Exposure of Aspergillus fumigatus to Klebsiella pneumoniae Culture Filtrate Inhibits Growth and Stimulates Gliotoxin Production
Abstract
1. Introduction
2. Materials and Methods
2.1. A. fumigatus Growth Conditions
2.2. Preparation of the Bacterial Culture Filtrate
2.3. Assessment of Fungal Biomass
2.4. Gliotoxin Extraction and Quantification by RP-HPLC
2.5. Extraction of Protein from K. pneumoniae Culture Filtrate
2.6. Protein Extraction from A. fumigatus Exposed to Bacterial Culture Filtrate
2.7. Label-Free Mass Spectrometry (LF/MS)
2.8. Mass Spectrometry and the Parameters for A. fumigatus and K. pneumoniae Culture Filtrate Proteomic Data Procurement
2.9. Data Analysis of A. fumigatus and K. pneumoniae Culture Filtrate Proteomes
3. Results
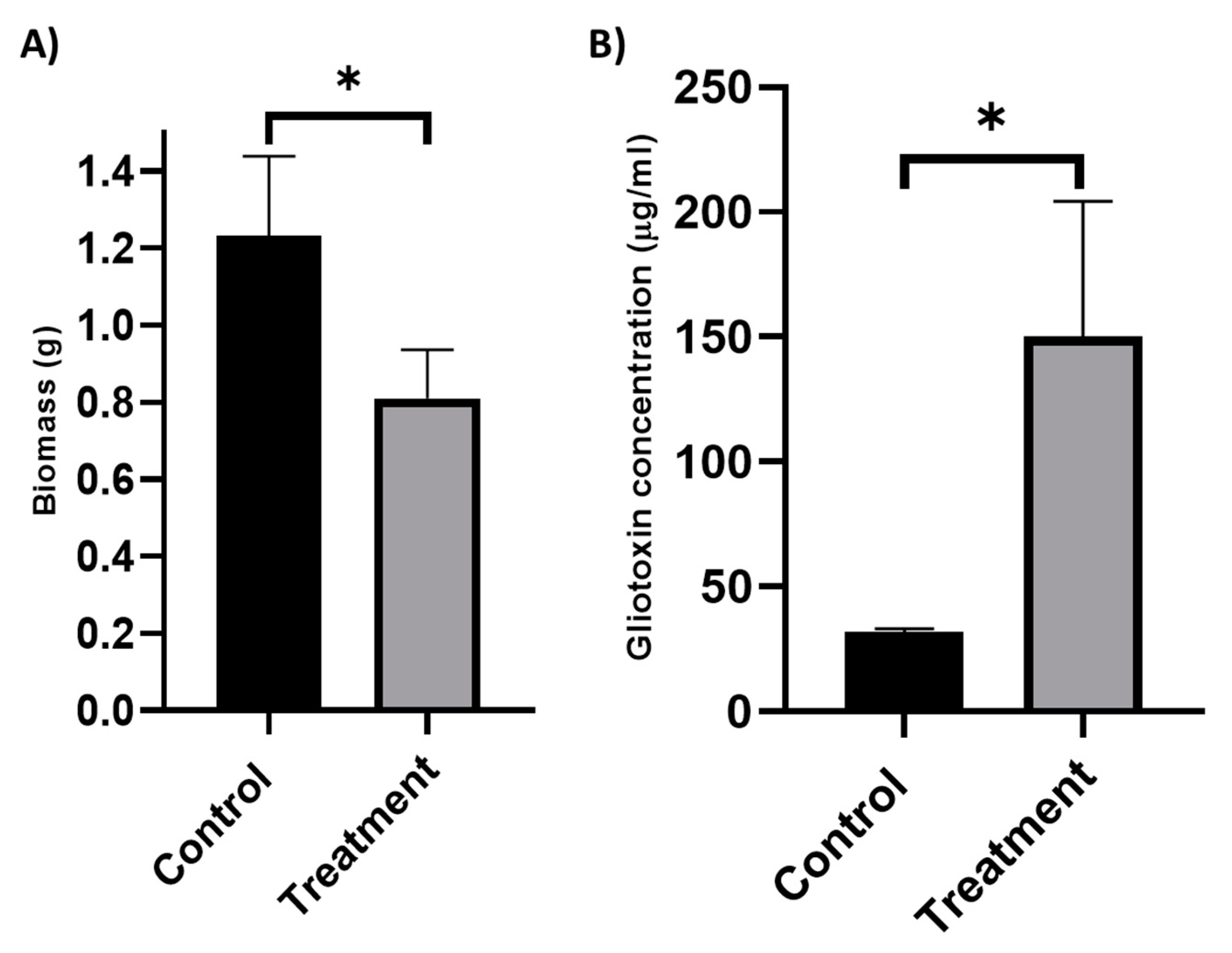
3.1. Analysis of the Effects of K. pneumoniae Culture Filtrate on A. fumigatus
3.2. Characterisation of K. pneumoniae Culture Filtrate
3.3. Analysis of the Effect of K. pneumoniae Culture Filtrate on the A. fumigatus Proteome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- O’Dwyer, D.N.; Dickson, R.P.; Moore, B.B. The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J. Immunol. 2016, 196, 4839–4847. [Google Scholar] [CrossRef]
- Leitao Filho, F.S.; Alotaibi, N.M.; Ngan, D.; Tam, S.; Yang, J.; Hollander, Z.; Chen, V.; FitzGerald, J.M.; Nislow, C.; Leung, J.M.; et al. Sputum Microbiome Is Associated with 1-Year Mortality after Chronic Obstructive Pulmonary Disease Hospitalizations. Am. J. Respir. Crit. Care Med. 2019, 199, 1205–1213. [Google Scholar] [CrossRef]
- Keown, K.; Reid, A.; Moore, J.E.; Taggart, C.C.; Downey, D.G. Coinfection with Pseudomonas aeruginosa and Aspergillus fumigatus in Cystic Fibrosis. Eur. Respir. Rev. 2020, 29, 200011. [Google Scholar] [CrossRef] [PubMed]
- LiPuma, J.J. The Changing Microbial Epidemiology in Cystic Fibrosis. Clin. Microbiol. Rev. 2010, 23, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A Major Worldwide Source and Shuttle for Antibiotic Resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Vieira, A.T.; Rocha, V.M.; Tavares, L.; Garcia, C.C.; Teixeira, M.M.; Oliveira, S.C.; Cassali, G.D.; Gamba, C.; Martins, F.S.; Nicoli, J.R. Control of Klebsiella pneumoniae Pulmonary Infection and Immunomodulation by Oral Treatment with the Commensal Probiotic Bifidobacterium Longum 51A. Microbes Infect. 2016, 18, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Fodah, R.A.; Scott, J.B.; Tam, H.-H.; Yan, P.; Pfeffer, T.L.; Bundschuh, R.; Warawa, J.M. Correlation of Klebsiella pneumoniae Comparative Genetic Analyses with Virulence Profiles in a Murine Respiratory Disease Model. PLoS ONE 2014, 9, e107394. [Google Scholar] [CrossRef]
