LtGAPR1 Is a Novel Secreted Effector from Lasiodiplodia theobromae That Interacts with NbPsQ2 to Negatively Regulate Infection
Abstract
1. Introduction
2. Results
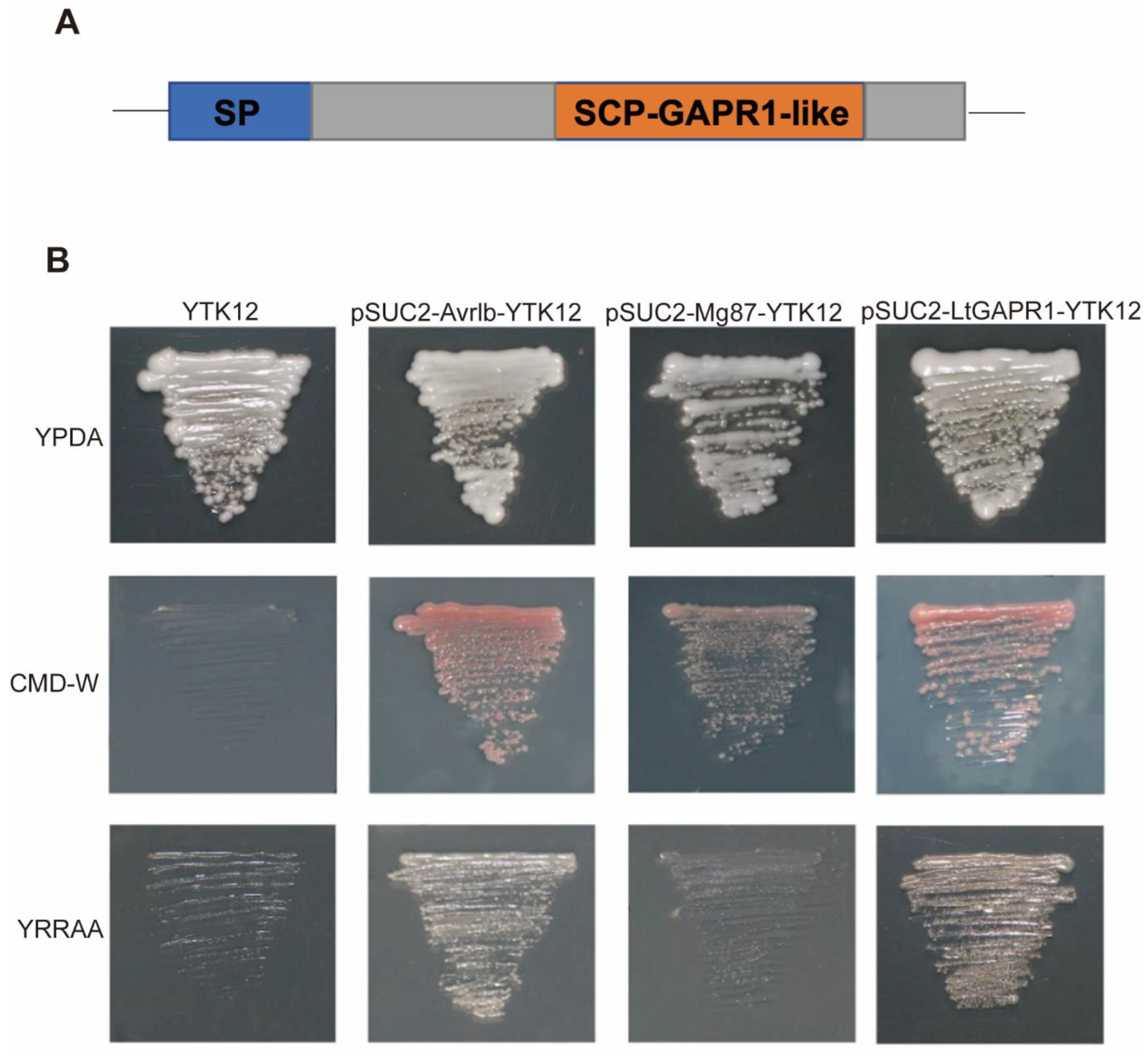
2.1. LtGAPR1 Is a Secreted Protein with an SCP-GAPR1-like Domain
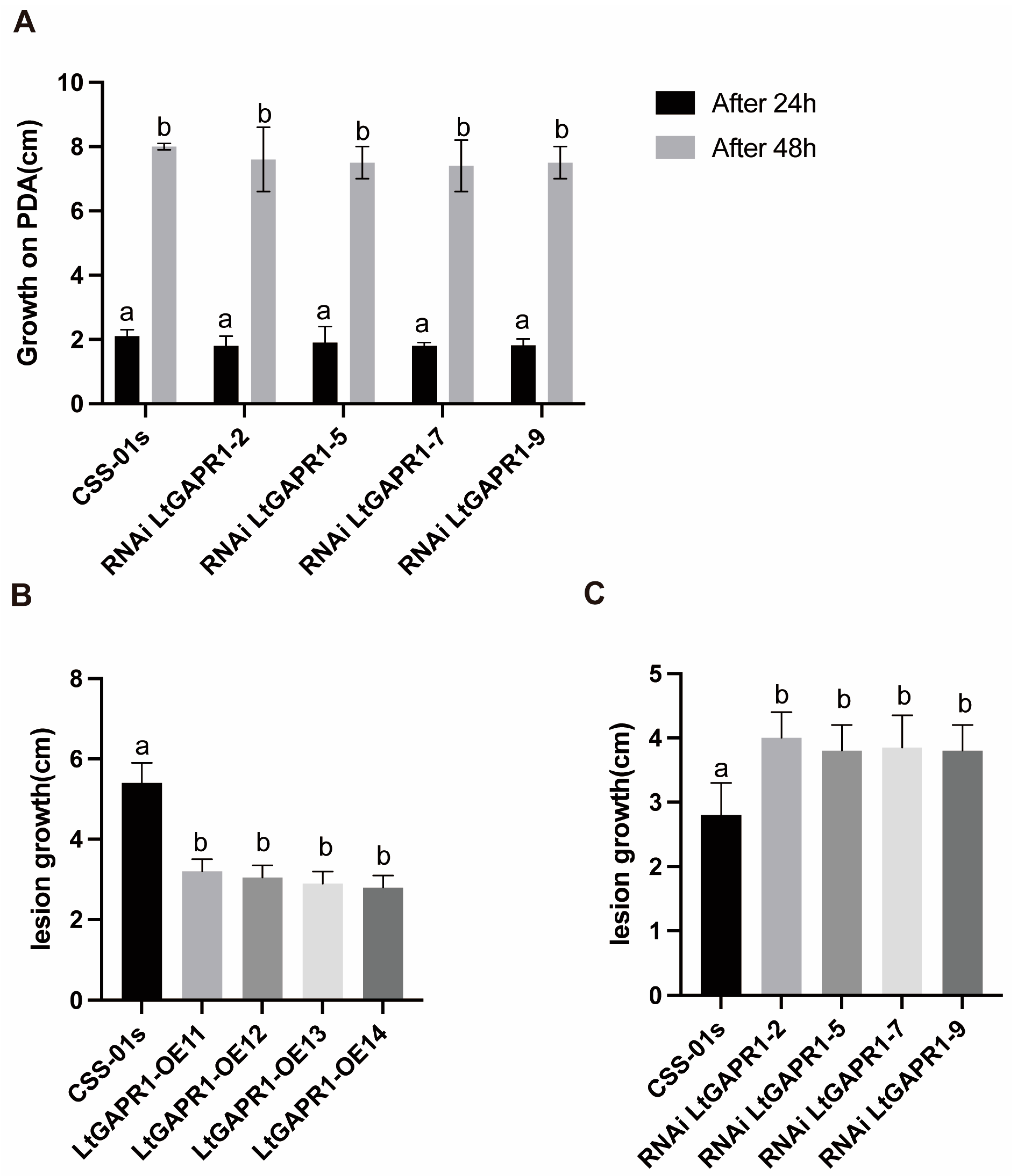
2.2. LtGAPR1 Is Not a Necessary Growth Factor but a Negative Virulence Factor of Vitis vinifera
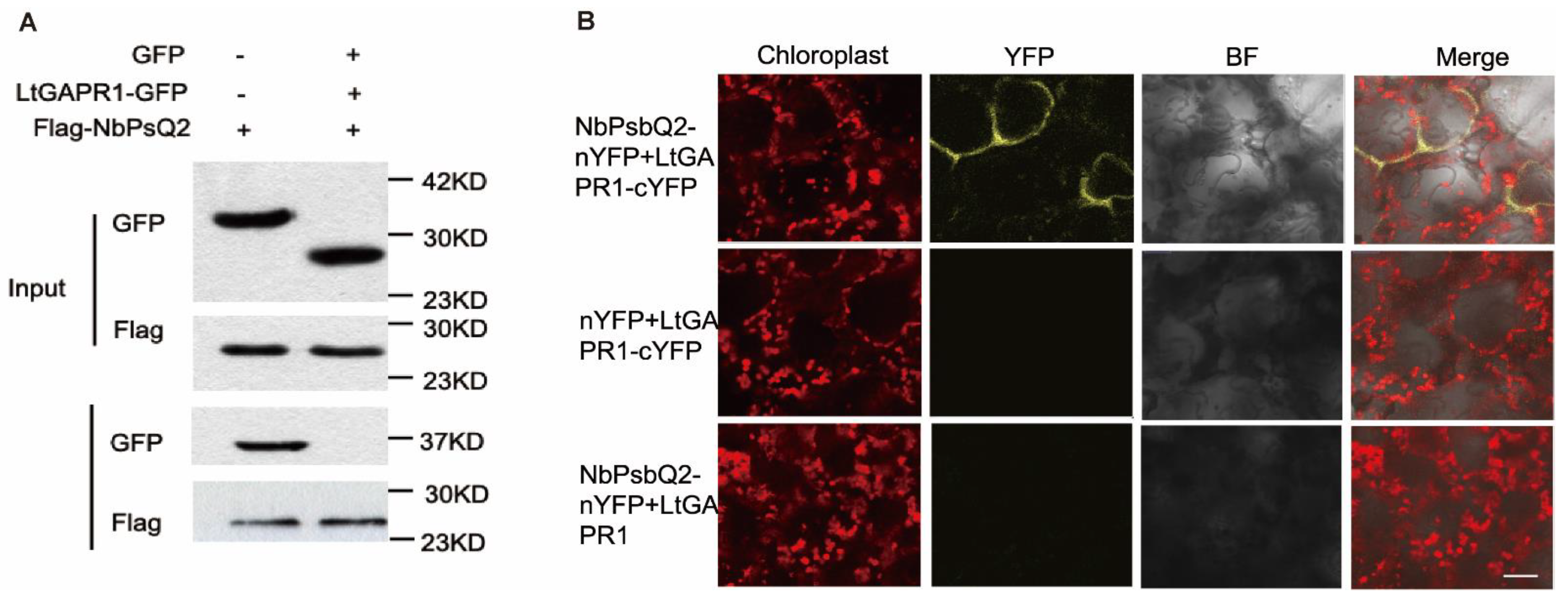
2.3. LtGAPR1 Interacts with NbPsQ2
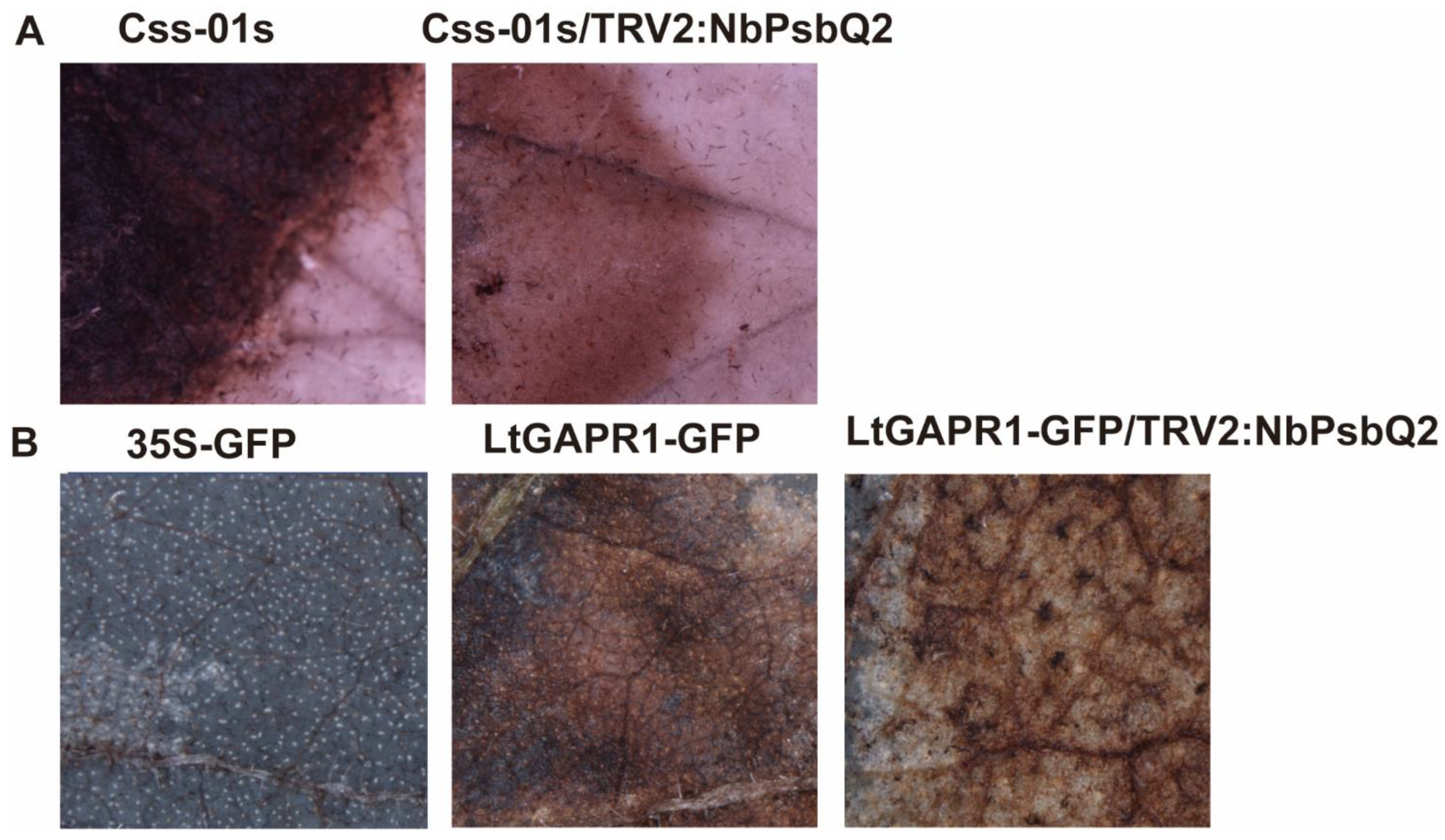
2.4. Overexpression of NbPsbQ2 Reduces Susceptibility to L. theobromae Infection, and Silencing NbPsbQ2 Enhances the Susceptibility of L. theobromae
2.5. LtGAPR1 Triggers ROS Production but Is Impaired in NbPsbQ2-Silenced Plants
3. Materials and Methods
3.1. Strain, Plant Materials, and Culture Conditions
3.2. Co-Immunoprecipitation Assay and Mass Spectrometry
3.3. Bimolecular Fluorescence Complementation Assay
3.4. Virus-Induced Gene Silencing
3.5. Agrobacterium-Mediated Transient Expression
3.6. Grapevine Inoculation Test of Overexpressed and RNAi Transformants of the LtGAPR1 Gene
3.7. Quantification of DAB Staining
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Macho, A.P.; Zipfel, C. Plant PRRs and the Activation of Innate Immune Signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [PubMed]
