Spatial Patterns of Planktonic Fungi Indicate Their Potential Contributions to Biological Carbon Pump and Organic Matter Remineralization in the Water Column of South China Sea
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Water Samples
2.2. Fungal and Bacterial Community Analysis
2.2.1. DNA Extraction
2.2.2. PCR Amplification
2.2.3. Downstream Processing of Sequencing Reads
2.3. Quantitative PCR Analysis
2.3.1. Fungal 18S rRNA Gene Copies
2.3.2. Bacterial 16S rRNA Gene Copies
2.4. Statistical Analysis
2.5. Prediction of Trophic Modes
2.6. Network Analysis
2.7. Identification of Metabolic Functions
3. Results
3.1. Environmental Characteristics in Relation to Various Water Zones
3.2. Distribution and Abundance Patterns
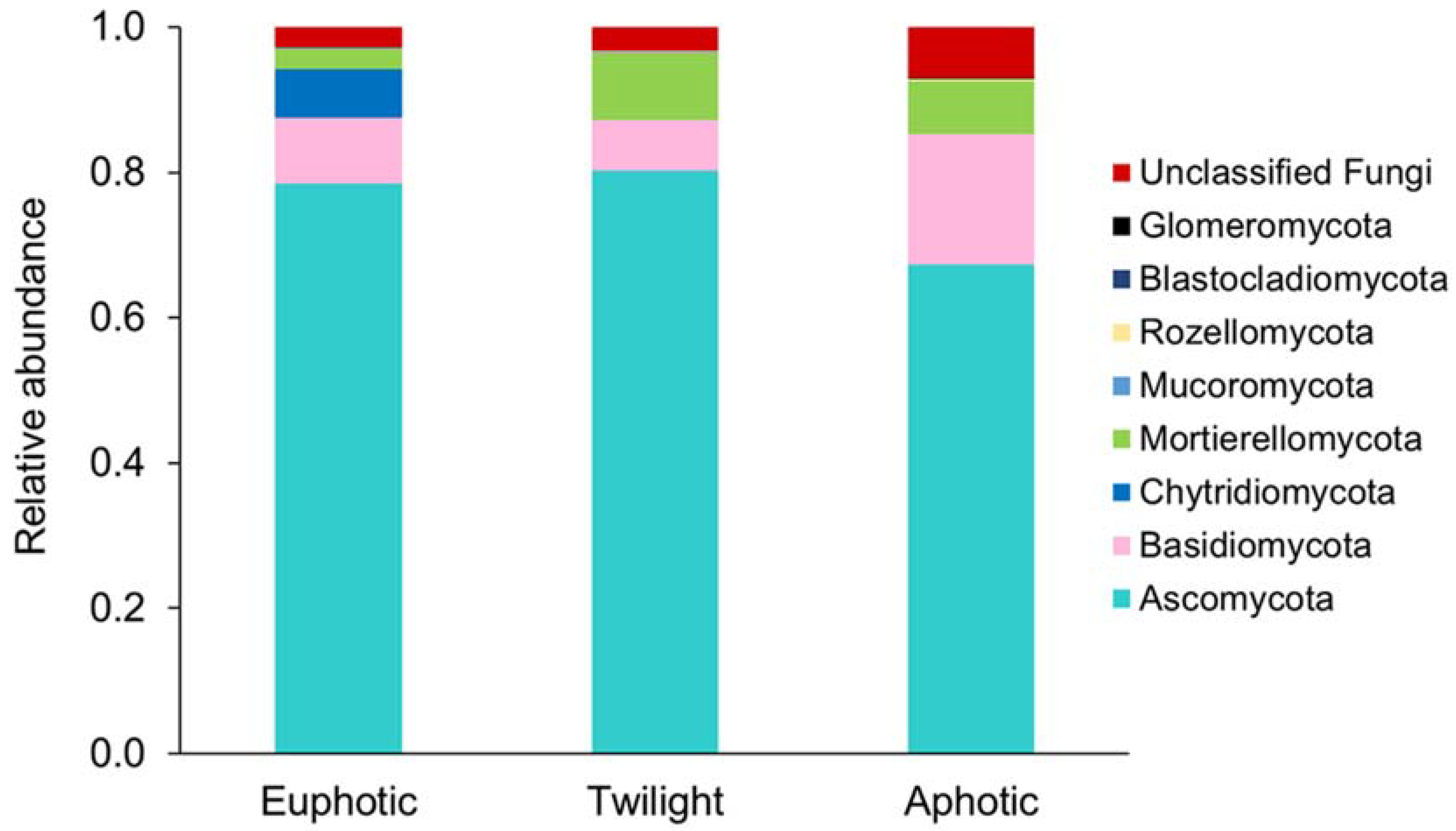
3.3. Variations in Community Composition Related to Various Water Zones
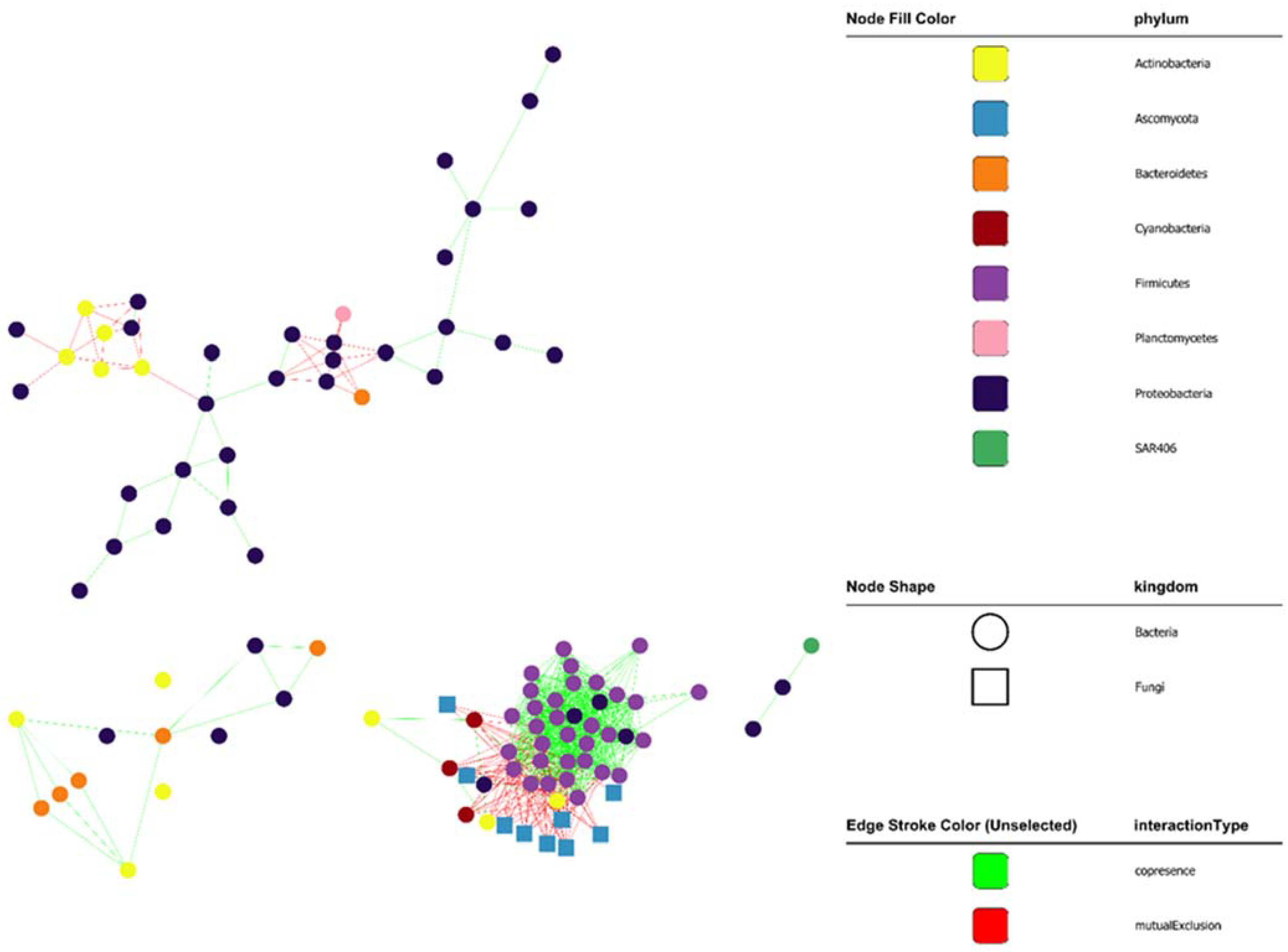
3.4. Interactions between Planktonic Communities
3.5. Trophic Modes of Mycoplankton
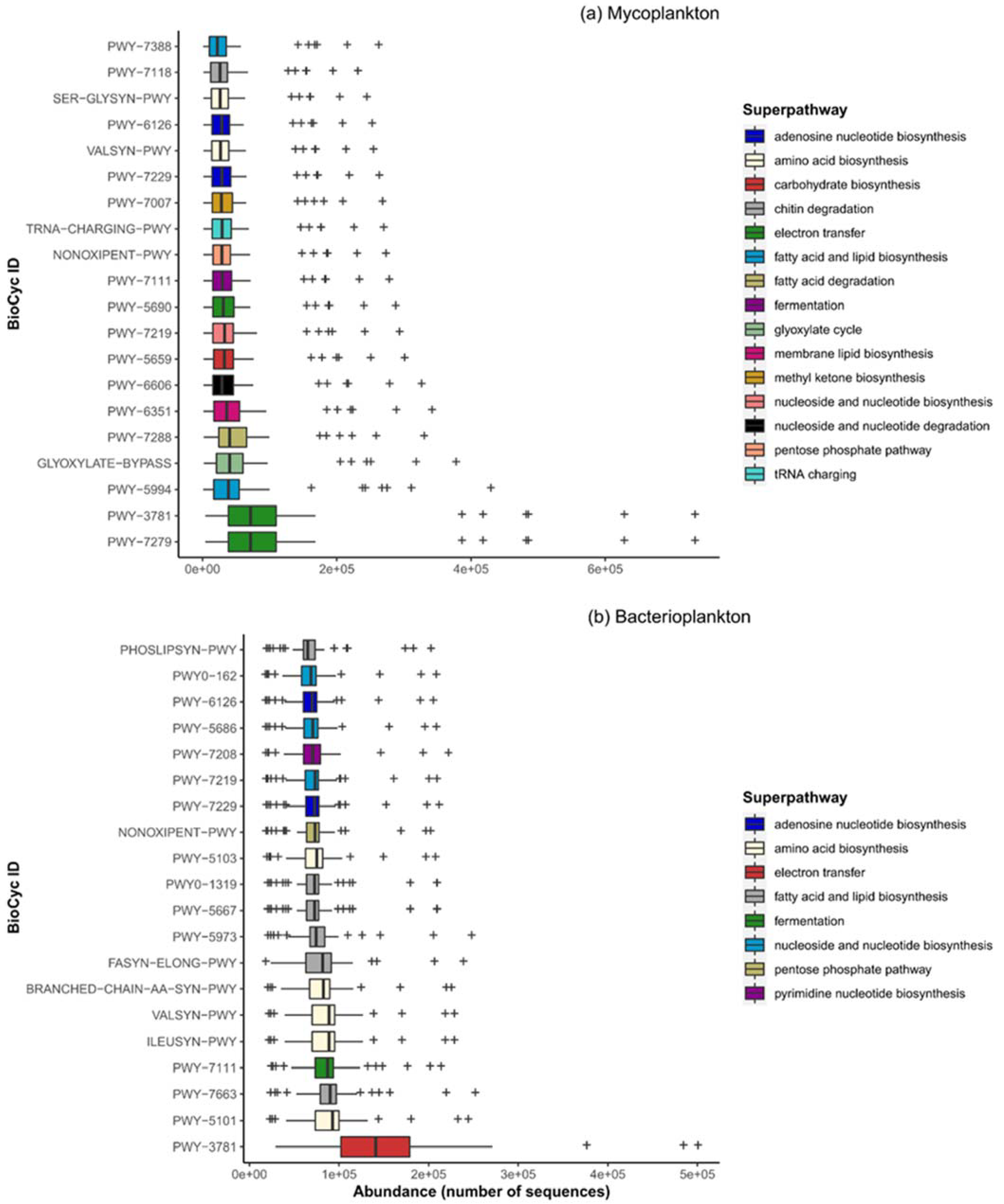
3.6. Metabolic Potential of Plankton Communities
4. Discussion
4.1. Fungal Abundance Patterns and Their Significance in the Water Column
4.2. Abiotic and Biotic Interactions and Their Implications
4.3. Implications of Trophic Modes and Metabolic Functions of Mycoplankton
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cunliffe, M.; Hollingsworth, A.; Bain, C.; Sharma, V.; Taylor, J.D. Algal polysaccharide utilisation by saprotrophic planktonic marine fungi. Fungal Ecol. 2017, 30, 135–138. [Google Scholar] [CrossRef]
- Duan, Y.; Xie, N.; Song, Z.; Ward, C.S.; Yung, C.-M.; Hunt, D.E.; Johnson, Z.I.; Wang, G. A High-resolution Time-series Reveals Seasonal Patterns of Planktonic Fungi at a Temperate Coastal Ocean Site (Beaufort, North Carolina, USA). Appl. Environ. Microbiol. 2018, 84, e00967-18. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M.H.; Pantoja, S.; Quiñones, R.A.; González, R.R. First record of filamentous fungi in the coastal upwelling ecosystem off central Chile. Gayana 2010, 74, 66–73. [Google Scholar]