- Delfino, E.; Giacobbe, D.R.; Del Bono, V.; Coppo, E.; Marchese, A.; Manno, G.; Morelli, P.; Minicucci, L.; Viscoli, C. First Report of Chronic Pulmonary Infection by KPC-3-Producing and Colistin-Resistant Klebsiella pneumoniae Sequence Type 258 (ST258) in an Adult Patient with Cystic Fibrosis. J. Clin. Microbiol. 2015, 53, 1442–1444. [Google Scholar] [CrossRef] [PubMed]
- Lattanzi, C.; Messina, G.; Fainardi, V.; Tripodi, M.C.; Pisi, G.; Esposito, S. Allergic Bronchopulmonary Aspergillosis in Children with Cystic Fibrosis: An Update on the Newest Diagnostic Tools and Therapeutic Approaches. Pathogens 2020, 9, 716. [Google Scholar] [CrossRef]
- Schweer, K.E.; Bangard, C.; Hekmat, K.; Cornely, O.A. Chronic Pulmonary Aspergillosis. Mycoses 2014, 57, 257–270. [Google Scholar] [CrossRef]
- Kradin, R.L.; Mark, E.J. The Pathology of Pulmonary Disorders Due to Aspergillus Spp. Arch. Pathol. Lab. Med. 2008, 132, 606–614. [Google Scholar] [CrossRef]
- Webb, B.J.; Ferraro, J.P.; Rea, S.; Kaufusi, S.; Goodman, B.E.; Spalding, J. Epidemiology and Clinical Features of Invasive Fungal Infection in a US Health Care Network. Open Forum Infect. Dis. 2018, 5, ofy187. [Google Scholar] [CrossRef]
- Bassetti, M.; Peghin, M.; Vena, A. Challenges and Solution of Invasive Aspergillosis in Non-Neutropenic Patients: A Review. Infect. Dis. Ther. 2018, 7, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Margalit, A.; Sheehan, D.; Carolan, J.C.; Kavanagh, K. Exposure to the Pseudomonas aeruginosa secretome alters the proteome and secondary metabolite production of Aspergillus fumigatus. Microbiology 2022, 168, 001164. [Google Scholar] [CrossRef] [PubMed]
- Margalit, A.; Carolan, J.C.; Sheehan, D.; Kavanagh, K. The Aspergillus fumigatus Secretome Alters the Proteome of Pseudomonas aeruginosa to Stimulate Bacterial Growth: Implications for Co-Infection. Mol. Cell. Proteom. 2020, 19, 1346–1359. [Google Scholar] [CrossRef]
- Dees, J.; Connell, J.; Stacy, A.; Turner, K.; Whiteley, M. Mechanisms of Synergy in Polymicrobial Infections. J. Microbiol. Seoul Korea 2014, 52, 188–199. [Google Scholar] [CrossRef]
- Nogueira, M.F.; Pereira, L.; Jenull, S.; Kuchler, K.; Lion, T. Klebsiella pneumoniae Prevents Spore Germination and Hyphal Development of Aspergillus Species. Sci. Rep. 2019, 9, 218. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2021, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Line, L.; Alhede, M.; Kolpen, M.; Kühl, M.; Ciofu, O.; Bjarnsholt, T.; Moser, C.; Toyofuku, M.; Nomura, N.; Høiby, N.; et al. Physiological Levels of Nitrate Support Anoxic Growth by Denitrification of Pseudomonas aeruginosa at Growth Rates Reported in Cystic Fibrosis Lungs and Sputum. Front. Microbiol. 2014, 5, 554. [Google Scholar] [CrossRef]
