Molecular Dissection of Crz1 and Its Dynamic Subcellular Localization in Cryptococcus neoformans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Genetic Manipulation
2.3. Microscopy
3. Results
3.1. The crz1Δ Mutant Displays Pleiotropic Growth Defects
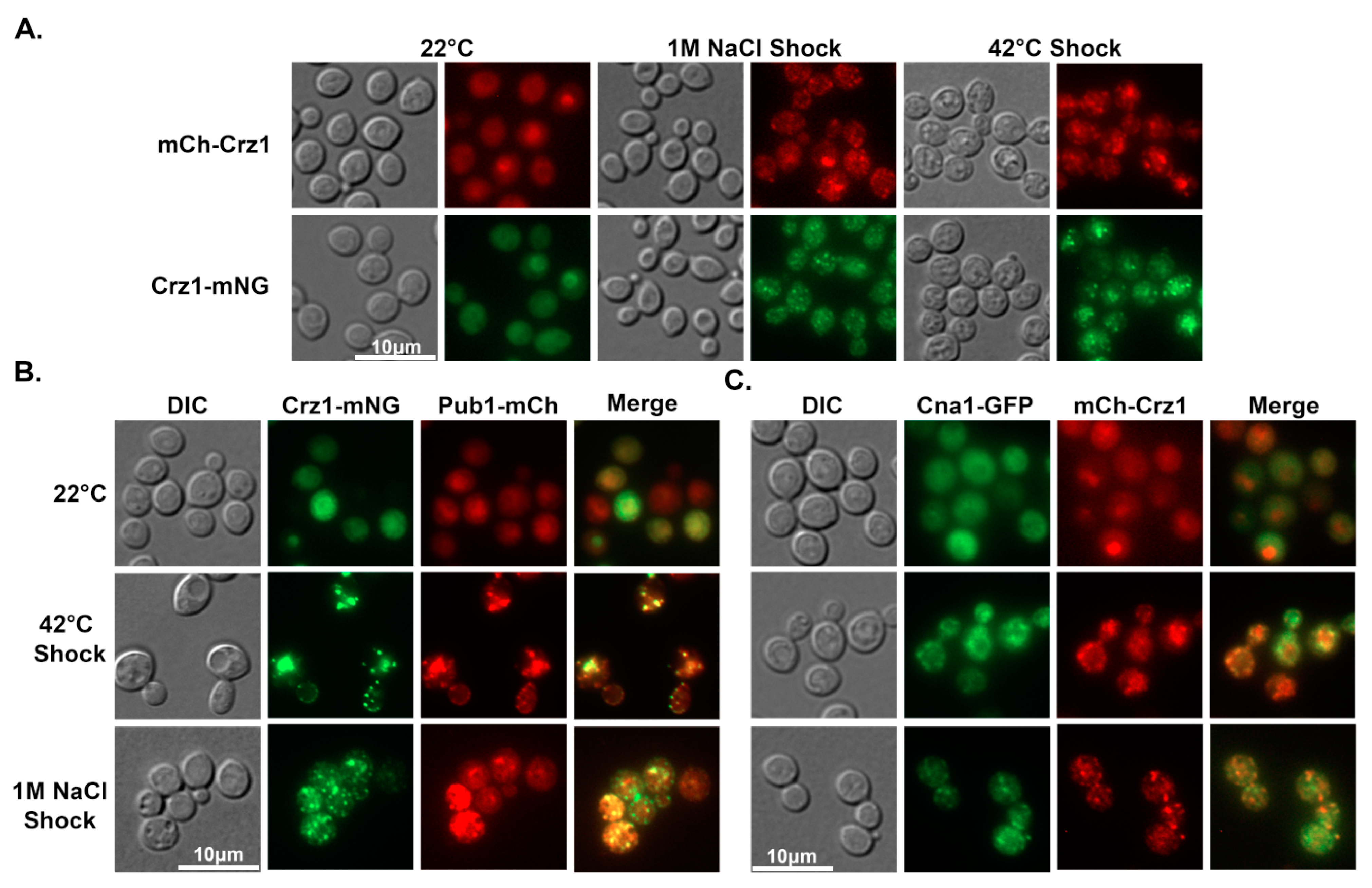
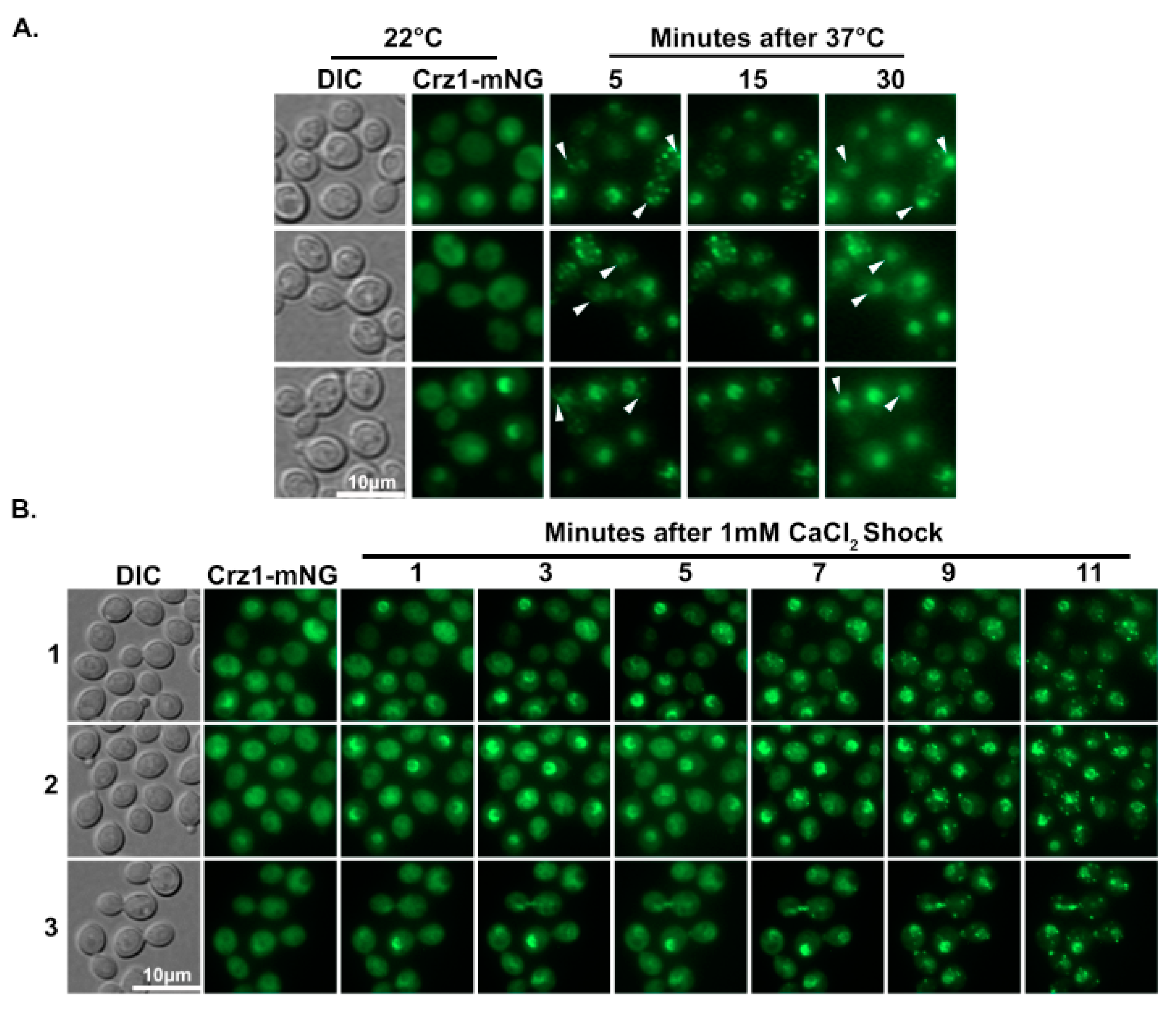
3.2. Crz1 Co-Localizes to Stress Granules with Pub1 and Calcineurin in Response to Heat or Salt Stress
3.3. Crz1 Localizes to Stress Granules at Host Physiological Conditions
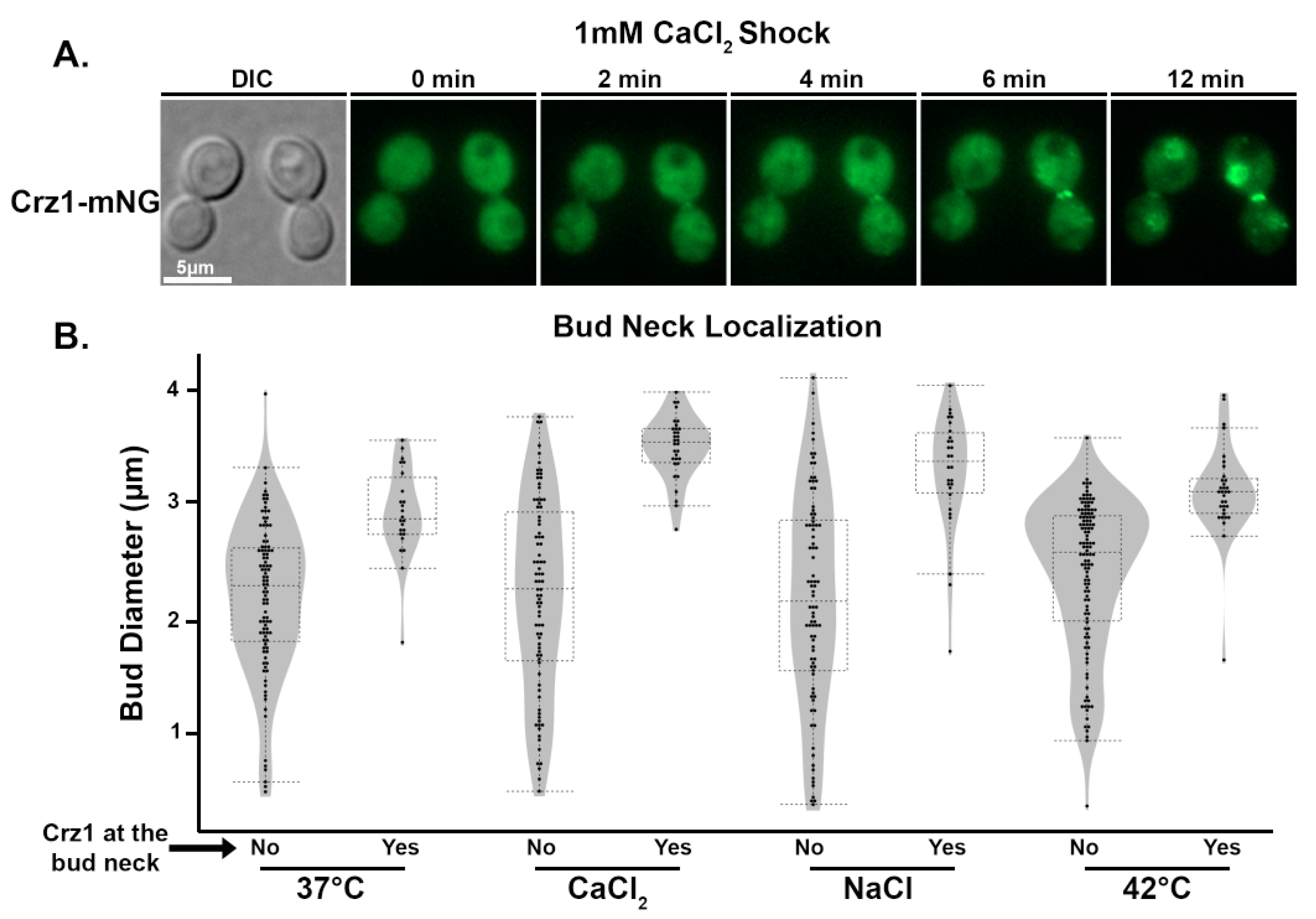
3.4. Crz1 Localizes to the Mother–Daughter Bud Neck in Response to Stress Conditions
3.5. Crz1 Truncation Analysis Reveals Functionally Critical Regions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thewes, S. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot. Cell 2014, 13, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Kim, Y.; Kim, S.; Park, J.; Lee, Y.H. MoCRZ1, a gene encoding a calcineurin-responsive transcription factor, regulates fungal growth and pathogenicity of Magnaporthe oryzae. Fungal Genet. Biol. 2009, 46, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Cramer, R.A., Jr.; Perfect, B.Z.; Pinchai, N.; Park, S.; Perlin, D.S.; Asfaw, Y.G.; Heitman, J.; Perfect, J.R.; Steinbach, W.J. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Shwab, E.K.; Juvvadi, P.R.; Waitt, G.; Soderblom, E.J.; Barrington, B.C.; Asfaw, Y.G.; Moseley, M.A.; Steinbach, W.J. Calcineurin-dependent dephosphorylation of the transcription factor CrzA at specific sites controls conidiation, stress tolerance, and virulence of Aspergillus fumigatus. Mol. Microbiol. 2019, 112, 62–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Yu, S.J.; Huang, H.Y.; Chang, Y.L.; Lehman, V.N.; Silao, F.G.; Bigol, U.G.; Bungay, A.A.; Averette, A.; Heitman, J. Calcineurin controls hyphal growth, virulence, and drug tolerance of Candida tropicalis. Eukaryot. Cell 2014, 13, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; de Larrinoa, I.F. