Inhibitory Effects of the Fungal Pigment Rubiginosin C on Hyphal and Biofilm Formation in Candida albicans and Candida auris
Abstract
1. Introduction
2. Materials and Methods
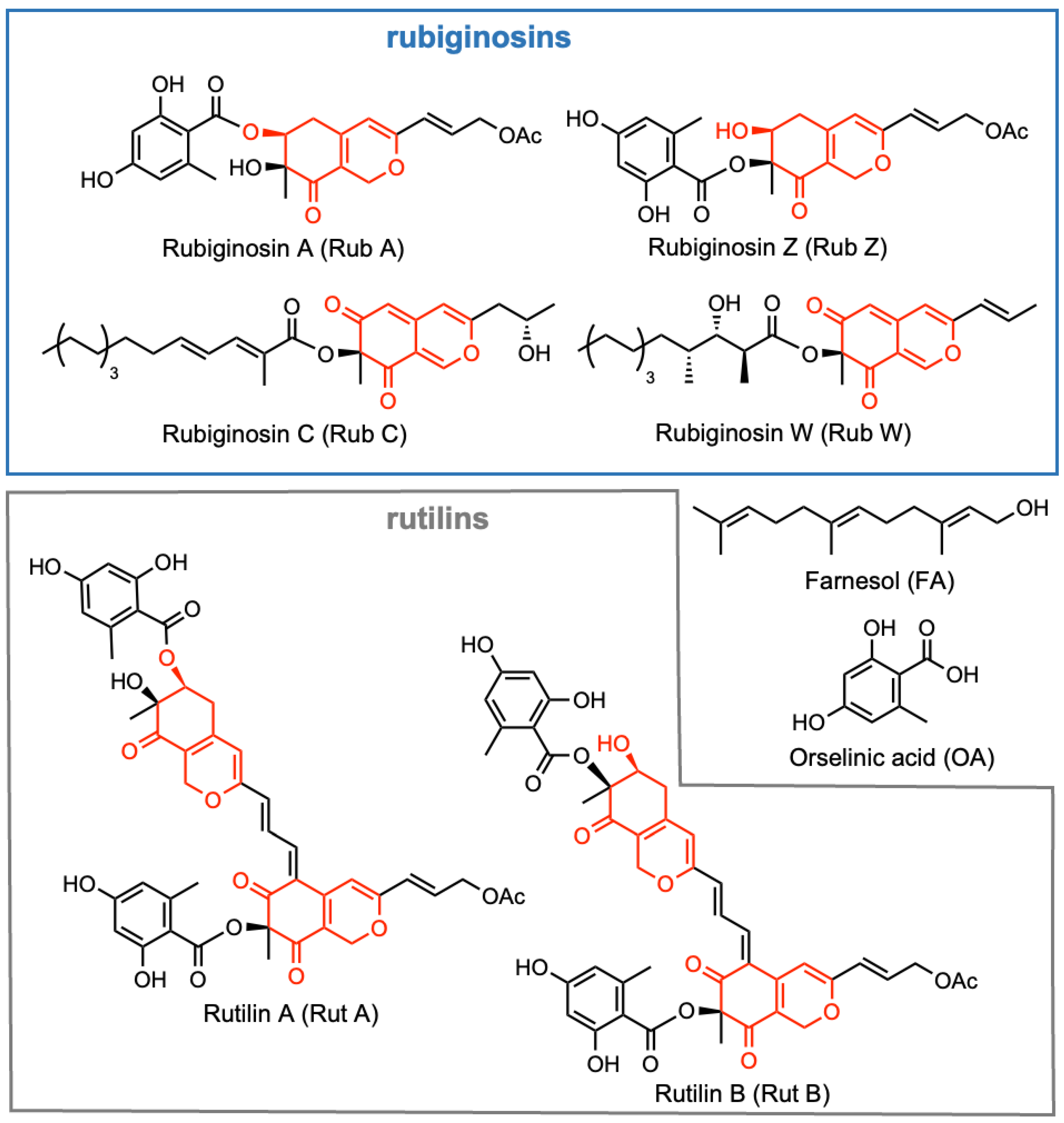
2.1. Isolation of Selected Azaphilones and Preparation of Pathogenic Strains
2.2. Determination of Minimum Inhibitory Concentration (MIC) and Cytotoxicity
2.3. Antibiofilm Assay with Crystal Violet
2.3.1. Biofilm Formation Assay of C. auris and C. albicans
2.3.2. Assay to Determine Rub C Effects on C. auris Biofilms of Various Ages
2.4. Observations of Biofilm by Confocal Laser Scanning Microscopy (CLSM)
2.5. Colony Forming Units (CFU) and the Growth Curve of Candida
2.5.1. CFU of C. auris
2.5.2. The Growth Curve of Candida
2.6. Observations of Candida Cells by Optical Microscopy
2.6.1. Visualization of C. auris and C. albicans Cells
2.6.2. Assay to Determine the Impact of Rub C on Hyphae
2.7. Observations of Candida Cells by Scanning Electron Microscopy (SEM)
2.8. Screening and Quantification of Hyphal Inhibitory Activities with β-Galactosidase Activity Assay
2.9. Statistical Analysis
3. Results
3.1. Determination of MIC and Cytotoxicity of Azaphilones
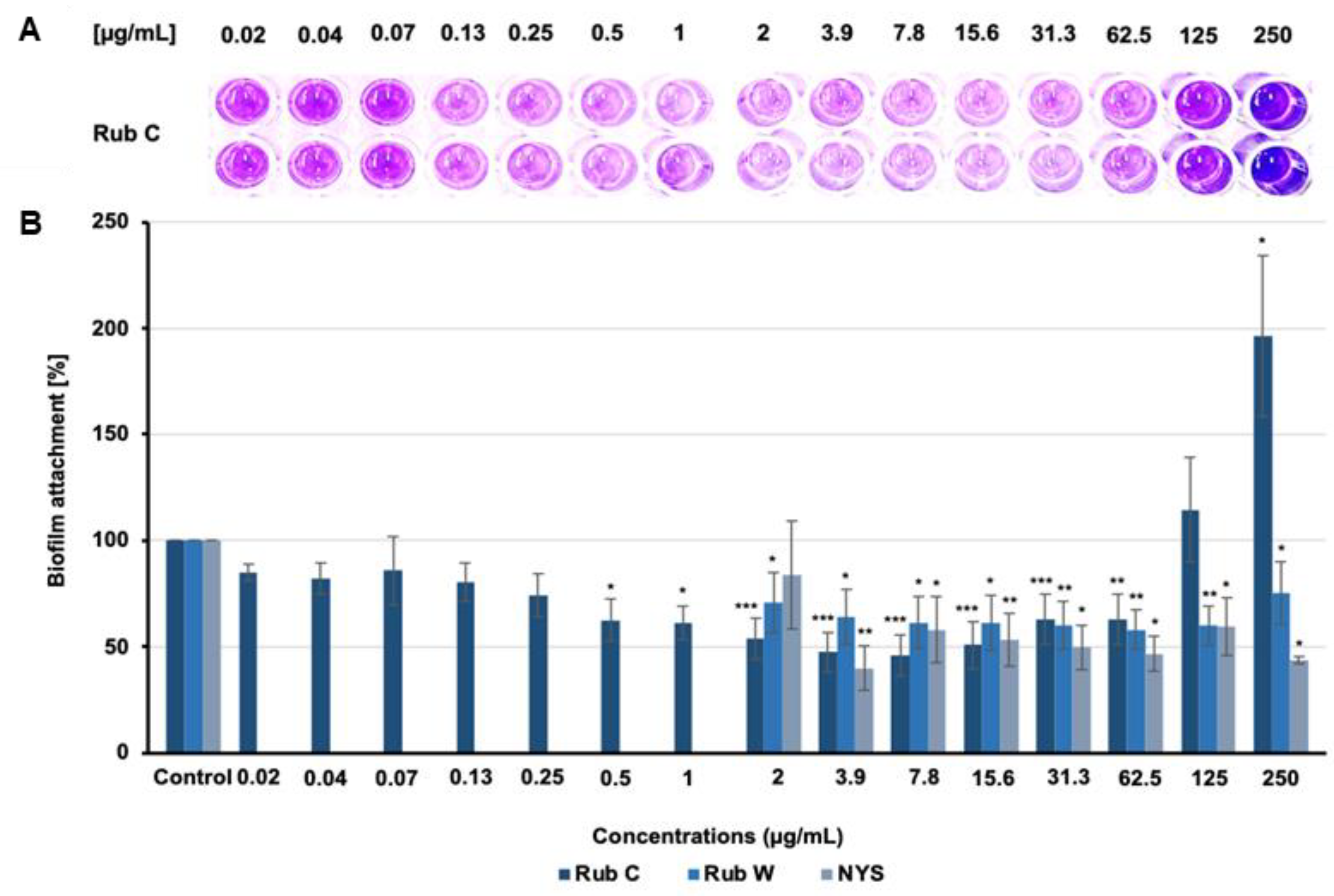
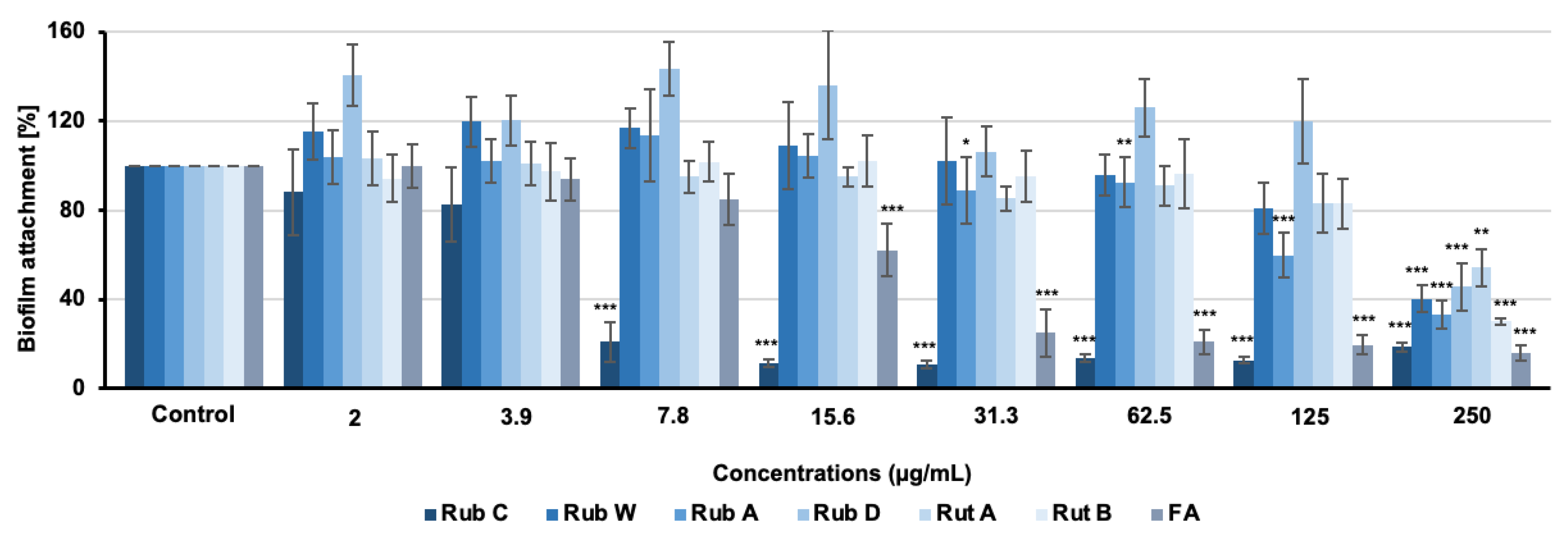
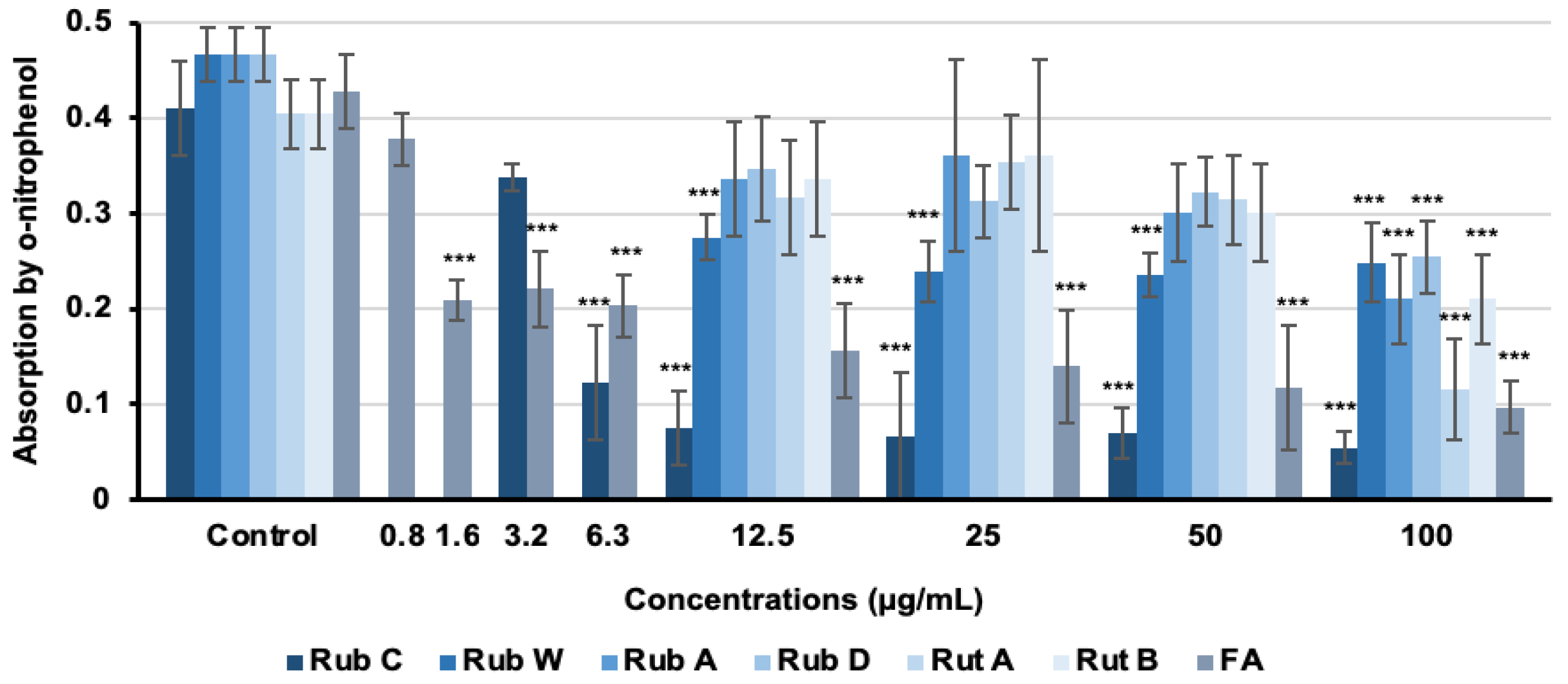
3.2. Inhibitory Effects of Azaphilone Pigments against Candida Biofilm Formation
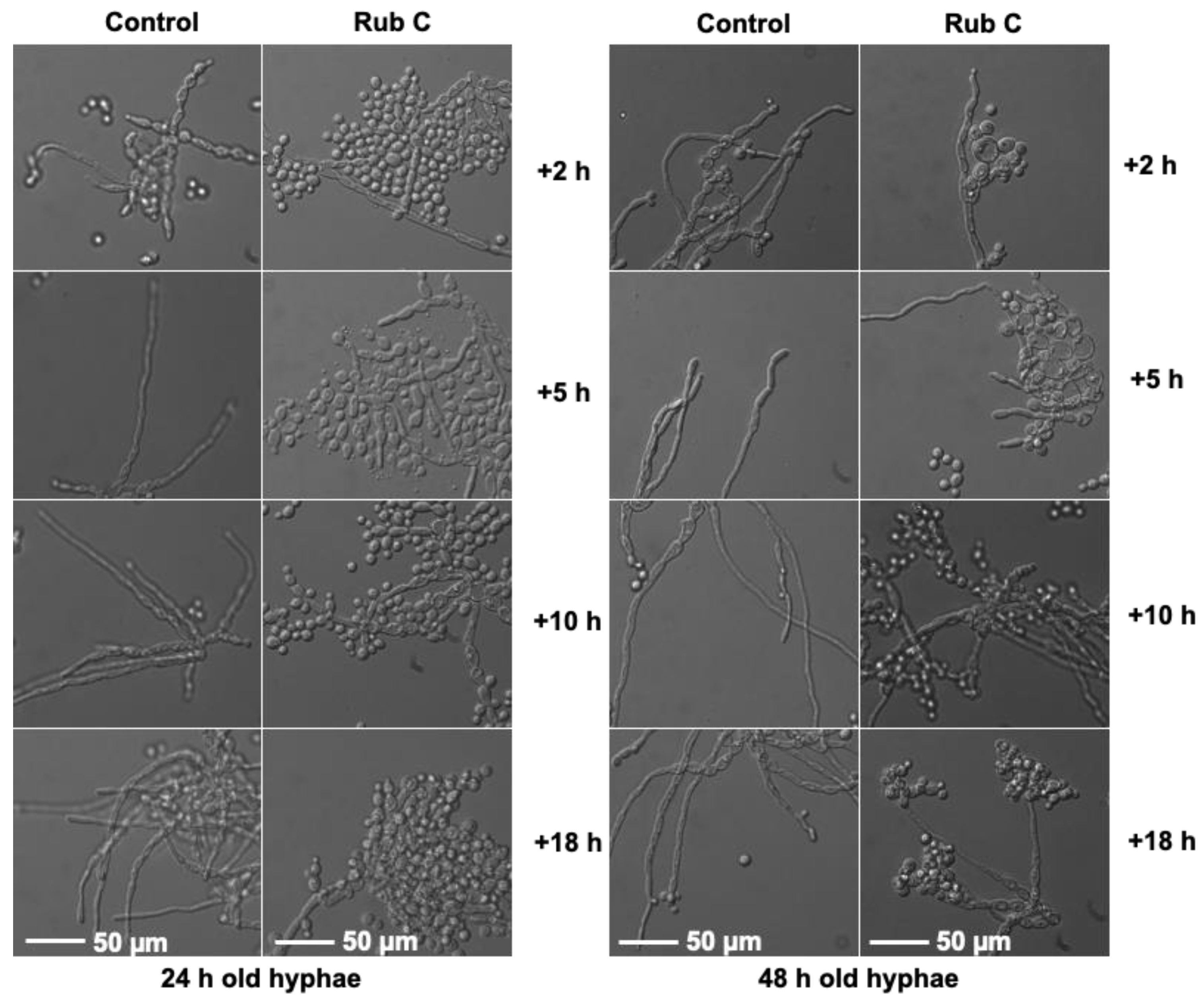
3.3. Rub C Activity against Different Maturation Stages of C. auris Biofilm
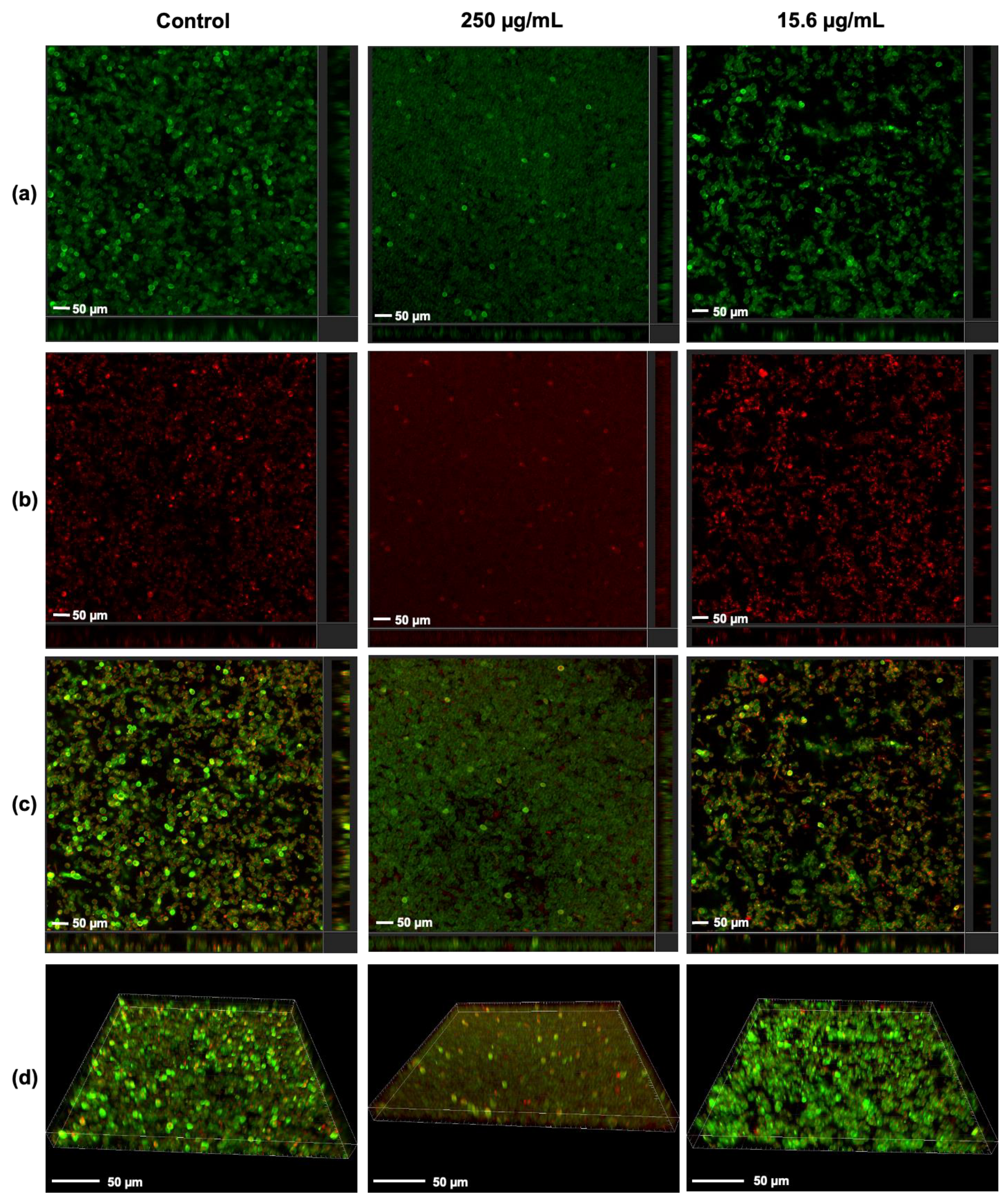
3.4. Visualization of the Effect of Rub C on C. auris Biofilms via CLSM
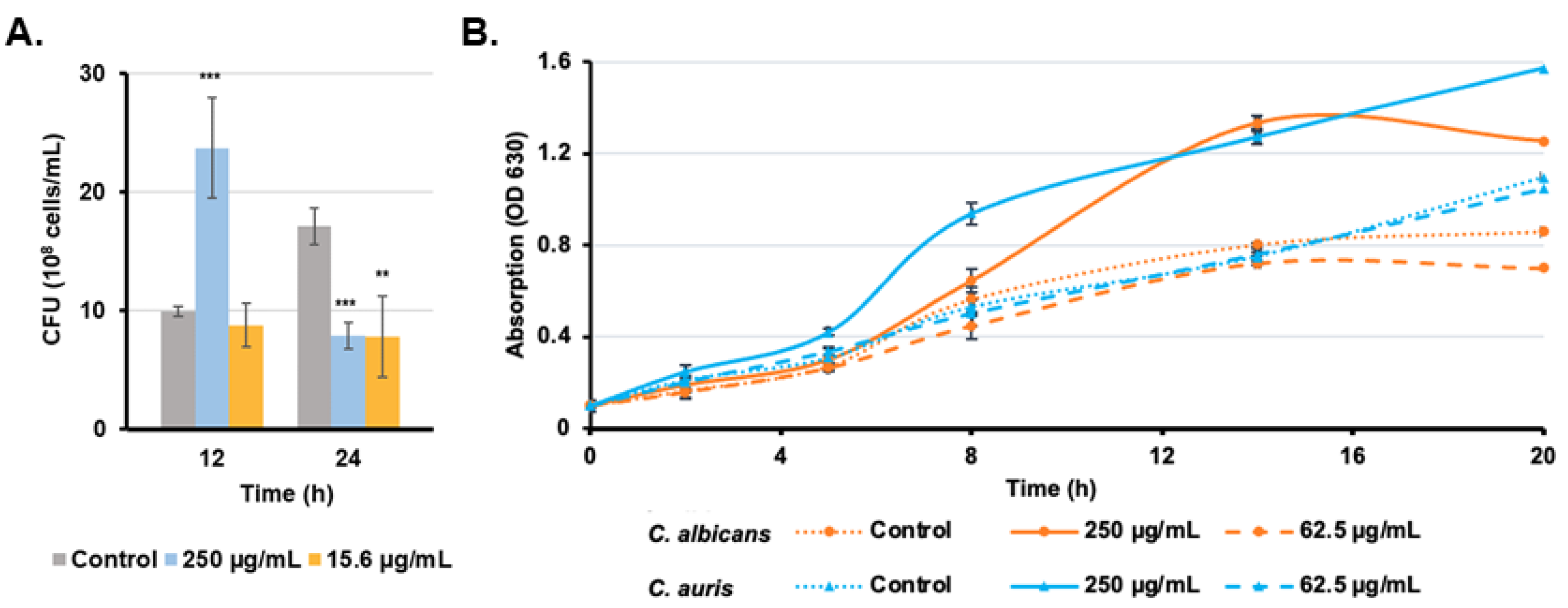
3.5. Candida Growth Is Promoted with High Rub C Concentrations
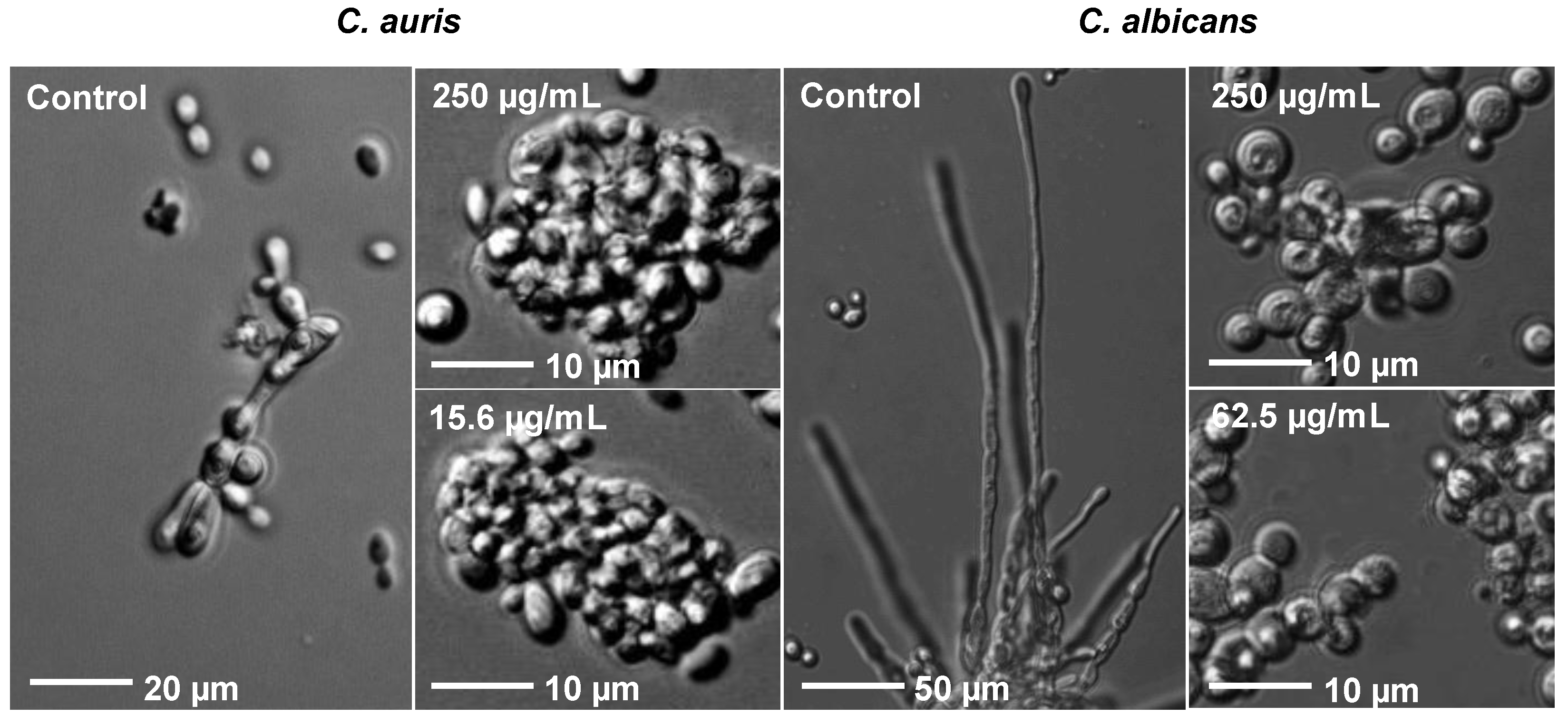
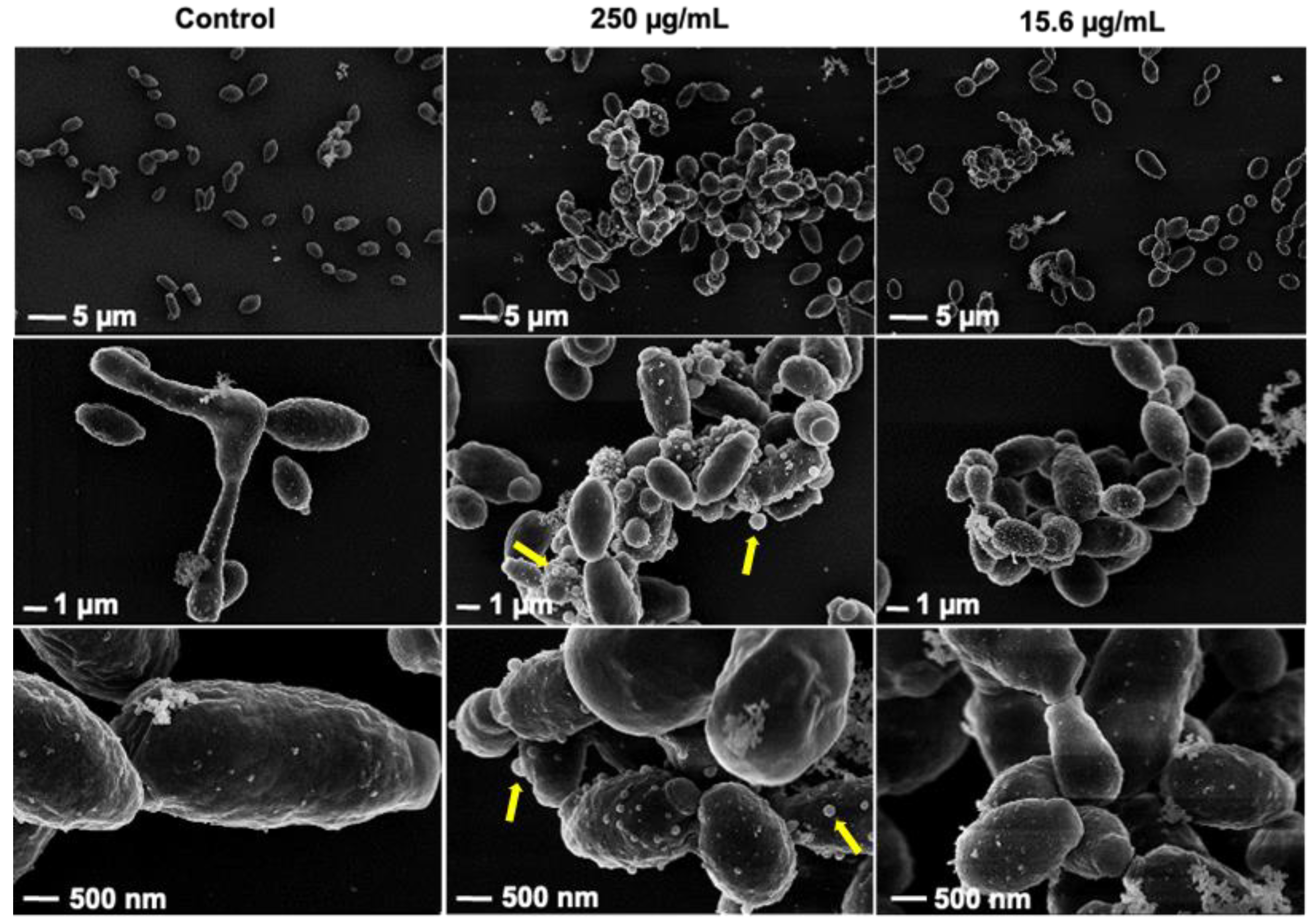
3.6. Rub C Induces Morphological Changes of C. auris and C. albicans Cells
3.7. Quantification of Inhibitory Activities of Azaphilones against C. albicans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Cavalheiro, M.; Teixeira, M.C. Candida Biofilms: Threats, challenges, and promising strategies. Front. Med. 2018, 5, 28. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Sardi, J.C.O.; Silva, D.R.; Mendes-Giannini, M.J.S.; Rosalen, P.L. Candida auris: Epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb. Pathog. 2018, 125, 116–121. [Google Scholar] [CrossRef]