- Boller, T.; Felix, G. A Renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–407. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Pathogen perception and responses in plant immunity. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar]
- Yan, J.Y.; Zhao, W.S.; Chen, Z.; Xing, Q.K.; Hang, Q.G.; Chethana, K.W.T.; Xue, M.F.; Xu, J.P.; Phillips, A.; Wang, Y.; et al. Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in woody plant degrading pathogens of botryosphaeriaceae. DNA Res. 2018, 25, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Thilini Chethana, K.W.; Peng, J.; Li, X.; Xing, Q.; Liu, M.; Zhang, W.; Hyde, K.D.; Zhao, W.; Yan, J. LtEPG1, a secretory endopolygalacturonase protein, regulates the virulence of Lasiodiplodia theobromae in Vitis vinifera and is recognized as a microbe-associated molecular patterns. Phytopathology 2020, 110, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Cao, Y.; Peng, J.; Zhang, W.; Wu, J.; Zhou, Y.; Li, X.; Yan, J. A putative effector LtCSEP1 from Lasiodiplodia theobromae inhibits BAX-triggered cell death and suppresses immunity responses in Nicotiana benthamiana. Plants 2022, 11, 1462. [Google Scholar] [CrossRef] [PubMed]
- Morales-Cruz, A.; Amrine, K.C.H.; Blanco-Ulate, B.; Lawrence, D.P.; Travadon, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC Genom. 2015, 16, 469. [Google Scholar]
