Tubulin Polymerization Promoting Proteins (TPPPs) of Aphelidiomycota: Correlation between the Incidence of p25alpha Domain and the Eukaryotic Flagellum
Abstract
1. Introduction
2. Materials and Methods
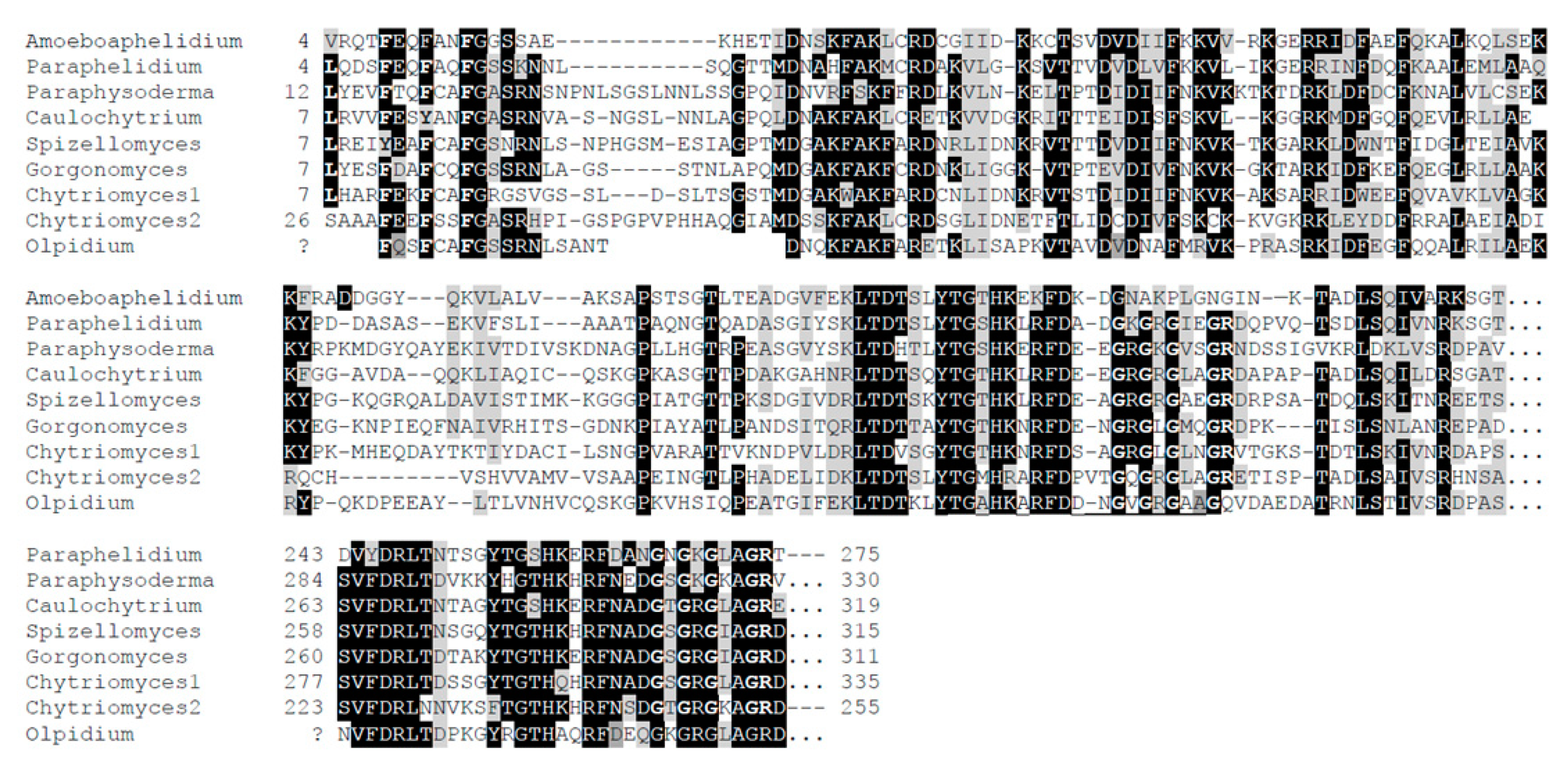
3. Results and Discussion
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Avidor-Reiss, T.; Maer, A.M.; Koundakjian, E.; Polyanovsky, A.; Keil, T.; Subramaniam, S.; Zuker, C.S. Decoding cilia function: Defining specialized genes required for compartmentalized cilia biogenesis. Cell 2004, 117, 527–539. [Google Scholar] [CrossRef]
- Klena, N.; Pigino, G. Structural biology of cilia and intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2022, 38, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Tirián, L.; Hlavanda, E.; Oláh, J.; Horváth, I.; Orosz, F.; Szabó, B.; Kovács, J.; Szabad, J.; Ovádi, J. TPPP/p25 promotes tubulin assemblies and blocks mitotic spindle formation. Proc. Nat. Acad. Sci. USA 2003, 100, 13976–13981. [Google Scholar] [CrossRef]
- Orosz, F.; Ovádi, J. TPPP orthologs are ciliary proteins. FEBS Lett. 2008, 582, 3757–3764. [Google Scholar] [CrossRef]
- Orosz, F. A new protein superfamily: TPPP-like proteins. PLoS ONE 2012, 7, e49276. [Google Scholar] [CrossRef]
- Orosz, F. On the TPPP-like proteins of flagellated Fungi. Fungal Biol. 2021, 125, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Tammana, D.; Tammana, T.V.S. Chlamydomonas FAP265 is a tubulin polymerization promoting protein, essential for flagellar reassembly and hatching of daughter cells from the sporangium. PLoS ONE 2017, 12, e0185108. [Google Scholar] [CrossRef]
- Zhang, C.; Li, D.; Meng, Z.; Zhou, J.; Min, Z.; Deng, S.; Shen, J.; Liu, M. Pyp25α is required for male gametocyte exflagellation. Pathog. Dis. 2022, 80, ftac043. [Google Scholar] [CrossRef]
- Orosz, F. On the TPPP protein of the enigmatic fungus, Olpidium—Correlation between the incidence of p25alpha domain and that of the eukaryotic flagellum. Int. J. Mol. Sci. 2022, 23, 13927. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Chang, Y.; Rochon, D.; Sekimoto, S.; Wang, Y.; Chovatia, M.; Sandor, L.; Salamov, A.; Grigoriev, I.V.; Stajich, J.E.; Spatafora, J.W. Genome-scale phylogenetic analyses confirm Olpidium as the closest living zoosporic fungus to the non-flagellated, terrestrial fungi. Sci. Rep. 2021, 11, 3217. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Torruella, G. Commun_Biol_Aphelid_Datasets. Figshare. Dataset. 2018. Available online: https://doi.org/10.6084/m9.figshare.7339469.v1 (accessed on 14 November 2022).