- Abirami, G.; Alexpandi, R.; Durgadevi, R.; Kannappan, A.; Ravi, A.V. Inhibitory effect of morin against Candida albicans pathogenicity and virulence factor production: An in vitro and in vivo approaches. Front. Microbiol. 2020, 11, 561298. [Google Scholar] [CrossRef]
- Gulati, M.; Nobile, C.J. Candida albicans biofilms: Development, regulation, and molecular mechanisms. Microbes Infect. 2016, 18, 310–321. [Google Scholar] [CrossRef]
- Horton, M.V.; Nett, J.E. Candida auris infection and biofilm formation: Going beyond the surface. Curr. Clin. Microbiol. Rep. 2020, 7, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Mukherjee, P.K. Candida Biofilms: Development, architecture, and resistance. Microbiol. Spectr. 2015, 3, 115–134. [Google Scholar] [CrossRef]
- Bapat, P.S.; Nobile, C.J. Photodynamic therapy is effective against Candida auris biofilms. Front. Cell. Infect. Microbiol. 2021, 11, 713092. [Google Scholar] [CrossRef] [PubMed]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Geremia, N.; Brugnaro, P.; Solinas, M.; Scarparo, C.; Panese, S. Candida auris as an emergent public health problem: A current update on european outbreaks and cases. Healthcare 2023, 2, 425. [Google Scholar] [CrossRef]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Abbasi, A.F.; Prakash, S.; Mangat, J.; Hosein, Z.; Haider, N.; Chan, J. Candida auris: An overview of the emerging drug-resistant fungal infection. Infect. Chemother. 2022, 54, 236–246. [Google Scholar] [CrossRef]
- Ademe, M.; Girma, F. Candida auris: From multidrug resistance to pan-resistant strains. Infect. Drug Resist. 2020, 5, 1287–1294. [Google Scholar] [CrossRef]
- Frías-De-León, M.G.; Hernández-Castro, R.; Vite-Garín, T.; Arenas, R.; Bonifaz, A.; Castañón-Olivares, L.; Acosta-Altamirano, G.; Martínez-Herrera, E. Antifungal resistance in Candida auris: Molecular determinants. Antibiotics 2020, 9, 568. [Google Scholar] [CrossRef] [PubMed]
- Rossato, L.; Colombo, A.L. Candida auris: What have we learned about its mechanisms of pathogenicity? Front. Microbiol. 2018, 9, 3081. [Google Scholar] [CrossRef]
