Production of L-Malic Acid by Metabolically Engineered Aspergillus nidulans Based on Efficient CRISPR–Cas9 and Cre-loxP Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Culture Conditions
2.2. Construction of Plasmids
2.3. Gene Editing
2.4. Western Blotting
2.5. RNA Extraction for qRT–PCR
2.6. Cre-LoxP System
2.7. Quantification of L-Malic Acid
3. Results and Discussion
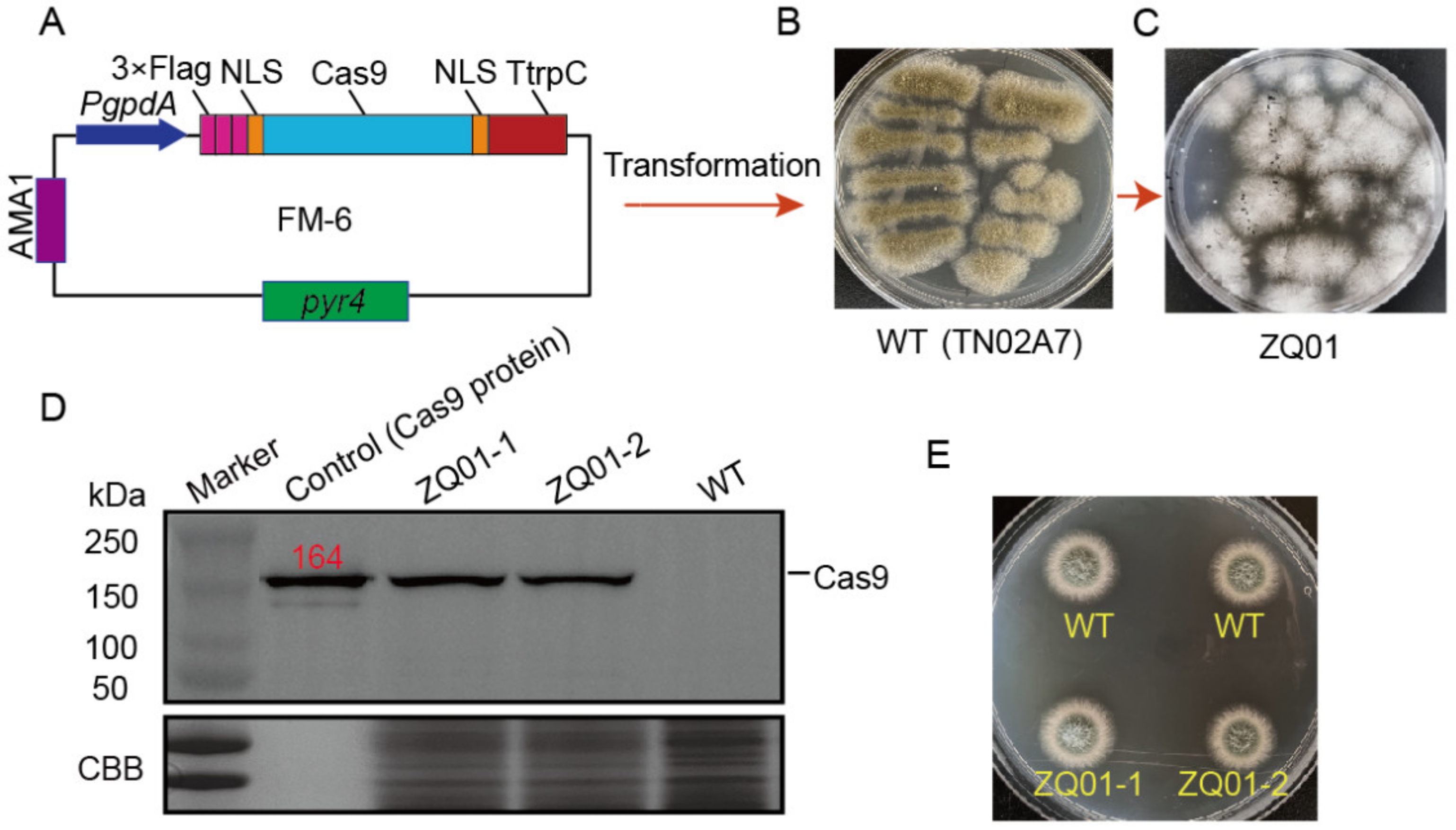
3.1. Expression of Cas9 in A. nidulans
3.2. Establishment of a CRISPR-Mediated Simultaneous Double-Gene-Editing System in A. nidulans
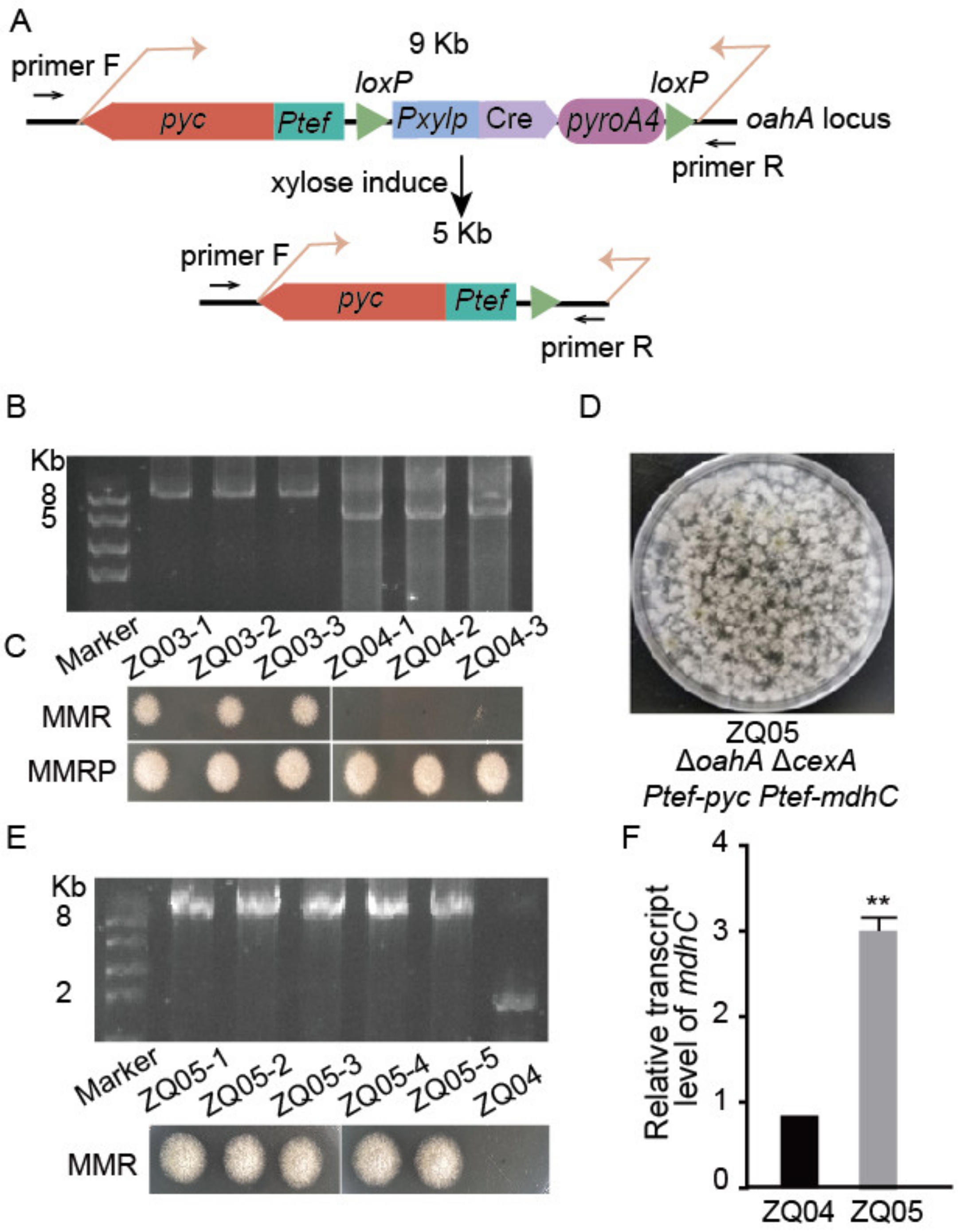
3.3. The Cre-loxP-Mediated Marker Recycling System for Multiple Gene Editing
3.4. Deleting the Putative Citric Acid Transporter Gene (cexA) and Overexpressing Malate Dehydrogenase Gene (mdhC) in A. nidulans
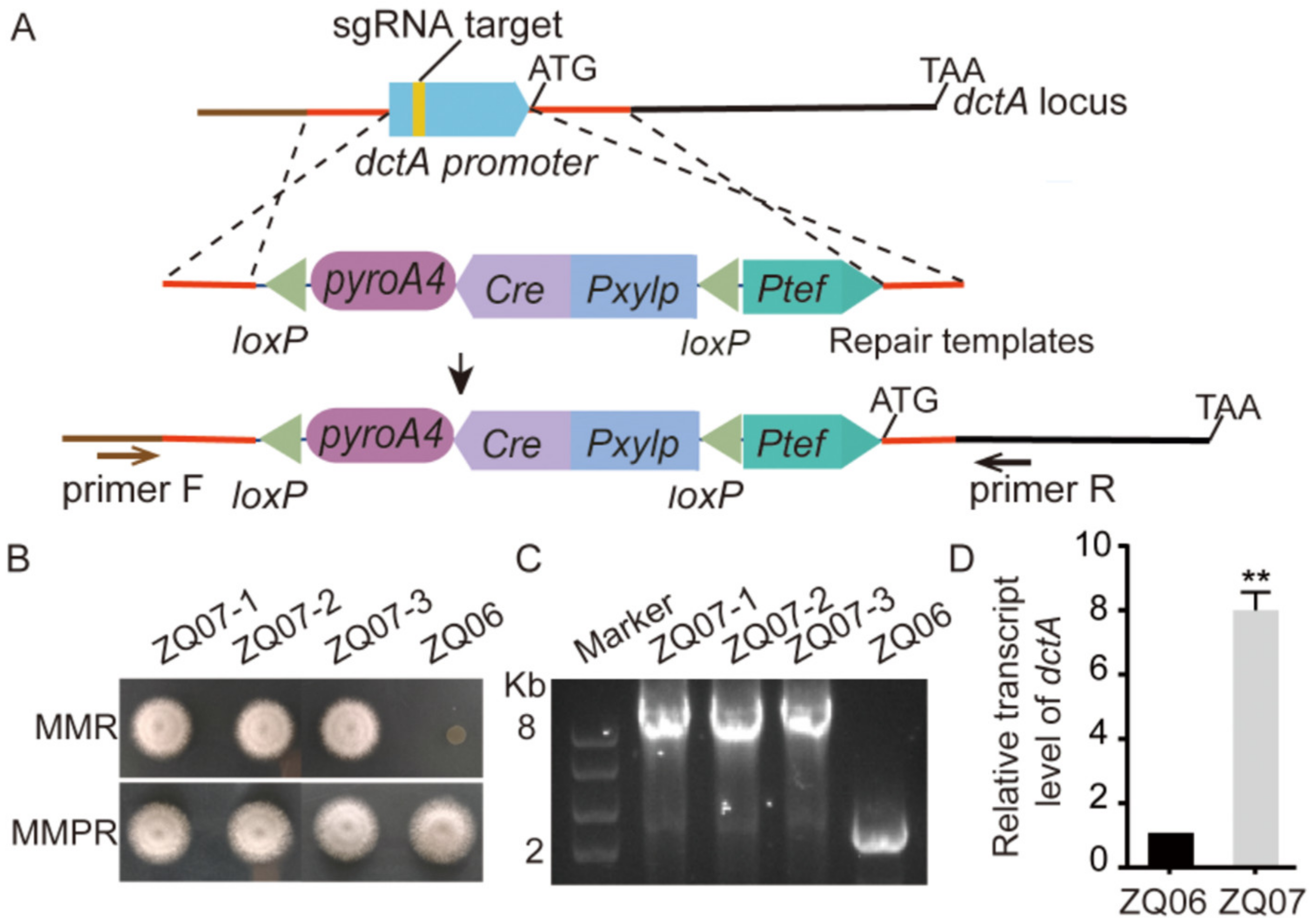
3.5. Overexpressing a Native Putative C4-dicarboxylate Transporter-DctA in A. nidulans
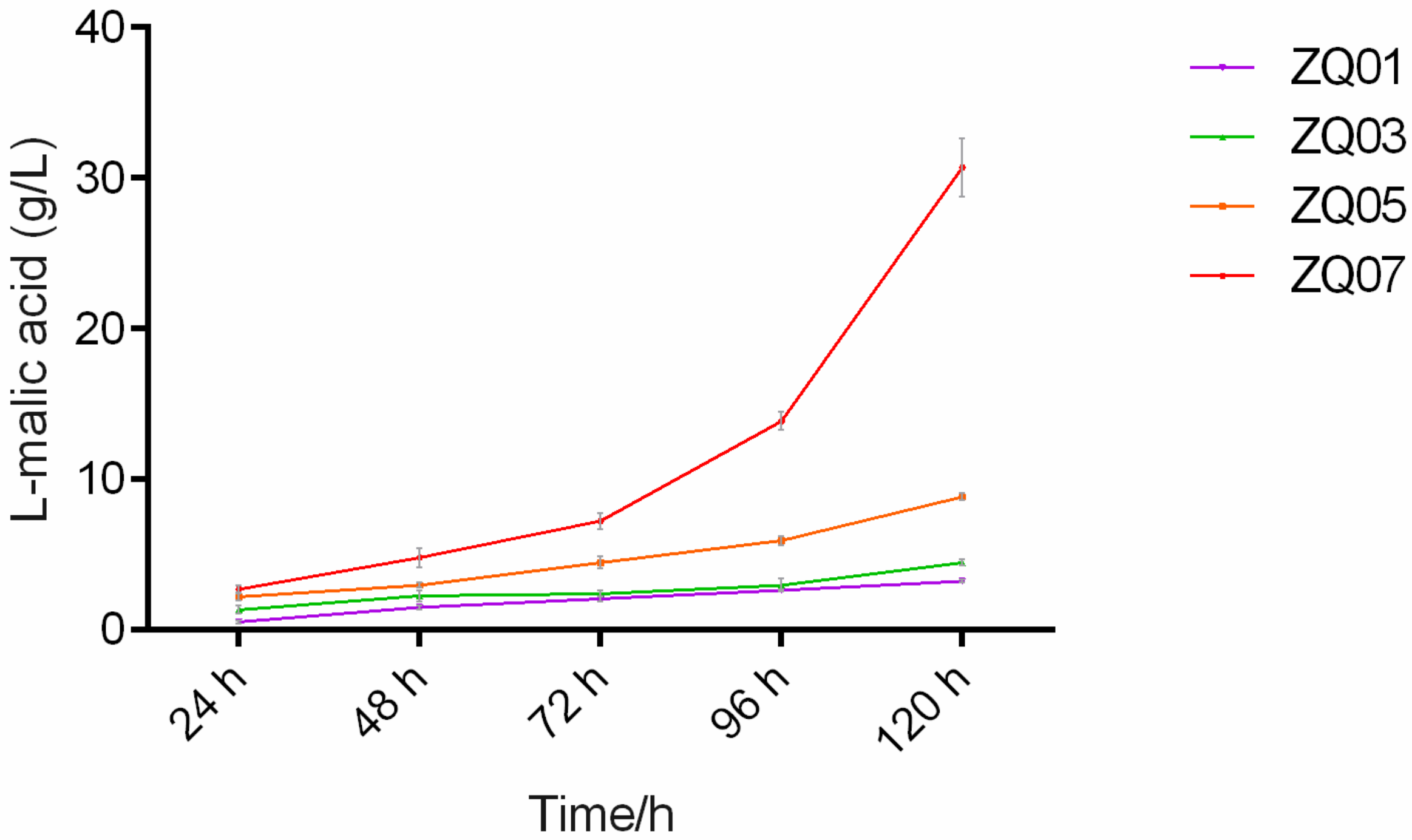
3.6. A. nidulans Has Good Potential to Produce L-malic Acid
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, Q.; Ye, C. Microbial cell factories based on filamentous bacteria, yeasts, and fungi. Microb. Cell Factories 2023, 22, 20. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.-M.; Lin, T.-S.; Wang, C.C.C. Total Heterologous Biosynthesis of Fungal Natural Products in Aspergillus nidulans. J. Nat. Prod. 2022, 85, 2484–2518. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.; Wu, B.; Ram, A.F.J.B.l. Aspergillus as a multi-purpose cell factory: Current status and perspectives. Biotechnol. Lett. 2011, 33, 469–476. [Google Scholar] [CrossRef]