- Sen, K.; Bai, M.; Sen, B.; Wang, G. Disentangling the structure and function of mycoplankton communities in the context of marine environmental heterogeneity. Sci. Total Environ. 2021, 766, 142635. [Google Scholar] [CrossRef]
- Taylor, J.D.; Cunliffe, M. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J. 2016, 10, 2118–2128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sen, K.; He, Y.; Xie, Y.; Wang, G. Impact of environmental gradients on the abundance and diversity of planktonic fungi across coastal habitats of contrasting trophic status. Sci. Total Environ. 2019, 683, 822–833. [Google Scholar] [CrossRef]
- Jeffries, T.C.; Curlevski, N.J.; Brown, M.V.; Harrison, D.P.; Doblin, M.A.; Petrou, K.; Ralph, P.J.; Seymour, J.R. Partitioning of fungal assemblages across different marine habitats. Environ. Microbiol. Rep. 2016, 8, 235–238. [Google Scholar] [CrossRef]
- Morales, S.; Biswas, A.; Herndl, G.; Baltar, F. Global Structuring of Phylogenetic and Functional Diversity of Pelagic Fungi by Depth and Temperature. Front. Mar. Sci. 2019, 6, 131. [Google Scholar] [CrossRef]
- Wang, X.; Singh, P.; Gao, Z.; Zhang, X.; Johnson, Z.I.; Wang, G.Y. Distribution and diversity of planktonic fungi in the West Pacific Warm Pool. PLoS ONE 2014, 9, e101523. [Google Scholar] [CrossRef]
- Sen, K.; Sen, B.; Wang, G. Diversity, Abundance, and Ecological Roles of Planktonic Fungi in Marine Environments. J. Fungi 2022, 8, 491. [Google Scholar] [CrossRef]
- Gutiérrez, M.H.; Pantoja, S.; Tejos, E.; Quiñones, R.A. The role of fungi in processing marine organic matter in the upwelling ecosystem off Chile. Mar. Biol. 2011, 158, 205–219. [Google Scholar] [CrossRef]
- Hassett, B.T.; Borrego, E.J.; Vonnahme, T.R.; Rämä, T.; Kolomiets, M.V.; Gradinger, R. Arctic marine fungi: Biomass, functional genes, and putative ecological roles. ISME J. 2019, 13, 1484–1496. [Google Scholar] [CrossRef]
- Raghukumar, S. The Role of Fungi in Marine Detrital Processes. In Marine Microbiology: Facets and Opportunities; Ramaiah, N., Ed.; National Institute of Oceanography: Goa, India, 2004; pp. 91–101. [Google Scholar]