- Cortegiani, A.; Misseri, G.; Fasciana, T.; Giammanco, A.; Giarratano, A.; Chowdhary, A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care 2018, 6, 69. [Google Scholar] [CrossRef]
- Welsh, R.M.; Bentz, M.L.; Shams, A.; Houston, H.; Lyons, A.; Rose, L.J.; Litvintseva, A.P. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J. Clin. Microbiol. 2017, 55, 2996–3005. [Google Scholar] [CrossRef]
- Larkin, E.; Hager, C.; Chandra, J.; Mukherjee, P.K.; Retuerto, M.; Salem, I.; Long, L. The emerging pathogen Candida auris: Growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob. Agents Chemother. 2017, 61, e02396-16. [Google Scholar] [CrossRef]
- Hernando-Ortiz, A.; Mateo, E.; Perez-Rodriguez, A.; de Groot, P.W.J.; Quindós, G.; Eraso, E. Virulence of Candida auris from different clinical origins in Caenorhabditis elegans and Galleria mellonella host models. Virulence 2021, 12, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Ramage, G.; Kean, R.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg. Infect. Dis. 2017, 23, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.M.; Yang, S.X.; Qin, J.C. Azaphilones: Chemistry and biology. Chem. Rev. 2013, 113, 4755–4811. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, L.P.S.; Gomes, D.C.; Cardoso, P.G.; Takahashi, J.A. Recent findings in azaphilone pigments. J. Fungi 2021, 7, 541. [Google Scholar] [CrossRef]
- Becker, K.; Pfütze, S.; Kuhnert, E.; Cox, R.J.; Stadler, M.; Surup, F. Hybridorubrins A–D: Azaphilone heterodimers from stromata of Hypoxylon fragiforme and insights into the biosynthetic machinery for azaphilone diversification. Chem. Eur. J. 2021, 27, 1438–1450. [Google Scholar] [CrossRef]
- Becker, K.; Stadler, M. Recent progress in biodiversity research on the Xylariales and their secondary metabolism. J. Antibiotics 2021, 74, 1–23. [Google Scholar] [CrossRef]
- Helaly, S.E.; Thongbai, B.; Stadler, M. Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Nat. Prod. Rep. 2018, 35, 992–1014. [Google Scholar] [CrossRef]
- Becker, K.; Kuhnert, E.; Cox, R.J.; Surup, F. Azaphilone pigments from Hypoxylon rubiginosum and H. texense: Absolute configuration, bioactivity, and biosynthesis. Eur. J. Org. Chem. 2021, 36, 5094–5103. [Google Scholar] [CrossRef]
