. 2015, 15, 9–17. [Google Scholar] [CrossRef]
- Mok, K.; Suratanon, N.; Roytrakul, S.; Charoenlappanit, S.; Patumcharoenpol, P.; Chatchatee, P.; Nakphaichit, M. ITS2 Sequencing and Targeted Meta-Proteomics of Infant Gut Mycobiome Reveal the Functional Role of Rhodotorula sp. during Atopic Dermatitis Manifestation. J. Fungi 2021, 7, 748. [Google Scholar] [CrossRef]
- Mar Rodríguez, M.; Pérez, D.; Javier Chaves, F.; Esteve, E.; Marin-Garcia, P.; Xifra, G.; Fernandez Real, J.M. Obesity changes the human gut mycobiome. Sci. Rep. 2015, 5, 14600. [Google Scholar] [CrossRef]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef]
- Pais, P.; Almeida, V.; Yılmaz, M.; Teixeira, M.C. Saccharomyces boulardii: What Makes It Tick as Successful Probiotic? J. Fungi 2020, 6, 78. [Google Scholar] [CrossRef]
- Liu, S.; Yang, L.; Zhang, Y.; Chen, H.; Li, X.; Xu, Z.; Liu, D. Review of yeast culture concerning the interactions between gut microbiota and young ruminant animals. Front. Vet. Sci. 2024, 11, 1335765. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Raoult, D. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Sheth, R.U.; Zhao, S.; Cohen, L.A.; Dabaghi, K.; Moody, T.; Wang, H.H. High-throughput microbial culturomics using automation and machine learning. Nat. Biotechnol. 2023, 41, 1424–1433. [Google Scholar] [CrossRef] [PubMed]
- Das, T.K.; Pradhan, S.; Chakrabarti, S.; Mondal, K.C.; Ghosh, K. Current status of probiotic and related health benefits. Appl. Food Res. 2022, 2, 100185. [Google Scholar] [CrossRef]
- Banjara, N.; Nickerson, K.W.; Suhr, M.J.; Hallen-Adams, H.E. Killer toxin from several food-derived Debaryomyces hansenii strains effective against pathogenic Candida yeasts. Int. J. Food Microbiol. 2016, 222, 23–29. [Google Scholar] [CrossRef]
- Shruthi, B.; Deepa, N.; Somashekaraiah, R.; Adithi, G.; Divyashree, S.; Sreenivasa, M.Y. Exploring biotechnological and functional characteristics of probiotic yeasts: A review. Biotechnol. Rep. 2022, 34, e00716. [Google Scholar] [CrossRef]
- Alkalbani, N.S.; Osaili, T.M.; Al-Nabulsi, A.A.; Olaimat, A.N.; Liu, S.Q.; Shah, N.P.; Ayyash, M.M. Assessment of Yeasts as Potential Probiotics: A Review of Gastrointestinal Tract Conditions and Investigation Methods. J. Fungi 2022, 8, 365. [Google Scholar] [CrossRef]
- Fan, Y.; Yin, C.; Xu, L.; Bai, R.; Wei, Z.; Gao, G.; Pi, Y. The Biological Functions of Yeast and Yeast Derivatives and Their Application in Swine Production: A Review. Microorganisms 2025, 13, 1669. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef]
- Xu, R.; Yu, Y.; Chen, T. Exploring the dark side of probiotics to pursue light: Intrinsic and extrinsic risks to be opportunistic pathogens. Curr. Res. Food Sci. 2025, 10, 101044. [Google Scholar] [CrossRef]
- Suravaram, S.; Hada, V.; Ahmed Siddiqui, I. Comparison of antimicrobial susceptibility interpretation among Enterobacteriaceae using CLSI and EUCAST breakpoints. Indian J. Med. Microbiol. 2021, 39, 315–319. [Google Scholar] [CrossRef]
- CLSI. Clinical and Laboratory Standards Institute; CLSI: Berwyn, PA, USA, 2020; Available online: https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf (accessed on 11 December 2025).