- El-Hawary, S.S.; Moawad, A.S.; Bahr, H.S.; Abdelmohsen, U.R.; Mohammed, R. Natural product diversity from the endophytic fungi of the genus Aspergillus. RSC Adv. 2020, 10, 22058–22079. [Google Scholar] [CrossRef]
- Kumar, A. Aspergillus nidulans: A Potential Resource of the Production of the Native and Heterologous Enzymes for Industrial Applications. Int. J. Microbiol. 2020, 2020, 8894215. [Google Scholar] [CrossRef]
- Caesar, L.K.; Kelleher, N.L.; Keller, N.P. In the fungus where it happens: History and future propelling Aspergillus nidulans as the archetype of natural products research. Fungal Genet. Biol. 2020, 144, 103477. [Google Scholar] [CrossRef] [PubMed]
- Jami, M.-S.; Martín, J.-F.; Barreiro, C.; Domínguez-Santos, R.; Vasco-Cárdenas, M.-F.; Pascual, M.; García-Estrada, C. Catabolism of phenylacetic acid in Penicillium rubens. Proteome-wide analysis in response to the benzylpenicillin side chain precursor. J. Proteom. 2018, 187, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; An, C.; Yao, Y.; Meng, X.; Gao, S.-S. De novo biosynthesis and gram-level production of m-cresol in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2021, 105, 6333–6343. [Google Scholar] [CrossRef]