- Lu, Z.; Huang, W.; Wang, L.; Xu, N.; Ding, Q.; Cao, C. Exhaled Nitric Oxide in Patients with Chronic Obstructive Pulmonary Disease: A Systematic Review and Meta-Analysis. Int. J. Chron. Obstruct. Pulmon. Dis. 2018, 13, 2695–2705. [Google Scholar] [CrossRef]
- Campos-Avelar, I.; Colas de la Noue, A.; Durand, N.; Fay, B.; Martinez, V.; Fontana, A.; Strub, C.; Schorr-Galindo, S. Minimizing Ochratoxin A Contamination through the Use of Actinobacteria and Their Active Molecules. Toxins 2020, 12, 296. [Google Scholar] [CrossRef]
- Campos-Avelar, I.; Colas de la Noue, A.; Durand, N.; Cazals, G.; Martinez, V.; Strub, C.; Fontana, A.; Schorr-Galindo, S. Aspergillus flavus Growth Inhibition and Aflatoxin B1 Decontamination by Streptomyces Isolates and Their Metabolites. Toxins 2021, 13, 340. [Google Scholar] [CrossRef] [PubMed]
- Gaddy, J.A.; Tomaras, A.P.; Actis, L.A. The Acinetobacter Baumannii 19606 OmpA Protein Plays a Role in Biofilm Formation on Abiotic Surfaces and in the Interaction of This Pathogen with Eukaryotic Cells. Infect. Immun. 2009, 77, 3150–3160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhou, H.; Jiang, Y.; He, J.; Yao, Y.; Wang, J.; Liu, X.; Leptihn, S.; Hua, X.; Yu, Y. Acinetobacter baumannii Outer Membrane Protein A Induces Pulmonary Epithelial Barrier Dysfunction and Bacterial Translocation Through The TLR2/IQGAP1 Axis. Front. Immunol. 2022, 13, 927955. [Google Scholar] [CrossRef]
- Chiu, K.-H.; Wang, L.-H.; Tsai, T.-T.; Lei, H.-Y.; Liao, P.-C. Secretomic Analysis of Host–Pathogen Interactions Reveals That Elongation Factor-Tu Is a Potential Adherence Factor of Helicobacter pylori during Pathogenesis. J. Proteome Res. 2017, 16, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.H.; Bolken, T.C.; Jones, K.F.; Zeller, G.O.; Hruby, D.E. Conserved DegP Protease in Gram-Positive Bacteria Is Essential for Thermal and Oxidative Tolerance and Full Virulence in Streptococcus pyogenes. Infect. Immun. 2001, 69, 5538–5545. [Google Scholar] [CrossRef]
- Zhang, S.; Cheng, Y.; Ma, J.; Wang, Y.; Chang, Z.; Fu, X. Degp Degrades a Wide Range of Substrate Proteins in Escherichia coli under Stress Conditions. Biochem. J. 2019, 476, 3549–3564. [Google Scholar] [CrossRef]
- Sabotič, J.; Kos, J. Microbial and Fungal Protease Inhibitors—Current and Potential Applications. Appl. Microbiol. Biotechnol. 2012, 93, 1351–1375. [Google Scholar] [CrossRef] [PubMed]