- Letcher, P.-M.; Powell, M.J. A taxonomic summary of Aphelidiaceae. IMA Fungus 2019, 10, 4. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixture models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Mikhailov, K.V.; Karpov, S.A.; Letcher, P.M.; Lee, P.A.; Logacheva, M.D.; Penin, A.A.; Nesterenko, M.A.; Pozdnyakov, I.R.; Potapenko, E.V.; Sherbakov Panchin, Y.V.; et al. Genomic analysis reveals cryptic diversity in aphelids and sheds light on the emergence of Fungi. Curr. Biol. 2022, 32, 4607–4619. [Google Scholar] [CrossRef] [PubMed]
- Torruella, G.; Grau-Bové, X.; Moreira, D.; Karpov, S.A.; Burns, J.A.; Sebé-Pedrós, A.; Völcker, E.; López-García, P. Global transcriptome analysis of the aphelid Paraphelidium tribonematis supports the phagotrophic origin of fungi. Commun. Biol. 2018, 1, 231. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Ahrendt, S.R.; Quandt, C.A.; Ciobanu, D.; Clum, A.; Salamov, A.; Andreopoulos, B.; Cheng, J.F.; Woyke, T.; Pelin, A.; Henrissat, B.; et al. Leveraging single-cell genomics to expand the fungal tree of life. Nat. Microbiol. 2018, 3, 1417–1428. [Google Scholar] [CrossRef]
- Amses, K.R.; Simmons, D.R.; Longcore, J.E.; Mondo, S.J.; Seto, K.; Jeronimo, G.H.; Bonds, A.E.; Quandt, C.A.; Davis, W.J.; Chang, Y.; et al. Diploid-dominant life cycles characterize the early evolution of Fungi. Proc. Nat. Acad. Sci. USA 2022, 119, e2116841119. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; Koonin, E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002, 18, 619–620. [Google Scholar] [CrossRef] [PubMed]
- Galindo, L.J.; Torruella, G.; López-García, P.; Ciobanu, M.; Gutiérrez-Preciado, A.; Karpov, S.A.; Moreira, D. Phylogenomics supports the monophyly of Aphelids and Fungi and identifies new molecular synapomorphies. Syst. Biol. 2022, online ahead of print. [CrossRef] [PubMed]
- Youssef, N.H.; Couger, M.B.; Struchtemeyer, C.G.; Liggenstoffer, A.S.; Prade, R.A.; Najar, F.Z.; Atiyeh, H.K.; Wilkins, M.R.; Elshahed, M.S. The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 2013, 79, 4620–4634. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- Karpov, S.A.; Mikhailov, K.V.; Mirzaeva, G.S.; Mirabdullaev, I.M.; Mamkaeva, K.A.; Titova, N.N.; Aleoshin, V.V. Obligately phagotrophic aphelids turned out to branch with the earliest-diverging fungi. Protist 2013, 164, 195–205. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orosz, F. Tubulin Polymerization Promoting Proteins (TPPPs) of Aphelidiomycota: Correlation between the Incidence of p25alpha Domain and the Eukaryotic Flagellum. J. Fungi 2023, 9, 376. https://doi.org/10.3390/jof9030376
Orosz F. Tubulin Polymerization Promoting Proteins (TPPPs) of Aphelidiomycota: Correlation between the Incidence of p25alpha Domain and the Eukaryotic Flagellum. Journal of Fungi. 2023; 9(3):376. https://doi.org/10.3390/jof9030376
Chicago/Turabian StyleOrosz, Ferenc. 2023. "Tubulin Polymerization Promoting Proteins (TPPPs) of Aphelidiomycota: Correlation between the Incidence of p25alpha Domain and the Eukaryotic Flagellum" Journal of Fungi 9, no. 3: 376. https://doi.org/10.3390/jof9030376
APA StyleOrosz, F. (2023). Tubulin Polymerization Promoting Proteins (TPPPs) of Aphelidiomycota: Correlation between the Incidence of p25alpha Domain and the Eukaryotic Flagellum. Journal of Fungi, 9(3), 376. https://doi.org/10.3390/jof9030376