- Romsdahl, J.; Wang, C.C.C. Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012–2018). MedChemComm 2019, 10, 840–866. [Google Scholar] [CrossRef]
- Segato, F.; Damásio, A.R.; Gonçalves, T.A.; de Lucas, R.C.; Squina, F.M.; Decker, S.R.; Prade, R.A. High-yield secretion of multiple client proteins in Aspergillus. Enzym. Microb. Technol. 2012, 51, 100–106. [Google Scholar] [CrossRef]
- Ballmann, P.; Lightfoot, J.; Müller, M.; Dröge, S.; Prade, R. Redesigning the Aspergillus nidulans xylanase regulatory pathway to enhance cellulase production with xylose as the carbon and inducer source. Microb. Cell Factories 2019, 18, 193. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.M.M.; Martins, M.; Goldbeck, R. Heterologous Expression of Lignocellulose-Modifying Enzymes in Microorganisms: Current Status. Mol. Biotechnol. 2021, 63, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Hillary, V.E.; Ceasar, S.A. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol. Biotechnol. 2022, 65, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Li, L.-X.; Yu, L.-Y.; Wang, B.; Pan, L. Impact of overexpressing NADH kinase on glucoamylase production in Aspergillus niger. J. Ind. Microbiol. Biotechnol. 2022, 49, kuac015. [Google Scholar] [CrossRef]
- Kun, R.S.; Meng, J.; Salazar-Cerezo, S.; Mäkelä, M.R.; de Vries, R.P.; Garrigues, S. CRISPR/Cas9 facilitates rapid generation of constitutive forms of transcription factors in Aspergillus niger through specific on-site genomic mutations resulting in increased saccharification of plant biomass. Enzym. Microb. Technol. 2020, 136, 109508. [Google Scholar] [CrossRef]