- van Loon, L.; van Strien, E. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar]
- Prados-Rosales, R.C.; Roldán-Rodríguez, R.; Serena, C.; López-Berges, M.S.; Guarro, J.; Martínez-Del-Pozo, Á.; Di Pietro, A. A PR-1-like Protein of Fusarium oxysporum Functions in Virulence on Mammalian Hosts. J. Biol. Chem. 2012, 287, 21970–21979. [Google Scholar] [CrossRef]
- Wangorsch, A.; Scheurer, S.; Blanca, M.; Blanca-Lopez, N.; Somoza, M.L.; Martín-Pedraza, L. Allergenic properties and molecular characteristics of PR-1 proteins. Front. Allergy 2022, 3, 1. [Google Scholar]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar]
- Gibbs, G.M.; Roelants, K.; O’bryan, M.K. The CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—Roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar]
- Yi, X.; Hargett, S.R.; Liu, H.; Frankel, L.K.; Bricker, T.M. The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in arabidopsis thaliana. J. Biol. Chem. 2007, 282, 24833–24841. [Google Scholar] [CrossRef]
- Roose, J.L.; Frankel, L.K.; Mummadisetti, M.P.; Bricker, T.M. The extrinsic proteins of photosystem II: Update. Planta 2016, 243, 889–908. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Herva, J.J.; González-Melendi, P.; Cuartas-Lanza, R.; Antúnez-Lamas, M.; Río-Alvarez, I.; Li, Z.; López-Torrejón, G.; Díaz, I.; del Pozo, J.C.; Chakravarthy, S.; et al. A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell. Microbiol. 2012, 14, 669–681. [Google Scholar] [PubMed]