- Damare, S.; Raghukumar, C. Fungi and macroaggregation in deep-sea sediments. Microb. Ecol. 2008, 56, 168–177. [Google Scholar] [CrossRef]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the Marine Environment: Open Questions and Unsolved Problems. mBio 2019, 10, e01189-18. [Google Scholar] [CrossRef]
- Bochdansky, A.B.; Clouse, M.A.; Herndl, G.J. Eukaryotic microbes, principally fungi and labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME J. 2017, 11, 362–373. [Google Scholar] [CrossRef]
- Gutiérrez, M.H.; Jara, A.M.; Pantoja, S. Fungal parasites infect marine diatoms in the upwelling ecosystem of the Humboldt current system off central Chile. Environ. Microbiol. 2016, 18, 1646–1653. [Google Scholar] [CrossRef] [PubMed]
- Worden, A.Z.; Follows, M.J.; Giovannoni, S.J.; Wilken, S.; Zimmerman, A.E.; Keeling, P.J. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 2015, 347, 1257594. [Google Scholar] [CrossRef]
- Banos, S.; Gysi, D.M.; Richter-Heitmann, T.; Glöckner, F.O.; Boersma, M.; Wiltshire, K.H.; Gerdts, G.; Wichels, A.; Reich, M. Seasonal Dynamics of Pelagic Mycoplanktonic Communities: Interplay of Taxon Abundance, Temporal Occurrence, and Biotic Interactions. Front. Microbiol. 2020, 11, 1305. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Kawamura, H.; Hong, H.; Qi, Y. A Review on the Currents in the South China Sea: Seasonal Circulation, South China Sea Warm Current and Kuroshio Intrusion. J. Oceanogr. 2000, 56, 607–624. [Google Scholar] [CrossRef]
- Li, J.; Li, N.; Li, F.; Zou, T.; Yu, S.; Wang, Y.; Qin, S.; Wang, G. Spatial Diversity of Bacterioplankton Communities in Surface Water of Northern South China Sea. PLoS ONE 2014, 9, e113014. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.; Dai, M.; Jiao, N.; Herndl, G.J. Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Mol. Ecol. 2014, 23, 2260–2274. [Google Scholar] [CrossRef]
- Arístegui, J.; Gasol, J.M.; Duarte, C.M.; Herndld, G.J. Microbial oceanography of the dark ocean’s pelagic realm. Limnol. Oceanogr. 2009, 54, 1501–1529. [Google Scholar] [CrossRef]