- Zhang, C.; Meng, X.; Wei, X.; Lu, L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 2016, 86, 47–57. [Google Scholar] [CrossRef]
- Wei, Z.; Xu, Y.; Xu, Q.; Cao, W.; Huang, H.; Liu, H. Microbial Biosynthesis of L-Malic Acid and Related Metabolic Engineering Strategies: Advances and Prospects. Front. Bioeng. Biotechnol. 2021, 9, 765685. [Google Scholar] [CrossRef]
- Brown, S.H.; Bashkirova, L.; Berka, R.; Chandler, T.; Doty, T.; McCall, K.; McCulloch, M.; McFarland, S.; Thompson, S.; Yaver, D.; et al. Metabolic engineering of Aspergillus oryzae NRRL 3488 for increased production of l-malic acid. Appl. Microbiol. Biotechnol. 2013, 97, 8903–8912. [Google Scholar] [CrossRef]
- Xu, Y.; Shan, L.; Zhou, Y.; Xie, Z.; Ball, A.S.; Cao, W.; Liu, H. Development of a Cre-loxP-based genetic system in Aspergillus niger ATCC1015 and its application to construction of efficient organic acid-producing cell factories. Appl. Microbiol. Biotechnol. 2019, 103, 8105–8114. [Google Scholar] [CrossRef]
- Liu, J.; Xie, Z.; Shin, H.-D.; Li, J.; Du, G.; Chen, J.; Liu, L. Rewiring the reductive tricarboxylic acid pathway and L-malate transport pathway of Aspergillus oryzae for overproduction of L-malate. J. Biotechnol. 