- Denoncin, K.; Collet, J.-F. Disulfide Bond Formation in the Bacterial Periplasm: Major Achievements and Challenges Ahead. Antioxid. Redox Signal. 2013, 19, 63–71. [Google Scholar] [CrossRef]
- Bernardo, P.H.; Brasch, N.; Chai, C.L.L.; Waring, P. A Novel Redox Mechanism for the Glutathione-Dependent Reversible Uptake of a Fungal Toxin in Cells. J. Biol. Chem. 2003, 278, 46549–46555. [Google Scholar] [CrossRef]
- Choi, H.S.; Shim, J.S.; Kim, J.-A.; Kang, S.W.; Kwon, H.J. Discovery of Gliotoxin as a New Small Molecule Targeting Thioredoxin Redox System. Biochem. Biophys. Res. Commun. 2007, 359, 523–528. [Google Scholar] [CrossRef]
- Lau, C.K.Y.; Krewulak, K.D.; Vogel, H.J. Bacterial Ferrous Iron Transport: The Feo System. FEMS Microbiol. Rev. 2016, 40, 273–298. [Google Scholar] [CrossRef]
- Neupane, D.P.; Kumar, S.; Yukl, E.T. Two ABC Transporters and a Periplasmic Metallochaperone Participate in Zinc Acquisition in Paracoccus Denitrificans. Biochemistry 2019, 58, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Traynor, A.M.; Owens, R.A.; Coughlin, C.M.; Holton, M.C.; Jones, G.W.; Calera, J.A.; Doyle, S. At the Metal-Metabolite Interface in Aspergillus fumigatus: Towards Untangling the Intersecting Roles of Zinc and Gliotoxin. Microbiol. Read. Engl. 2021, 167, 001106. [Google Scholar] [CrossRef] [PubMed]
- Vicentefranqueira, R.; Amich, J.; Marín, L.; Sánchez, C.I.; Leal, F.; Calera, J.A. The Transcription Factor ZafA Regulates the Homeostatic and Adaptive Response to Zinc Starvation in Aspergillus fumigatus. Genes 2018, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Reeves, E.P.; Murphy, T.; Daly, P.; Kavanagh, K. Amphotericin B Enhances the Synthesis and Release of the Immunosuppressive Agent Gliotoxin from the Pulmonary Pathogen Aspergillus fumigatus. J. Med. Microbiol. 2004, 53, 719–725. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Gautam, P.; Pandit, H.; Singh, Y.; Basir, S.F.; Madan, T. Identification of Fibrinogen-Binding Proteins of Aspergillus Fumigatus Using Proteomic Approach. Mycopathologia 2012, 173, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Wasylnka, J.A.; Moore, M.M. Adhesion of Aspergillus Species to Extracellular Matrix Proteins: Evidence for Involvement of Negatively Charged Carbohydrates on the Conidial Surface. Infect. Immun. 2000, 68, 3377–3384. [Google Scholar] [CrossRef]
- Lee, M.J.; Geller, A.M.; Bamford, N.C.; Liu, H.; Gravelat, F.N.; Snarr, B.D.; Le Mauff, F.; Chabot, J.; Ralph, B.; Ostapska, H.; et al. Deacetylation of Fungal Exopolysaccharide Mediates Adhesion and Biofilm Formation. mBio 2016, 7, e00252-16. [Google Scholar] [CrossRef]