- Robinson, C.; Steinberg, D.K.; Anderson, T.R.; Arístegui, J.; Carlson, C.A.; Frost, J.R.; Ghiglione, J.-F.; Hernández-León, S.; Jackson, G.A.; Koppelmann, R.; et al. Mesopelagic zone ecology and biogeochemistry—A synthesis. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2010, 57, 1504–1518. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Srinivasan, S.; Hoffman, N.G.; Morgan, M.T.; Matsen, F.A.; Fiedler, T.L.; Hall, R.W.; Ross, F.J.; McCoy, C.O.; Bumgarner, R.; Marrazzo, J.M.; et al. Bacterial communities in women with bacterial vaginosis: High resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS ONE 2012, 7, e37818. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr. 2018, 6, e27295v27291. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Dollive, S.; Peterfreund, G.L.; Sherrill-Mix, S.; Bittinger, K.; Sinha, R.; Hoffmann, C.; Nabel, C.S.; Hill, D.A.; Artis, D.; Bachman, M.A.; et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012, 13, R60. [Google Scholar] [CrossRef]
- Wang, Y.; Sen, B.; He, Y.; Xie, N.; Wang, G. Spatiotemporal Distribution and Assemblages of Planktonic Fungi in the Coastal Waters of the Bohai Sea. Front. Microbiol. 2018, 9, 584. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Faust, K.; Sathirapongsasuti, J.F.; Izard, J.; Segata, N.; Gevers, D.; Raes, J.; Huttenhower, C. Microbial Co-occurrence Relationships in the Human Microbiome. PLoS Comput. Biol. 2012, 8, e1002606. [Google Scholar] [CrossRef] [PubMed]
- Debroas, D.; Domaizon, I.; Humbert, J.F.; Jardillier, L.; Lepere, C.; Oudart, A.; Taib, N. Overview of freshwater microbial eukaryotes diversity: A first analysis of publicly available metabarcoding data. FEMS Microbiol. Ecol. 2017, 93, fix023. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Burgaud, G.; Woehlke, S.; Rédou, V.; Orsi, W.; Beaudoin, D.; Barbier, G.; Biddle, J.F.; Edgcomb, V.P. Deciphering the presence and activity of fungal communities in marine sediments using a model estuarine system. Aquat. Microb. Ecol. 2013, 70, 45–62. [Google Scholar] [CrossRef]
- Vargas-Gastélum, L.; Riquelme, M. The Mycobiota of the Deep Sea: What Omics Can Offer. Life 2020, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Hassett, B.T.; Gradinger, R. Chytrids dominate arctic marine fungal communities. Environ. Microbiol. 2016, 18, 2001–2009. [Google Scholar] [CrossRef]