2017, 253, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, H.; Zhan, T.; Qin, Y.; Fan, F.; Zhang, X. Metabolic engineering of the acid-tolerant yeast Pichia kudriavzevii for efficient L-malic acid production at low pH. Metab. Eng. 2023, 75, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, Q.; Kong, X.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Ledesma-Amaro, R.; Chen, J.; Liu, L. Highly efficient neutralizer-free l-malic acid production using engineered Saccharomyces cerevisiae. Bioresour. Technol. 2023, 370, 128580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lu, L. Precise and Efficient In-Frame Integration of an Exogenous GFP Tag in Aspergillus fumigatus by a CRISPR System. Methods Mol. Biol. 2017, 1625, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ban, A.; Ebara, N.; Mizutani, O.; Tanaka, M.; Shintani, T.; Gomi, K. Self-excising Cre/mutant lox marker recycling system for multiple gene integrations and consecutive gene deletions in Aspergillus oryzae. J. Biosci. Bioeng. 2017, 123, 403–411. [Google Scholar] [CrossRef]
- Hartmann, T.; Dümig, M.; Jaber, B.M.; Szewczyk, E.; Olbermann, P.; Morschhäuser, J.; Krappmann, S. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system. Appl. Environ. Microbiol. 2010, 76, 6313–6317. [Google Scholar] [CrossRef]
- Nandakumar, M.P.; Shen, J.; Raman, B.; Marten, M.R. Solubilization of Trichloroacetic Acid (TCA) Precipitated Microbial Proteins via NaOH for Two-Dimensional Electrophoresis. J. Proteome Res. 2003, 2, 89–93. [Google Scholar] [CrossRef]