- Quang, D.N.; Hashimoto, T.; Stadler, M.; Asakawa, Y. New azaphilones from the inedible mushroom Hypoxylon rubiginosum. J. Nat. Prod. 2004, 67, 1152–1155. [Google Scholar] [CrossRef]
- Surup, F.; Narmani, A.; Wendt, L.; Pfütze, S.; Kretz, R.; Becker, K.; Menbrivès, C.; Giosa, A.; Elliott, M.; Petit, C.; et al. Identification of fungal fossils and novel azaphilone pigments in ancient carbonised specimens of Hypoxylon fragiforme from forest soils of Châtillon-sur-Seine (Burgundy). Fungal Divers. 2018, 92, 345–356. [Google Scholar] [CrossRef]
- Wendt, L.; Benjamin, E.S.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018, 17, 115–154. [Google Scholar] [CrossRef]
- Kuhnert, E.; Navarro-Muñoz, J.C.; Becker, K.; Stadler, M.; Collemare, J.; Cox, R.J. Secondary metabolite biosynthetic diversity in the fungal family Hypoxylaceae and Xylaria hypoxylon. Stud. Mycol. 2021, 99, 100118. [Google Scholar] [CrossRef] [PubMed]
- Hogan, D.A.; Vik, Å.; Kolter, R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004, 54, 1212–1223. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Wessel, A.-C.; Luangsa-ard, J.J.; Stadler, M. Viridistratins A–C, Antimicrobial and cytotoxic benzo[j]fluoranthenes from stromata of Annulohypoxylon viridistratum (Hypoxylaceae, Ascomycota). Biomolecules 2020, 10, 805. [Google Scholar] [CrossRef]
- Humberto, H.L.; Liliana, I.; Miguel, J.Y.; Jose, L. Inhibition of Candida auris biofilm formation on medical and environmental surfaces by silver nanoparticles. ACS Appl. Mater. Interfaces 2020, 12, 21183–21191. [Google Scholar] [CrossRef]
- Srivastava, V.; Ahmad, A. Abrogation of pathogenic attributes in drug resistant Candida auris strains by farnesol. PLoS ONE 2020, 15, e0233102. [Google Scholar] [CrossRef]
- Güttlein, P.; Schrey, H.; Zeng, H.; Schobert, R. Syntheses and biological effects of natural Morinda lactone and derivatives. Org. Biomol. Chem. 2022, 20, 4794–4802. [Google Scholar] [CrossRef]
- Gillsch, F.; Zeng, H.; Bär, S.I.; Schrey, H.; Schobert, R. Synthesis and bioactivity of ophiofuranones A and B. J. Org. Chem. 2022, 87, 6520–6523. [Google Scholar] [CrossRef]