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr. Genet. 2005, 48, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Moranova, Z.; Virtudazo, E.; Hricova, K.; Ohkusu, M.; Kawamoto, S.; Husickova, V.; Raclavsky, V. The CRZ1/SP1-like gene links survival under limited aeration, cell integrity and biofilm formation in the pathogenic yeast Cryptococcus neoformans. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czechoslov. 2014, 158, 212–220. [Google Scholar] [CrossRef]
- Lev, S.; Desmarini, D.; Chayakulkeeree, M.; Sorrell, T.C.; Djordjevic, J.T. The Crz1/Sp1 transcription factor of Cryptococcus neoformans is activated by calcineurin and regulates cell wall integrity. PLoS ONE 2012, 7, e51403. [Google Scholar] [CrossRef]
- Adler, A.; Park, Y.D.; Larsen, P.; Nagarajan, V.; Wollenberg, K.; Qiu, J.; Myers, T.G.; Williamson, P.R. A novel specificity protein 1 (SP1)-like gene regulating protein kinase C-1 (Pkc1)-dependent cell wall integrity and virulence factors in Cryptococcus neoformans. J. Biol. Chem. 2011, 286, 20977–20990. [Google Scholar] [CrossRef]
- Chow, E.W.; Clancey, S.A.; Billmyre, R.B.; Averette, A.F.; Granek, J.A.; Mieczkowski, P.; Cardenas, M.E.; Heitman, J. Elucidation of the calcineurin-Crz1 stress response transcriptional network in the human fungal pathogen Cryptococcus neoformans. PLoS Genet. 2017, 13, e1006667. [Google Scholar] [CrossRef]
- Xu, X.; Lin, J.; Zhao, Y.; Kirkman, E.; So, Y.-S.; Bahn, Y.-S.; Lin, X. Glucosamine stimulates pheromone-independent dimorphic transition in Cryptococcus neoformans by promoting Crz1 nuclear translocation. PLoS Genet. 2017, 13, e1006982. [Google Scholar] [CrossRef] [PubMed]