- Kurth, E.G.; Peremyslov, V.V.; Prokhnevsky, A.I.; Kasschau, K.D.; Miller, M.; Carrington, J.C.; Dolja, V.V. Virus-derived gene expression and RNA interference vector for grapevine. J. Virol. 2012, 86. [Google Scholar] [CrossRef]
- Kong, L.; Wu, J.; Lu, L.; Xu, Y.; Zhou, X. Interaction between rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol. Plant 2014, 7, 691–708. [Google Scholar]
- Gnanasekaran, P.; Ponnusamy, K.; Chakraborty, S. A geminivirus betasatellite encoded βC1 protein interacts with PsbP and subverts PsbP-mediated antiviral defence in plants. Mol. Plant Pathol. 2019, 20, 943–960. [Google Scholar] [CrossRef] [PubMed]
- Fang, A.; Han, Y.; Zhang, N.; Zhang, M.; Liu, L.; Li, S.; Lu, F.; Sun, W. Identification and characterization of plant cell death–inducing secreted proteins from Ustilaginoidea virens. Mol. Plant-Microbe Interact. 2016, 29, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.A.; Collins-Racie, L.A.; Colbert, M.; Duckett, M.; Golden-Fleet, M.; Kelleher, K.; Kriz, R.; LaVallie, E.R.; Merberg, D.; Spaulding, V.; et al. A genetic selection for isolating cDNAs encoding secreted proteins. Gene 1997, 198, 289–296. [Google Scholar] [PubMed]