- Treiber, L.; Pezolt, C.; Zeng, H.; Schrey, H.; Jungwirth, S.; Shekhar, A.; Stadler, M.; Bilitewski, U.; Erb-Brinkmann, M.; Schobert, R. Dual Agents: Fungal macrocidins and synthetic analogues with herbicidal and antibiofilm activities. Antibiotics 2021, 10, 1022. [Google Scholar] [CrossRef]
- Kean, R.; Delaney, C.; Sherry, L.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R.; Williams, C.; Ramage, G. Transcriptome assembly and profiling of Candida auris reveals novel insights into biofilm-mediated resistance. mSphere 2018, 3, e00334-18. [Google Scholar] [CrossRef]
- Müsken, M.; Pawar, V.; Schwebs, T.; Bähre, H.; Felgner, S.; Weiss, S.; Häussler, S. Breaking the vicious cycle of antibiotic killing and regrowth of biofilm-residing Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e01635-18. [Google Scholar] [CrossRef] [PubMed]
- Vittal, R.; Haudenshield, J.S.; Hartman, G.L. A multiplexed immunofluorescence method identifies Phakopsora pachyrhizi urediniospores and determines their viability. Phytopathology 2012, 102, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Simm, C.; Weerasinghe, H.; Thomas, D.R.; Harrison, P.F.; Newton, H.J.; Beilharz, T.H.; Traven, A. Disruption of iron homeostasis and mitochondrial metabolism are promising targets to inhibit Candida auris. Microbiol. Spectr. 2022, 10, e00100-22. [Google Scholar] [CrossRef] [PubMed]
- Vediyappan, G.; Dumontet, V.; Pelissier, F.; d’Enfert, C. Gymnemic acids inhibit hyphal growth and virulence in Candida albicans. PLoS ONE 2013, 8, e74189. [Google Scholar] [CrossRef] [PubMed]
- Bense, S.; Witte, J.; Preuße, M.; Koska, M.; Pezoldt, L.; Dröge, A.; Hartmann, O.; Müsken, M.; Schulze, J.; Fiebig, T.; et al. Pseudomonas aeruginosa post-translational responses to elevated c-di-GMP levels. Mol. Microbiol. 2022, 117, 1213–1226. [Google Scholar] [CrossRef]
- Nobile, C.J.; Nett, J.E.; Andes, D.R.; Mitchell, A.P. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 2006, 5, 1604–1610. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Eickhoff, H.; Hohn, E.; Bilitewski, U. Identification of inhibitors of yeast-to-hyphae transition in Candida albicans by a reporter screening assay. J. Biotechnol. 2013, 164, 137–142. [Google Scholar] [CrossRef]