- Gastebois, A.; Fontaine, T.; Latgé, J.-P.; Mouyna, I. β(1-3)Glucanosyltransferase Gel4p Is Essential for Aspergillus fumigatus. Eukaryot. Cell 2010, 9, 1294–1298. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Fuoli, L.; Mellado, E.; Garcia-Effron, G.; Lopez, J.F.; Grimalt, J.O.; Cuenca-Estrella, J.M.; Rodriguez-Tudela, J.L. Ergosterol Biosynthesis Pathway in Aspergillus fumigatus. Steroids 2008, 73, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Fuoli, L.; Mellado, E. Ergosterol Biosynthesis in Aspergillus fumigatus: Its Relevance as an Antifungal Target and Role in Antifungal Drug Resistance. Front. Microbiol. 2013, 3, 439. [Google Scholar] [CrossRef]
- Dayton, J.S.; Means, A.R. Ca(2+)/Calmodulin-Dependent Kinase Is Essential for Both Growth and Nuclear Division in Aspergillus Nidulans. Mol. Biol. Cell 1996, 7, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Vorapreeda, T.; Khongto, B.; Thammarongtham, C.; Srisuk, T.; Laoteng, K. Metabolic Regulation of Sugar Assimilation for Lipid Production in Aspergillus oryzae BCC7051 through Comparative Transcriptome Perspective. Biology 2021, 10, 885. [Google Scholar] [CrossRef]
- Keller, N.; Bok, J.; Chung, D.; Perrin, R.M.; Keats Shwab, E. LaeA, a Global Regulator of Aspergillus Toxins. Med. Mycol. 2006, 44, S83–S85. [Google Scholar] [CrossRef]
- Lin, H.-C.; Chooi, Y.-H.; Dhingra, S.; Xu, W.; Calvo, A.M.; Tang, Y. The Fumagillin Biosynthetic Gene Cluster in Aspergillus fumigatus Encodes a Cryptic Terpene Cyclase Involved in the Formation of β-Trans-Bergamotene. J. Am. Chem. Soc. 2013, 135, 4616–4619. [Google Scholar] [CrossRef]


| Protein Name | Uniprot Code | Unique Peptides | % Coverage | Function |
|---|---|---|---|---|
| Outer membrane protein A | B5XY48 | 8 | 22.28 | Membrane proteins |
| Outer membrane protein X | B5XYT0 | 7 | 39.76 | |
| Outer membrane protein C | B5XNZ9 | 7 | 21.48 | |
| Penicillin-binding protein activator LpoA | B5XSZ6 | 5 | 9.54 | |
| OmpA family protein | B5XN00 | 3 | 24.54 | |
| Penicillin-binding protein activator LpoB | B5XXG6 | 2 | 16.74 | |
| OmpA family protein | B5XVK9 | 3 | 28.75 | |
| LPS-assembly protein LptD | B5Y1Z1 | 3 | 5.37 | |
| MrkF protein | B5XUK5 | 2 | 16.33 | |
| LPS-assembly lipoprotein LptE | B5XZR3 | 2 | 13.26 | |
| Elongation factor Tu | B5XN88 | 10 | 40.35 | virulence |
| Elongation factor Ts | B5Y1K1 | 2 | 12.01 | |
| Tol-Pal system protein TolB | B5XZC1 | 6 | 24.18 | |
| Pectinesterase | B5XZ84 | 7 | 22.01 | Enzymatic activity |
| Autonomous glycyl radical cofactor | B5XNF9 | 6 | 51.18 | |