- Harishchandra, D.L.; Zhang, W.; Li, X.; Chethana, K.W.T.; Hyde, K.D.; Brooks, S.; Yan, J.; Peng, J. A LysM domain-containing protein LtLysM1 is important for vegetative growth and pathogenesis in woody plant pathogen Lasiodiplodia theobromae. Plant Pathol. J. 2020, 36, 323–334. [Google Scholar] [CrossRef]
- Peng, J.; Li, X.; Li, Y.; Zhang, W.; Zhou, Y.; Yan, J. Lasiodiplodia theobromae protein LtScp1 contributes to fungal virulence and protects fungal mycelia against hydrolysis by grapevine chitinase. Environ. Microbiol. 2022, 24, 4670–4683. [Google Scholar] [CrossRef] [PubMed]
- Félix, C.; Meneses, R.; Gonçalves, M.F.; Tilleman, L.; Duarte, A.S.; Jorrín-Novo, J.V.; Van de Peer, Y.; Deforce, D.; Van Nieuwerburgh, F.; Esteves, A.C.; et al. A multi-omics analysis of the grapevine pathogen Lasiodiplodia theobromae reveals that temperature affects the expression of virulence- and pathogenicity-related genes. Sci. Rep. 2019, 9, 13144. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, C.; Duan, G.; Wang, Y.; Zhang, Y.; Yang, J. Overexpression of magnaporthe oryzae systemic defense trigger 1 (MoSDT1) confers improved rice blast resistance in rice. Int. J. Mol. Sci. 2019, 20, 4762. [Google Scholar] [CrossRef]