- Bunbury-Blanchette, A.L.; Walker, A.K. Occurrence and Distribution of Fungi in Saline Environments. In Microorganisms in Saline Environments: Strategies and Function; Giri, B., Varma, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 19–38. [Google Scholar]
- Hyde, K.D.; Jones, E.B.G.; Leaño, E.; Pointing, S.B.; Poonyth, A.D.; Vrijmoed, L.L.P. Role of fungi in marine ecosystems. Biodivers. Conserv. 1998, 7, 1147–1161. [Google Scholar] [CrossRef]
- Turner, J.T. Zooplankton fecal pellets, marine snow, phytodetritus and the ocean’s biological pump. Prog. Oceanogr. 2015, 130, 205–248. [Google Scholar] [CrossRef]
- Duret, M.T.; Lampitt, R.S.; Lam, P. Eukaryotic influence on the oceanic biological carbon pump in the Scotia Sea as revealed by 18S rRNA gene sequencing of suspended and sinking particles. Limnol. Oceanogr. 2020, 65, S49–S70. [Google Scholar] [CrossRef]
- Jiao, N.; Herndl, G.J.; Hansell, D.A.; Benner, R.; Kattner, G.; Wilhelm, S.W.; Kirchman, D.L.; Weinbauer, M.G.; Luo, T.; Chen, F.; et al. Microbial production of recalcitrant dissolved organic matter: Long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 2010, 8, 593. [Google Scholar] [CrossRef]
- Clipson, N.; Otte, M.; Landy, E.; Gadd, G.M. Biogeochemical roles of fungi in marine and estuarine habitats. In Fungi in Biogeochemical Cycles; Cambridge University Press: Cambridge, UK, 2006; pp. 436–461. [Google Scholar]
- Raghukumar, S. Speculations on niches occupied by fungi in the sea with relation to bacteria. Proc. Indian Acad. Sci. (Plant Sci.) 1990, 100, 129–138. [Google Scholar] [CrossRef]
- Wymore, A.S.; Compson, Z.G.; Liu, C.M.; Price, L.B.; Whitham, T.G.; Keim, P.; Marks, J.C. Contrasting rRNA gene abundance patterns for aquatic fungi and bacteria in response to leaf-litter chemistry. Freshw. Sci. 2013, 32, 663–672. [Google Scholar] [CrossRef]