- Stathopoulos-Gerontides, A.; Guo, J.J.; Cyert, M.S. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 1999, 13, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, S.; Jana, T.; Barkai, N. Order through disorder: The role of intrinsically disordered regions in transcription factor binding specificity. Curr. Opin. Struct. Biol. 2021, 71, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, S.; Jana, T.; Mittelman, K.; Chapal, M.; Kumar, D.K.; Carmi, M.; Barkai, N. Intrinsically Disordered Regions Direct Transcription Factor In Vivo Binding Specificity. Mol. Cell 2020, 79, 459–471.e4. [Google Scholar] [CrossRef]
- Yang, P.; Mathieu, C.; Kolaitis, R.-M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e8. [Google Scholar] [CrossRef]
- Protter, D.S.W.; Rao, B.S.; Van Treeck, B.; Lin, Y.; Mizoue, L.; Rosen, M.K.; Parker, R. Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep. 2018, 22, 1401–1412. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, C.; Duan, Y.; Liu, T.; Liu, H.; Su, C.; Lu, Y. The intrinsically disordered region from PP2C phosphatases functions as a conserved CO2 sensor. Nat. Cell Biol. 2022, 24, 1029–1037. [Google Scholar] [CrossRef]
- Fan, Y.; Lin, X. Multiple Applications of a Transient CRISPR-Cas9 Coupled with Electroporation (TRACE) System in the Cryptococcus neoformans Species Complex. Genetics 2018, 208, 1357–1372. [Google Scholar] [CrossRef]
- Lin, J.; Fan, Y.; Lin, X. Transformation of Cryptococcus neoformans by electroporation using a transient CRISPR-Cas9 expression (TRACE) system. Fungal Genet. Biol. FG B 2020, 138, 103364. [Google Scholar] [CrossRef]
- Upadhya, R.; Lam, W.C.; Maybruck, B.T.; Donlin, M.J.; Chang, A.L.; Kayode, S.; Ormerod, K.L.; Fraser, J.A.; Doering, T.L.; Lodge, J.K. A fluorogenic C. neoformans reporter strain with a robust expression of m-cherry expressed from a safe haven site in the genome. Fungal Genet. Biol. FG B 2017, 108, 13–25. [Google Scholar] [CrossRef]