- Angchuan, J.; Khunnamwong, P.; Wongpanit, K.; Limtong, S.; Srisuk, N. Yeasts Associated with the Small-Intestinal Contents and Epithelium of Pon Yang Kham (Charolais Crossbred) Fattening Beef Cattle. Microorganisms 2021, 9, 1444. [Google Scholar] [CrossRef] [PubMed]
- Mok, K.; Honwichit, O.; Funnuam, T.; Charoensiddhi, S.; Nitisinprasert, S.; Nielsen, D.S.; Nakphaichit, M. Synergistic activity of Limosilactobacillus reuteri KUB-AC5 and water-based plants against Salmonella challenge in a human in vitro gut model. Sci. Rep. 2024, 14, 4730. [Google Scholar] [CrossRef] [PubMed]
- Huseyin, C.E.; Rubio, R.C.; O’Sullivan, O.; Cotter, P.D.; Scanlan, P.D. The Fungal Frontier: A Comparative Analysis of Methods Used in the Study of the Human Gut Mycobiome. Front. Microbiol. 2017, 8, 1432. [Google Scholar] [CrossRef] [PubMed]
- Dione, N.; Khelaifia, S.; La Scola, B.; Lagier, J.C.; Raoult, D. A quasi-universal medium to break the aerobic/anaerobic bacterial culture dichotomy in clinical microbiology. Clin. Microbiol. Infect. 2016, 22, 53–58. [Google Scholar] [CrossRef]
- Chang, H.-W.; Nam, Y.-D.; Sung, Y.; Kim, K.-H.; Roh, S.W.; Yoon, J.-H.; Bae, J.W. Quantitative real time PCR assays for the enumeration of Saccharomyces cerevisiae and the Saccharomyces sensu stricto complex in human feces. J. Microbiol. Methods 2007, 71, 191–201. [Google Scholar] [CrossRef]
- Zhang, J.; Hung, G.C.; Nagamine, K.; Li, B.; Tsai, S.; Lo, S.C. Development of Candida-Specific Real-Time PCR Assays for the Detection and Identification of Eight Medically Important Candida Species. Microbiol. Insights 2016, 9, 21–28. [Google Scholar] [CrossRef]
- de Melo Riceto, É.B.; de Paula Menezes, R.; Penatti, M.P.A.; dos Santos Pedroso, R. Enzymatic and hemolytic activity in different Candida species. Rev. Iberoam. De Micol. 2015, 32, 79–82. [Google Scholar] [CrossRef]
- Goldstein, J.; Roberts, J.W. Microtube coagulase test for detection of coagulase-positive staphylococci. J. Clin. Microbiol. 1982, 15, 848–851. [Google Scholar] [CrossRef]
- Woo, P.C.; Leung, A.S.; Leung, K.W.; Yuen, K.Y. Identification of slide coagulase positive, tube coagulase negative Staphylococcus aureus by 16S ribosomal RNA gene sequencing. Mol. Pathol. 2001, 54, 244–247. [Google Scholar] [CrossRef]
- Hassanzadazar, H.; Ehsani, A.; Mardani, K.; Hesari, J. Investigation of antibacterial, acid and bile tolerance properties of lactobacilli isolated from Koozeh cheese. Vet. Res. Forum 2012, 3, 181–185. [Google Scholar]
- Shijila Rani, A.S.; Babu, S.; Anbukumaran, A.; Veeramani, S.; Ambikapathy, V.; Gomathi, S.; Senthilkumar, G. Chapter 21—Prospective Approaches of Pseudonocardia alaniniphila Hydrobionts for Litopenaeus vannamei. In Advances in Probiotics; Dhanasekaran, D., Sankaranarayanan, A., Eds.; Academic Press: New York, NY, USA, 2021; pp. 327–348. [Google Scholar]