- Li, W.; Wang, M.; Burgaud, G.; Yu, H.; Cai, L. Fungal Community Composition and Potential Depth-Related Driving Factors Impacting Distribution Pattern and Trophic Modes from Epi- to Abyssopelagic Zones of the Western Pacific Ocean. Microb. Ecol. 2019, 78, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, T. Yeast Biodiversity in Freshwater, Marine and Deep-Sea Environments. In Biodiversity and Ecophysiology of Yeasts; Péter, G., Rosa, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 241–262. [Google Scholar]
- Pernice, M.C.; Giner, C.R.; Logares, R.; Perera-Bel, J.; Acinas, S.G.; Duarte, C.M.; Gasol, J.M.; Massana, R. Large variability of bathypelagic microbial eukaryotic communities across the world’s oceans. ISME J. 2016, 10, 945–958. [Google Scholar] [CrossRef]
- Bass, D.; Howe, A.; Brown, N.; Barton, H.; Demidova, M.; Michelle, H.; Li, L.; Sanders, H.; Watkinson, S.C.; Willcock, S.; et al. Yeast forms dominate fungal diversity in the deep oceans. Proc. R. Soc. B Biol. Sci. 2007, 274, 3069–3077. [Google Scholar] [CrossRef]
- Mille-Lindblom, C.; Tranvik, L.J. Antagonism between bacteria and fungi on decomposing aquatic plant litter. Microb. Ecol. 2003, 45, 173–182. [Google Scholar] [CrossRef]
- Romaní, A.M.; Fischer, H.; Mille-Lindblom, C.; Tranvik, L.J. Interactions of Bacteria and Fungi on Decomposing Litter: Differential Extracellular Enzyme Activities. Ecology 2006, 87, 2559–2569. [Google Scholar] [CrossRef]
- Voronin, L.V. Terrigenous micromycetes in freshwater ecosystems (review). Inland Water Biol. 2014, 7, 352–356. [Google Scholar] [CrossRef]
- Gajbhiye, M.; Kapadnis, B. Bio-efficiency of Antifungal Lactic Acid Bacterial Isolates for Pomegranate Fruit Rot Management. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 1477–1488. [Google Scholar] [CrossRef]
- Leyva Salas, M.; Mounier, J.; Valence, F.; Coton, M.; Thierry, A.; Coton, E. Antifungal Microbial Agents for Food Biopreservation-A Review. Microorganisms 2017, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Perini, L.; Gostinčar, C.; Anesio, A.M.; Williamson, C.; Tranter, M.; Gunde-Cimerman, N. Darkening of the Greenland Ice Sheet: Fungal Abundance and Diversity Are Associated With Algal Bloom. Front. Microbiol. 2019, 10, 557. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Gao, Y.-H.; Gong, L.-F.; Li, M.; Pang, K.-L.; Luo, Z.-H. Fungal diversity in the deep-sea hadal sediments of the Yap Trench by cultivation and high throughput sequencing methods based on ITS rRNA gene. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2019, 145, 125–136. [Google Scholar] [CrossRef]