- Park, H.-S.; Chow, E.W.L.; Fu, C.; Soderblom, E.J.; Moseley, M.A.; Heitman, J.; Cardenas, M.E. Calcineurin Targets Involved in Stress Survival and Fungal Virulence. PLoS Pathog. 2016, 12, e1005873. [Google Scholar] [CrossRef]
- Kozubowski, L.; Aboobakar, E.F.; Cardenas, M.E.; Heitman, J. Calcineurin colocalizes with P-bodies and stress granules during thermal stress in Cryptococcus neoformans. Eukaryot. Cell 2011, 10, 1396–1402. [Google Scholar] [CrossRef]
- Kroschwald, S.; Munder, M.C.; Maharana, S.; Franzmann, T.M.; Richter, D.; Ruer, M.; Hyman, A.A.; Alberti, S. Different Material States of Pub1 Condensates Define Distinct Modes of Stress Adaptation and Recovery. Cell Rep. 2018, 23, 3327–3339. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Occhipinti, P.; Gladfelter, A.S. PolyQ-dependent RNA-protein assemblies control symmetry breaking. J. Cell Biol. 2015, 208, 533–544. [Google Scholar] [CrossRef]
- Marcelo, A.; Koppenol, R.; de Almeida, L.P.; Matos, C.A.; Nóbrega, C. Stress granules, RNA-binding proteins and polyglutamine diseases: Too much aggregation? Cell Death Dis. 2021, 12, 592. [Google Scholar] [CrossRef]
- Buchan, J.R.; Muhlrad, D.; Parker, R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008, 183, 441–455. [Google Scholar] [CrossRef]
- Mitchell, S.F.; Jain, S.; She, M.; Parker, R. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013, 20, 127–133. [Google Scholar] [CrossRef]
- Fox, D.S.; Cox, G.M.; Heitman, J. Phospholipid-binding protein Cts1 controls septation and functions coordinately with calcineurin in Cryptococcus neoformans. Eukaryot. Cell 2003, 2, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Aboobakar, E.F.; Wang, X.; Heitman, J.; Kozubowski, L. The C2 domain protein Cts1 functions in the calcineurin signaling circuit during high-temperature stress responses in Cryptococcus neoformans. Eukaryot. Cell 2011, 10, 1714–1723. [Google Scholar] [CrossRef] [PubMed]