- Wongrattanapipat, S.; Chiracharoenchitta, A.; Choowongwitthaya, B.; Komsathorn, P.; La-ongkham, O.; Nitisinprasert, S.; Nakphaichit, M. Selection of potential probiotics with cholesterol-lowering properties for probiotic yoghurt production. Food Sci. Technol. Int. 2022, 28, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Seelbinder, B.; Chen, J.; Brunke, S.; Vazquez-Uribe, R.; Santhaman, R.; Meyer, A.C.; Panagiotou, G. Antibiotics create a shift from mutualism to competition in human gut communities with a longer-lasting impact on fungi than bacteria. Microbiome 2020, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L. Effect of antibiotics on bacterial populations: A multi-hierachical selection process. F1000Research 2017, 6, 51. [Google Scholar] [CrossRef]
- Abid, R.; Waseem, H.; Ali, J.; Ghazanfar, S.; Muhammad Ali, G.; Elasbali, A.M.; Alharethi, S.H. Probiotic Yeast Saccharomyces: Back to Nature to Improve Human Health. J. Fungi 2022, 8, 444. [Google Scholar] [CrossRef]
- Hossain, M.N.; Afrin, S.; Humayun, S.; Ahmed, M.M.; Saha, B.K. Identification and Growth Characterization of a Novel Strain of Saccharomyces boulardii Isolated from Soya Paste. Front. Nutr. 2020, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Ingber, D.E. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- de Carvalho, B.T.; Subotić, A.; Vandecruys, P.; Deleu, S.; Vermeire, S.; Thevelein, J.M. Enhancing probiotic impact: Engineering Saccharomyces boulardii for optimal acetic acid production and gastric passage tolerance. Appl. Environ. Microbiol. 2024, 90, e0032524. [Google Scholar] [CrossRef]
- Macura, A.B.; Witalis, J. Fungi isolated from the stool in patients with gastrointestinal disorders in 2005–2009. Prz. Epidemiol. 2010, 64, 313–317. [Google Scholar]
- Psomas, E.; Fletouris, D.; Litopoulou-Tzanetaki, E.; Tzanetakis, N. Assimilation of cholesterol by yeast strains isolated from infant feces and Feta cheese. J. Dairy Sci. 2003, 86, 3416–3422. [Google Scholar] [CrossRef]
- Perna, N.T.; Glasner, J.D.; Burland, V.; Plunkett, G. Chapter 1—The Genomes of Escherichia coli K-12 and Pathogenic E. coli. In Escherichia Coli; Donnenberg, M.S., Ed.; Academic Press: San Diego, CA, USA, 2002; pp. 3–53. [Google Scholar]
- Im, E.J.; Lee, H.H.; Kim, M.; Kim, M.K. Evaluation of Enterococcal Probiotic Usage and Review of Potential Health Benefits, Safety, and Risk of Antibiotic-Resistant Strain Emergence. Antibiotics 2023, 12, 1327. [Google Scholar] [CrossRef]
- Vepštaitė-Monstavičė, I.; Lukša-Žebelovič, J.; Apšegaitė, V.; Mozūraitis, R.; Lisicinas, R.; Stanevičienė, R.; Servienė, E. Profiles of Killer Systems and Volatile Organic Compounds of Rowanberry and Rosehip-Inhabiting Yeasts Substantiate Implications for Biocontrol. Foods 2025, 14, 288. [Google Scholar] [CrossRef]