- Van Duyne, G.D. Cre Recombinase. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Yamada, K.; Yamamoto, T.; Uwasa, K.; Osakabe, K.; Takano, Y. The establishment of multiple knockout mutants of Colletotrichum orbiculare by CRISPR-Cas9 and Cre-loxP systems. Fungal Genet. Biol. 2023, 165, 103777. [Google Scholar] [CrossRef]
- Cao, W.; Yan, L.; Li, M.; Liu, X.; Xu, Y.; Xie, Z.; Liu, H. Identification and engineering a C4-dicarboxylate transporter for improvement of malic acid production in Aspergillus niger. Appl. Microbiol. Biotechnol. 2020, 104, 9773–9783. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, Y.; Cao, W.; Liu, H. Improved Production of Malic Acid in Aspergillus niger by Abolishing Citric Acid Accumulation and Enhancing Glycolytic Flux. ACS Synth. Biol. 2020, 9, 1418–1425. [Google Scholar] [CrossRef] [PubMed]

| Strain | Genotype | Source |
|---|---|---|
| TN02A7/WT | pyrG89; pyroA4; nkuA::argB2; riboB2; veA1 | FGSC |
| ZQ01/TN02A7cas9 | pyrG89; pyroA4; nkuA::argB2; riboB2; veA1; pyr4::cas9 | This study |
| ZQ02/ΔoahA Pgpd-pyc | ZQ01, ΔoahA::Pgpd-pyc-loxP-Pxylp-Cre-pyroA-loxP | This study |
| ZQ03/ΔoahA Ptef-pyc | ZQ01, ΔoahA::Ptef-pyc-loxP-Pxylp-Cre-pyroA-loxP | This study |
| ZQ04/ΔoahA ΔpyroA Ptef-pyc | ZQ01, ΔoahA::Ptef-pyc, | This study |
| ZQ05/ΔoahA ΔcexA Ptef-pyc Ptef-mdhC | ZQ04, ΔcexA::Ptef-mdhC-loxP-Pxylp-Cre-pyroA-loxP | This study |