- Juvvadi, P.R.; Steinbach, W.J. Calcineurin Orchestrates Hyphal Growth, Septation, Drug Resistance and Pathogenesis of Aspergillus fumigatus: Where Do We Go from Here? Pathogens 2015, 4, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Juvvadi, P.R.; Fortwendel, J.R.; Rogg, L.E.; Burns, K.A.; Randell, S.H.; Steinbach, W.J. Localization and activity of the calcineurin catalytic and regulatory subunit complex at the septum is essential for hyphal elongation and proper septation in Aspergillus fumigatus. Mol. Microbiol. 2011, 82, 1235–1259. [Google Scholar] [CrossRef] [PubMed]
- Juvvadi, P.R.; Fortwendel, J.R.; Pinchai, N.; Perfect, B.Z.; Heitman, J.; Steinbach, W.J. Calcineurin localizes to the hyphal septum in Aspergillus fumigatus: Implications for septum formation and conidiophore development. Eukaryot. Cell 2008, 7, 1606–1610. [Google Scholar] [CrossRef]
- Lu, Y.; Sugiura, R.; Yada, T.; Cheng, H.; Sio, S.O.; Shuntoh, H.; Kuno, T. Calcineurin is implicated in the regulation of the septation initiation network in fission yeast. Genes Cells Devoted Mol. Cell. Mech. 2002, 7, 1009–1019. [Google Scholar] [CrossRef]
- Nielsen, K.; Cox, G.M.; Wang, P.; Toffaletti, D.L.; Perfect, J.R.; Heitman, J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 2003, 71, 4831–4841. [Google Scholar] [CrossRef]


| 22 °C | 37 °C | GlcN | NaCl | CaCl2 | FK506 | 42 °C | 42 °C + FK506 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | N | N | N | N | G | N | N | G | N | G | N | G |
| WT | 13% | 99% | 78% | 1% | 99% | 97% | 0% | 5% | 0% | 100% | 0% | 100% |
| 7S -> A | 37% | 100% | 96% | 0% | 100% | 100% | 1% | 0% | 100% | 0% | 0% | 100% |