| Protease VII | B5RKF2 | 6 | 25.72 | |
| Periplasmic serine endoprotease DegP-like | B5Y1K8 | 6 | 15.93 | |
| Enolase | B5XV19 | 6 | 21.06 | |
| Endolytic peptidoglycan transglycosylase RlpA | B5XZS1 | 5 | 15.06 | |
| Beta-lactamase | B5XQY6 | 4 | 11.88 | |
| Prephenate dehydratase/arogenate dehydratase | B5XVG4 | 3 | 16.60 | |
| Metal-binding protein | B5XZ21 | 6 | 39.35 | Metal binding |
| High-affinity zinc uptake system protein ZnuA | B5XQ08 | 5 | 25.47 | |
| Copper homeostasis protein CutF | B5Y1H9 | 3 | 16.81 | |
| Iron uptake system component EfeO | B5XXM1 | 3 | 10.4 | |
| Thiol:disulfide interchange protein | B5XZJ6 | 4 | 31.40 | |
| Alkyl hydroperoxide reductase C | B5XZT7 | 4 | 35.82 | Detoxification |
| Thioredoxin | B5XYY8 | 5 | 51.37 | |
| Acriflavine resistance protein A | B5Y0P5 | 3 | 9.57 | |
| Thiol peroxidase | B5XRV9 | 3 | 32.14 | |
| Alkyl hydroperoxide reductase C | B5Y0Z0 | 3 | 35.82 | |
| Glutathione ABC transporter, periplasmic glutathione-binding protein | B5XYQ7 | 2 | 2.92 | |
| Inhibitor of vertebrate lysozyme | B5Y004 | 2 | 15.54 | |
| Putative lipoprotein | B5XUP5 | 1 | 36.1 | Structurally bound |
| Outer membrane protein A | B5XY48 | 6 | 19.1 | |
| Chaperone protein DnaK | B5Y242 | 2 | 2.7 |
| Number | Fold Change | Protein Name | Protein IDs | Unique Peptides | Sequence Coverage [%] |
|---|---|---|---|---|---|
| 1 | 63.39 | Fibrinogen C-terminal domain-containing protein | Q4W8X0 | 3 | 38.5 |
| 2 | 7.88 | SGL domain-containing protein | Q4WP91 | 4 | 15 |
| 3 | 6.93 | Polysaccharide deacetylase family protein | Q4WUN9 | 9 | 48.7 |
| 4 | 6.17 | Glutathione S-transferase gliG | A4GYZ0 | 18 | 73.3 |
| 5 | 5.81 | ABM domain-containing protein | Q4WG08 | 2 | 24.3 |
| 6 | 5.80 | Endonuclease/exonuclease/phosphatase family | Q4WKR6 | 8 | 31.9 |
| 7 | 5.04 | D-xylose reductase (NAD(P)H) | Q4WI64 | 10 | 38.6 |
| 8 | 4.96 | Ribonuclease mitogillin | P67875 | 6 | 44.9 |
| 9 | 4.86 | Amine oxidase | Q4WFX6 | 16 | 46 |
| 10 | 4.79 | DUF4468 domain-containing protein | Q4WMI8 | 9 | 49.7 |
| 11 | 4.42 | DUF907 domain protein | Q4WHA4 | 1 | 3 |
| 12 | 3.67 | Nonribosomal peptide synthetase gliP | Q4WMJ7 | 14 | 14.2 |
| 13 | 3.51 | O-methyltransferase gliM | Q4WMJ5 | 12 | 32.9 |
| 14 | 2.98 | Oxidoreductase, short-chain | Q4WUP1 | 5 | 40.5 |
| 15 | 2.89 | Gamma-glutamyl cyclotransferase gliK | E9R9Y3 | 2 | 16.8 |
| 16 | 2.50 | GPI anchored serine-threonine rich protein | Q4WTF2 | 2 | 34.5 |
| 17 | 2.50 | Phosphatidylglycerol/phosphatidylinositol transfer | Q4X136 | 11 | 46.9 |
| 18 | 2.35 | Cache_2 domain-containing protein | Q4WYY2 | 4 | 50.8 |