- Miceli, M.H.; Bernardo, S.M.; Ku, T.S.; Walraven, C.; Lee, S.A. In vitro analyses of the effects of heparin and parabens on Candida albicans biofilms and planktonic cells. Antimicrob. Agents Chemother. 2012, 56, 148–153. [Google Scholar] [CrossRef]
- Bashyal, B.P.; Wijeratne, E.M.; Faeth, S.H.; Gunatilaka, A.A. Globosumones A–C, cytotoxic orsellinic acid esters from the Sonoran desert endophytic fungus Chaetomium globosum. J. Nat. Prod. 2005, 68, 724–728. [Google Scholar] [CrossRef]
- Yang, N.J.; Hinner, M.J. Getting across the cell membrane: An overview for small molecules, peptides, and proteins. Methods Mol. Biol. 2015, 1266, 29–53. [Google Scholar] [CrossRef]
- Shchepin, R.; Hornby, J.M.; Burger, E.; Niessen, T.; Dussault, P.; Nickerson, K.W. Quorum sensing in Candida albicans: Probing farnesol’s mode of action with 40 natural and synthetic farnesol analogs. Chem. Biol 2003, 10, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Zarnowski, R.; Jaromin, A.; Zagórska, A.; Dominguez, E.G.; Sidoryk, K.; Gubernator, J.; Andes, D.R. A label-free cellular proteomics approach to decipher the antifungal action of DiMIQ, a potent indolo[2,3-b] quinoline agent, against Candida albicans biofilms. Int. J. Mol. Sci. 2021, 22, 108. [Google Scholar] [CrossRef] [PubMed]
- Hornby, J.M.; Jensen, E.C.; Lisec, A.D.; Tasto, J.J.; Jahnke, B.; Shoemaker, R.D.; Nickerson, K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef]
- Riekhof, W.R.; Nickerson, K.W. Quorum sensing in Candida albicans: Farnesol versus farnesoic acid. FEBS Lett. 2017, 591, 1637–1640. [Google Scholar] [CrossRef]
- Ramage, G.; Saville, S.P.; Wickes, B.L.; López-Ribot, J.L. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 2002, 68, 5459–5463. [Google Scholar] [CrossRef] [PubMed]
- Paramonova, E.; Krom, B.P.; van der Mei, H.C.; Busscher, H.J.; Sharma, P.K. Hyphal content determines the compression strength of Candida albicans biofilms. Microbiology 2009, 155, 1997–2003. [Google Scholar] [CrossRef]
- Mohammad, H.; Eldesouky, H.E.; Hazbun, T.; Abdelrahman, S.M.; Seleem, N. Identification of a phenylthiazole small molecule with dual antifungal and antibiofilm activity against Candida albicans and Candida auris. Sci. Rep. 2019, 9, 18941. [Google Scholar] [CrossRef]