- Hatoum, R.; Labrie, S.; Fliss, I. Antimicrobial and probiotic properties of yeasts: From fundamental to novel applications. Front. Microbiol. 2012, 3, 421. [Google Scholar] [CrossRef]


| Antibiotic | Concentration (µg/mL) | ||
|---|---|---|---|
| A (Low) | B (Moderate) | C (High) | |
| Colistin | 1 | 2 | 4 |
| Chloramphenicol | 8 | 16 | 32 |
| Vancomcyin | 4 | 8 | 16 |
| Tested Strain | Dose | ||
|---|---|---|---|
| Low (cm) | Moderate (cm) | High (cm) | |
| Escherichia coli ATCC 8739 | 3.17 ± 0.15 | 4.13 ± 0.12 | 4.70 ± 0.10 |
| Salmonella enteritidis S003 | 3.23 ± 0.15 | 4.03 ± 0.12 | 4.67 ± 0.06 |
| Staphylococcus aureus ATCC 6538P | 2.10 ± 0.10 | 3.00 ± 0.10 | 3.50 ± 0.26 |
| Rhodotorula mucilaginosa TBRC-4420 | ND | ND | 1.37 ± 0.12 |
| Candida albicans PCJ94-2 | ND | ND | ND |
| Candida tropicalis PYJ100-2 | ND | ND | ND |
| Saccharomyces cerevisiae YC1 | ND | ND | ND |
| Source of Variation | % Var | SS | DF | MS | F | p-Value |
|---|---|---|---|---|---|---|
| Temperature | 16.0800 | 2.4810 | 1 | 2.4810 | 598.000 | <0.0001 |
| Medium Type | 15.2100 | 2.3470 | 1 | 2.3470 | 624.1000 | <0.0001 |
| Antibiotic Dose | 54.1500 | 8.3530 | 1 | 8.3530 | 1899.000 | <0.0001 |
| Temperature × Medium Type | 0.0590 | 0.0090 | 1 | 0.0090 | 2.4360 | 0.1936 |
| Temperature × Antibiotic Dose | 4.1060 | 0.6330 | 1 | 0.6330 | 144.0000 | 0.0003 |
| Medium Type × Antibiotic Dose | 9.5530 | 1.4740 | 1 | 1.4740 | 345.8000 | <0.0001 |
| Temperature × Medium Type × Antibiotic Dose | 0.4100 | 0.0630 | 1 | 0.0630 | 14.8500 | 0.0182 |
| Volunteers | 0.1080 | 0.0170 | 4 | 0.0040 | ||
| Volunteers × Medium Type | 0.0980 | 0.0150 | 4 | 0.0040 | ||
| Volunteers × Antibiotic Dose | 0.0110 | 0.0180 | 4 | 0.0040 | ||
| Residual | 0.0170 | 4 | 0.0040 |
| Isolate Codes | Inhibition Size Clear Zone (cm) | |||||
|---|---|---|---|---|---|---|
| S. aureus ATCC 6538P | E. coli ATCC 8739 | E. coli O157:H7 | S. typhimurium DMST 48437 | S. enteritidis S003 | Spectrum | |
| Y6 | 1.9 ± 0.0 | 1.4 ± 0.0 | 1.1 ± 0.0 | 1.6 ± 0.0 | 1.5 ± 0.0 | Broad Strong |
| Y22 | 1.7 ± 0.0 | 1.4 ± 0.0 | 1.2 ± 0.0 | 1.3 ± 0.0 | 1.2 ± 0.1 | Broad Strong |
| Y42 | 1.6 ± 0.1 | 1.6 ± 0.1 | 1.8 ± 0.0 | 1.2 ± 0.1 | 1.9 ± 0.1 | Broad Strong |
| Y48 | 1.9 ± 0.0 | 0.0 ± 0.0 | 1.4 ± 0.1 | 1.2 ± 0.0 | 1.8 ± 0.1 | Broad Strong |
| Y55 | 1.4 ± 0.0 | 1.8 ± 0.0 | 0.0 ± 0.0 | 1.4 ± 0.1 | 0.5 ± 0.0 | Broad Strong |
| Y56 | 1.1 ± 0.1 | 2.0 ± 0.0 | 0.9 ± 0.0 | 1.1 ± 0.1 | 1.2 ± 0.0 | Broad Strong |
| Y73 | 1.4 ± 0.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 1.9 ± 0.1 | 1.3 ± 0.1 | Broad Strong |
| Y105 | 1.6 ± 0.0 | 1.4 ± 0.0 | 1.8 ± 0.0 | 0.8 ± 0.0 | 0.9 ± 0.1 | Broad Strong |