| ZQ06/ΔoahA ΔcexA ΔpyroA Ptef-pyc Ptef-mdhC | ZQ04, ΔcexA::Ptef-mdhC, | This study |
| ZQ07/ΔoahA ΔcexA Ptef-pyc Ptef-mdhC Ptef-dctA | ZQ06, pryoA::Ptef-dctA | This study |
| Target Gene | The Target Regions of sgRNA | PAM Sequence |
|---|---|---|
| oahA | 5′-GGCGGAGTTTGGAGGCAG-3′ | 5′-CGG-3′ |
| cexA | 5′-GGACCTAGGATGTGGAAC-3′ | 5′-TGG-3′ |
| dctA promoter | 5′- GGGATTCGAAGCTGAGGC-3′ | 5′-AGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Zhang, C.; Pei, L.; Qian, Q.; Lu, L. Production of L-Malic Acid by Metabolically Engineered Aspergillus nidulans Based on Efficient CRISPR–Cas9 and Cre-loxP Systems. J. Fungi 2023, 9, 719. https://doi.org/10.3390/jof9070719
Chen Z, Zhang C, Pei L, Qian Q, Lu L. Production of L-Malic Acid by Metabolically Engineered Aspergillus nidulans Based on Efficient CRISPR–Cas9 and Cre-loxP Systems. Journal of Fungi. 2023; 9(7):719. https://doi.org/10.3390/jof9070719
Chicago/Turabian StyleChen, Ziqing, Chi Zhang, Lingling Pei, Qi Qian, and Ling Lu. 2023. "Production of L-Malic Acid by Metabolically Engineered Aspergillus nidulans Based on Efficient CRISPR–Cas9 and Cre-loxP Systems" Journal of Fungi 9, no. 7: 719. https://doi.org/10.3390/jof9070719
APA StyleChen, Z., Zhang, C., Pei, L., Qian, Q., & Lu, L. (2023). Production of L-Malic Acid by Metabolically Engineered Aspergillus nidulans Based on Efficient CRISPR–Cas9 and Cre-loxP Systems. Journal of Fungi, 9(7), 719. https://doi.org/10.3390/jof9070719