- Wang, X.; Bing, J.; Zheng, Q.; Zhang, F.; Liu, J.; Yue, H.; Tao, L.; Du, H.; Wang, Y.; Wang, H.; et al. The first isolate of Candida auris in China: Clinical and biological aspects. Emerg. Microbes Infect. 2018, 7, 93. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, X.; Ren, B.; Cheng, L. The regulation of hyphae growth in Candida albicans. Virulence 2020, 11, 337–348. [Google Scholar] [CrossRef]
- Uwamahoro, N.; Verma-Gaur, J.; Shen, H.H.; Qu, Y.; Lewis, R.; Lu, J.; Bambery, K.; Masters, S.L.; Vince, J.E.; Naderer, T.; et al. The pathogen Candida albicans hijacks pyroptosis for escape from macrophages. mBio 2014, 5, e00003-14. [Google Scholar] [CrossRef]
- Brand, A. Hyphal growth in human fungal pathogens and its role in virulence. Int. J. Microbiol. 2012, 2012, 517529. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Bamunuarachchi, N.I.; Tabassum, N.; Jo, D.M.; Khan, M.M.; Kim, Y.M. Suppression of hyphal formation and virulence of Candida albicans by natural and synthetic compounds. Biofouling 2021, 37, 626–655. [Google Scholar] [CrossRef] [PubMed]
- Miramón, P.; Kasper, L.; Hube, B. Thriving within the host: Candida spp. interactions with phagocytic cells. Med. Microbiol. Immunol. 2016, 202, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Zamith-Miranda, D.; Heyman, H.M.; Couvillion, S.P.; Cordero, R.J.B.; Rodrigues, M.L.; Nimrichter, L.; Casadevall, A.; Amatuzzi, R.F.; Alves, L.R.; Nakayasu, E.S.; et al. Comparative molecular and immunoregulatory analysis of extracellular vesicles from Candida albicans and Candida auris. mSystems 2021, 6, e00822-21. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, Z.; Liu, Y.; Liao, B.; Zong, Y.; Shi, Y.; Liao, M.; Wang, J.; Zhou, X.; Cheng, L.; et al. Extracellular vesicles of Candida albicans regulate its own growth through the L-arginine/nitric oxide pathway. Appl. Microbiol. Biotechnol. 2023, 107, 355–367. [Google Scholar] [CrossRef]
- Honorato, L.; de Araujo, J.F.D.; Ellis, C.C.; Piffer, A.C.; Pereira, Y.; Frases, S.; de Sousa Araújo, G.R.; Pontes, B.; Mendes, M.T.; Pereira, M.D.; et al. Extracellular vesicles regulate biofilm formation and yeast-to-hyphae differentiation in Candida albicans. mBio 2022, 13, e00301-22. [Google Scholar] [CrossRef]