| Y127 | 1.7 ± 0.0 | 1.2 ± 0.1 | 1.5 ± 0.0 | 0.8 ± 0.0 | 1.9 ± 0.0 | Broad Strong |
| Y44 | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.5 ± 0.0 | Broad Weak |
| Y59 | 0.4 ± 0.0 | 0.5 ± 0.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | Broad Weak |
| Y7 | 0.2 ± 0.1 | 0.6 ± 0.0 | 0.4 ± 0.0 | 0.5 ± 0.0 | 0.3 ± 0.0 | Broad Weak |
| Y8 | 0.2 ± 0.0 | 1.9 ± 0.0 | 0.6 ± 0.0 | 0.9 ± 0.0 | 0.0 ± 0.0 | Broad Weak |
| Y34 | 0.5 ± 0.0 | 0.6 ± 0.1 | 0.8 ± 0.0 | 0.7 ± 0.1 | 0.0 ± 0.0 | Broad Weak |
| Y69 | 0.0 ± 0.0 | 0.6 ± 0.1 | 0.0 ± 0.0 | 1.1 ± 0.0 | 1.4 ± 0.0 | Narrow |
| Y74 | 0.0 ± 0.0 | 1.9 ± 0.1 | 0.6 ± 0.0 | 1.7 ± 0.0 | 1.6 ± 0.0 | Narrow |
| Y101 | 0.0 ± 0.0 | 1.5 ± 0.0 | 0.4 ± 0.1 | 1.1 ± 0.1 | 1.2 ± 0.1 | Narrow |
| Y109 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.2 ± 0.1 | 0.8 ± 0.0 | 0.0 ± 0.0 | Narrow |
| Y111 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | Narrow |
| Y115 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.2 | 1.3 ± 0.0 | Narrow |
| Y130 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | Narrow |
| Y192 | 0.0 ± 0.0 | 1.4 ± 0.1 | 0.2 ± 0.0 | 1.9 ± 0.0 | 0.0 ± 0.0 | Narrow |
| Y200 | 0.0 ± 0.0 | 1.4 ± 0.0 | 1.6 ± 0.0 | 1.6 ± 0.1 | 0.4 ± 0.1 | Narrow |
| Y201 | 0.0 ± 0.0 | 0.8 ± 0.0 | 0.5 ± 0.0 | 0.9 ± 0.0 | 1.2 ± 0.0 | Narrow |
| Y5 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.3 ± 0.0 | Narrow |
| Y10 | 0.0 ± 0.0 | 0.8 ± 0.0 | 0.7 ± 0.0 | 0.4 ± 0.0 | 1.6 ± 0.1 | Narrow |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mok, K.; Popitool, K.; Songla, A.; Pongsuwanporn, T.; Torrungruang, P.; Nitisinprasert, S.; Nakayama, J.; Nakphaichit, M. Optimized Selective Media Enhance the Isolation and Characterization of Gut-Derived Probiotic Yeasts. J. Fungi 2025, 11, 885. https://doi.org/10.3390/jof11120885
Mok K, Popitool K, Songla A, Pongsuwanporn T, Torrungruang P, Nitisinprasert S, Nakayama J, Nakphaichit M. Optimized Selective Media Enhance the Isolation and Characterization of Gut-Derived Probiotic Yeasts. Journal of Fungi. 2025; 11(12):885. https://doi.org/10.3390/jof11120885
Chicago/Turabian StyleMok, Kevin, Kwantida Popitool, Areerat Songla, Tawisa Pongsuwanporn, Pitchsupang Torrungruang, Sunee Nitisinprasert, Jiro Nakayama, and Massalin Nakphaichit. 2025. "Optimized Selective Media Enhance the Isolation and Characterization of Gut-Derived Probiotic Yeasts" Journal of Fungi 11, no. 12: 885. https://doi.org/10.3390/jof11120885
APA StyleMok, K., Popitool, K., Songla, A., Pongsuwanporn, T., Torrungruang, P., Nitisinprasert, S., Nakayama, J., & Nakphaichit, M. (2025). Optimized Selective Media Enhance the Isolation and Characterization of Gut-Derived Probiotic Yeasts. Journal of Fungi, 11(12), 885. https://doi.org/10.3390/jof11120885