- Xu, W.; Gong, L.-f.; Pang, K.-L.; Luo, Z.-H. Fungal diversity in deep-sea sediments of a hydrothermal vent system in the Southwest Indian Ridge. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2018, 131, 16–26. [Google Scholar] [CrossRef]
- Krause, E.; Wichels, A.; Giménez, L.; Gerdts, G. Marine fungi may benefit from ocean acidification. Aquat. Microb. Ecol. 2013, 69, 59–67. [Google Scholar] [CrossRef]
- Singh, P.; Raghukumar, C.; Verma, P.; Shouche, Y. Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Divers. 2010, 40, 89–102. [Google Scholar] [CrossRef]
- Wang, M.; Mara, P.; Burgaud, G.; Edgcomb, V.; Long, X.; Yang, H.; Cai, L.; Li, W. Metatranscriptomics and metabarcoding reveal spatiotemporal shifts in fungal communities and their activities in Chinese coastal waters. Mol. Ecol. 2023, 32, 2750–2765. [Google Scholar] [CrossRef]
- Orsi, W.; Biddle, J.F.; Edgcomb, V. Deep sequencing of subseafloor eukaryotic rRNA reveals active Fungi across marine subsurface provinces. PLoS ONE 2013, 8, e56335. [Google Scholar] [CrossRef]
- Chrismas, N.; Cunliffe, M. Depth-dependent mycoplankton glycoside hydrolase gene activity in the open ocean—Evidence from the Tara Oceans eukaryote metatranscriptomes. ISME J. 2020, 14, 2361–2365. [Google Scholar] [CrossRef]
- Breyer, E.; Zhao, Z.; Herndl, G.J.; Baltar, F. Global contribution of pelagic fungi to protein degradation in the ocean. Microbiome 2022, 10, 143. [Google Scholar] [CrossRef]
- Orsi, W.D.; Vuillemin, A.; Coskun, O.K.; Rodriguez, P.; Oertel, Y.; Niggemann, J.; Mohrholz, V.; Gomez-Saez, G.V. Carbon assimilating fungi from surface ocean to subseafloor revealed by coupled phylogenetic and stable isotope analysis. ISME J. 2022, 16, 1245–1261. [Google Scholar] [CrossRef] [PubMed]
- Baltar, F.; Zhao, Z.; Herndl, G.J. Potential and expression of carbohydrate utilization by marine fungi in the global ocean. Microbiome 2021, 9, 106. [Google Scholar] [CrossRef] [PubMed]