- Vargas-Blanco, D.; Lynn, A.; Rosch, J.; Noreldin, R.; Salerni, A.; Lambert, C.; Rao, R.P. A pre-therapeutic coating for medical devices that prevents the attachment of Candida albicans. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 41. [Google Scholar] [CrossRef]
- Fazly, A.; Jain, C.; Dehner, A.C.; Issi, L.; Lilly, E.A.; Ali, A.; Cao, H.; Fidel, P.L., Jr.; Rao, R.P.; Kaufman, P.D. Chemical screening identifies filastatin, a small molecule inhibitor of Candida albicans adhesion, morphogenesis, and pathogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 13594–13599. [Google Scholar] [CrossRef]

| Tested Organisms | Strain No. | MIC [μg/mL] | ||||||
|---|---|---|---|---|---|---|---|---|
| Rub | Rut | Nystatin | ||||||
| A | C | W | Z | A | B | |||
| C. albicans [28] | DSM 1665 | – | – | – | – | – | – | 8.3 |
| C. albicans | DSM 11225 | – | – | – | – | – | – | 8.3 |
| C. albicans CAI-4 HWP1-lacZ | ZK3379 | – | – | – | – | – | – | 8.3 |
| C. auris | DSM 21092 | – | – | – | – | – | – | 31.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, H.; Stadler, M.; Abraham, W.-R.; Müsken, M.; Schrey, H. Inhibitory Effects of the Fungal Pigment Rubiginosin C on Hyphal and Biofilm Formation in Candida albicans and Candida auris. J. Fungi 2023, 9, 726. https://doi.org/10.3390/jof9070726
Zeng H, Stadler M, Abraham W-R, Müsken M, Schrey H. Inhibitory Effects of the Fungal Pigment Rubiginosin C on Hyphal and Biofilm Formation in Candida albicans and Candida auris. Journal of Fungi. 2023; 9(7):726. https://doi.org/10.3390/jof9070726
Chicago/Turabian StyleZeng, Haoxuan, Marc Stadler, Wolf-Rainer Abraham, Mathias Müsken, and Hedda Schrey. 2023. "Inhibitory Effects of the Fungal Pigment Rubiginosin C on Hyphal and Biofilm Formation in Candida albicans and Candida auris" Journal of Fungi 9, no. 7: 726. https://doi.org/10.3390/jof9070726
APA StyleZeng, H., Stadler, M., Abraham, W.-R., Müsken, M., & Schrey, H. (2023). Inhibitory Effects of the Fungal Pigment Rubiginosin C on Hyphal and Biofilm Formation in Candida albicans and Candida auris. Journal of Fungi, 9(7), 726. https://doi.org/10.3390/jof9070726