| 19 | 2.34 | Thioredoxin reductase gliT | E9RAH5 | 20 | 85.9 |
| 20 | 2.33 | Cell wall protein PhiA | Q4WF87 | 6 | 73 |
| Number | Fold Change | Protein Name | Protein IDs | Unique Peptides | Sequence Coverage [%] |
|---|---|---|---|---|---|
| 1 | −6.30 | HYPK_UBA domain-containing protein | Q4WPC3 | 2 | 17.7 |
| 2 | −5.49 | Methyltransferase | Q4X081 | 4 | 30.3 |
| 3 | −4.20 | Calcium/calmodulin dependent protein kinase | Q4WXH7 | 7 | 25.1 |
| 4 | −4.05 | Elongation of fatty acids protein | Q4WEE9 | 4 | 16 |
| 5 | −3.97 | 1,3-beta-glucanosyltransferase | Q4WBF7 | 4 | 8.9 |
| 6 | −3.96 | Protein DOM34 homolog | Q4WI62 | 7 | 26.6 |
| 7 | −3.86 | Short chain dehydrogenase helC | Q4WR19 | 7 | 37.7 |
| 8 | −3.84 | Polyketide transferase af380 | Q4WAY4 | 8 | 47.9 |
| 9 | −3.42 | BTB/POZ domain protein | Q4WFH8 | 6 | 32 |
| 10 | −3.02 | Methyltransferase psoC | Q4WB00 | 21 | 71.2 |
| 11 | −2.86 | Methylsterol monooxygenase erg25B | Q4W9I3 | 3 | 13.9 |
| 12 | −2.86 | DUF948 domain-containing protein | Q4WXM0 | 4 | 42.3 |
| 13 | −2.85 | Cytochrome P450 monooxygenase helB1 | Q4WR17 | 9 | 20.5 |
| 14 | −2.83 | Aspartyl aminopeptidase | Q4WX56 | 5 | 23.4 |
| 15 | −2.80 | Protostadienol synthase helA | Q4WR16 | 14 | 28.2 |
| 16 | −2.80 | Signal recognition particle 54 kDa protein | Q4WEQ8 | 3 | 11.5 |
| 17 | −2.79 | Tripeptidyl-peptidase sed4 | Q4WQU0 | 3 | 8.1 |
| 18 | −2.64 | 60S ribosomal protein L22, putative | Q4WYA0 | 7 | 52.1 |
| 19 | −2.58 | Amino acid permease (Gap1), putative | Q4WG99 | 12 | 19 |
| 20 | −2.46 | 3-ketosteroid 1-dehydrogenase helE | Q4WR24 | 6 | 18.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Curtis, A.; Ryan, M.; Kavanagh, K. Exposure of Aspergillus fumigatus to Klebsiella pneumoniae Culture Filtrate Inhibits Growth and Stimulates Gliotoxin Production. J. Fungi 2023, 9, 222. https://doi.org/10.3390/jof9020222
Curtis A, Ryan M, Kavanagh K. Exposure of Aspergillus fumigatus to Klebsiella pneumoniae Culture Filtrate Inhibits Growth and Stimulates Gliotoxin Production. Journal of Fungi. 2023; 9(2):222. https://doi.org/10.3390/jof9020222
Chicago/Turabian StyleCurtis, Aaron, Michelle Ryan, and Kevin Kavanagh. 2023. "Exposure of Aspergillus fumigatus to Klebsiella pneumoniae Culture Filtrate Inhibits Growth and Stimulates Gliotoxin Production" Journal of Fungi 9, no. 2: 222. https://doi.org/10.3390/jof9020222
APA StyleCurtis, A., Ryan, M., & Kavanagh, K. (2023). Exposure of Aspergillus fumigatus to Klebsiella pneumoniae Culture Filtrate Inhibits Growth and Stimulates Gliotoxin Production. Journal of Fungi, 9(2), 222. https://doi.org/10.3390/jof9020222