| polyQ -> A | 11% | 32% | 94% | 0% | 100% | 98% | 2% | 2% | 0% | 100% | 0% | 92% |
| ∆N 1-257 | 74% | 95% | 99% | 10% | 90% | 100% | 0% | 4% | 51% | 36% | 1% | 57% |
| ∆N 1-368 | 92% | 100% | 97% | 100% | 90% | 97% | 95% | 0% | 97% | 0% | 99% | 0% |
| ∆N 1-451 | 100% | 100% | 100% | 15% | 85% | 98% | 2% | 0% | 100% | 0% | 13% | 87% |
| ∆N 1-705 | 54% | 87% | 87% | 100% | 23% | 100% | 91% | 0% | 100% | 0% | 100% | 0% |
| ∆N 1-802 | 100% | 100% | 62% | 100% | 100% | 98% | 94% | 0% | 100% | 0% | 95% | 0% |
| ∆N 1-823 | 97% | 100% | 100% | 97% | 94% | 100% | 85% | 0% | 91% | 0% | 100% | 0% |
| ∆NC 946-1030 | 85% | 94% | 52% | 100% | 0% | 98% | 84% | 0% | 100% | 0% | 96% | 0% |
| ∆NC 833-926 | 73% | 98% | 100% | 100% | 100% | 100% | 100% | 0% | 100% | 0% | 100% | 0% |
| ∆NC 625-665 | 23% | 82% | 97% | 5% | 84% | 85% | 0% | 23% | 17% | 0% | 0% | 20% |
| % Different from WT | 0–20 | >20 | >40 | >60 | >80 | 100 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chadwick, B.J.; Ross, B.E.; Lin, X. Molecular Dissection of Crz1 and Its Dynamic Subcellular Localization in Cryptococcus neoformans. J. Fungi 2023, 9, 252. https://doi.org/10.3390/jof9020252
Chadwick BJ, Ross BE, Lin X. Molecular Dissection of Crz1 and Its Dynamic Subcellular Localization in Cryptococcus neoformans. Journal of Fungi. 2023; 9(2):252. https://doi.org/10.3390/jof9020252
Chicago/Turabian StyleChadwick, Benjamin J., Brittain Elizabeth Ross, and Xiaorong Lin. 2023. "Molecular Dissection of Crz1 and Its Dynamic Subcellular Localization in Cryptococcus neoformans" Journal of Fungi 9, no. 2: 252. https://doi.org/10.3390/jof9020252
APA StyleChadwick, B. J., Ross, B. E., & Lin, X. (2023). Molecular Dissection of Crz1 and Its Dynamic Subcellular Localization in Cryptococcus neoformans. Journal of Fungi, 9(2), 252. https://doi.org/10.3390/jof9020252