| Zones | Mycoplankton (18S rRNA Gene Copies/Ng DNA) | Bacterioplankton (16S rRNA Gene Copies/Ng DNA) |
|---|---|---|
| Euphotic | 3588 ± 1585 b | (9.09 ± 3.1) × 107 b |
| Twilight | 923 ± 241 c | (8.67 ± 2.95) × 106 a |
| Aphotic | 194 ± 122 a | (1.09 ± 0.87) × 106 a |
| Trophic Modes | Euphotic | Twilight | Aphotic |
|---|---|---|---|
| Pathotroph | 1471 ± 518 a | 3040 ± 934 a | 811 ± 248 a |
| Saprotroph | 1003 ± 263 b | 6116 ± 1675 a | 2038 ± 535 a |
| Symbiotroph | 80.7 ± 25.6 a | 335 ± 131 a | 162 ± 84.5 a |
| Pathotroph–Saprotroph | 699 ± 179 a | 1419 ± 372 a | 514 ± 149 a |
| Pathotroph–Symbiotroph | 14.4 ± 9.36 a | 348 ± 145 b | 32.8 ± 22.9 ab |
| Saprotroph–Symbiotroph | 18.5 ± 8.24 a | 23.2 ± 17.5 a | 11.0 ± 9.11 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sen, K.; Bai, M.; Li, J.; Ding, X.; Sen, B.; Wang, G. Spatial Patterns of Planktonic Fungi Indicate Their Potential Contributions to Biological Carbon Pump and Organic Matter Remineralization in the Water Column of South China Sea. J. Fungi 2023, 9, 640. https://doi.org/10.3390/jof9060640
Sen K, Bai M, Li J, Ding X, Sen B, Wang G. Spatial Patterns of Planktonic Fungi Indicate Their Potential Contributions to Biological Carbon Pump and Organic Matter Remineralization in the Water Column of South China Sea. Journal of Fungi. 2023; 9(6):640. https://doi.org/10.3390/jof9060640
Chicago/Turabian StyleSen, Kalyani, Mohan Bai, Jiaqian Li, Xueyan Ding, Biswarup Sen, and Guangyi Wang. 2023. "Spatial Patterns of Planktonic Fungi Indicate Their Potential Contributions to Biological Carbon Pump and Organic Matter Remineralization in the Water Column of South China Sea" Journal of Fungi 9, no. 6: 640. https://doi.org/10.3390/jof9060640
APA StyleSen, K., Bai, M., Li, J., Ding, X., Sen, B., & Wang, G. (2023). Spatial Patterns of Planktonic Fungi Indicate Their Potential Contributions to Biological Carbon Pump and Organic Matter Remineralization in the Water Column of South China Sea. Journal of Fungi, 9(6), 640. https://doi.org/10.3390/jof9060640