- Ifuku, K.; Yamamoto, Y.; Ono, T.-A.; Ishihara, S.; Sato, F. PsbP protein, but not psbq protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol. 2005, 139, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Hargett, S.R.; Frankel, L.K.; Bricker, T.M. The PsbQ protein is required in arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J. Biol. Chem. 2006, 281, 26260–26267. [Google Scholar] [CrossRef]
- Naidu, R.; Krishnan, M.; Nayudu, M.; Gnanam, A. Studies on peanut green mosaic virus infected peanut (Arachis hypogaea L.) leaves. III. Changes in the polypeptides of photosystem II particles. Physiol. Mol. Plant Pathol. 1986, 29, 53–58. [Google Scholar] [CrossRef]
- Abbink, T.E.; Peart, J.R.; Mos, T.N.; Baulcombe, D.C.; Bol, J.F.; Linthorst, H.J. Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology 2002, 295, 307–319. [Google Scholar]
- Wang, S.; Li, Q.-P.; Wang, J.; Yan, Y.; Zhang, G.-L.; Zhang, H.; Wu, J.; Chen, F.; Wang, X.; Kang, Z.; et al. YR36/WKS1-mediated phosphorylation of PsbO, an extrinsic member of photosystem II, inhibits photosynthesis and confers stripe rust resistance in wheat. Mol. Plant 2019, 12, 1639–1650. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Peng, J.; Zhang, W.; Chethana, T.; Wang, X.; Wang, H.; Yan, J. LtGAPR1 Is a Novel Secreted Effector from Lasiodiplodia theobromae That Interacts with NbPsQ2 to Negatively Regulate Infection. J. Fungi 2023, 9, 188. https://doi.org/10.3390/jof9020188
Huang C, Peng J, Zhang W, Chethana T, Wang X, Wang H, Yan J. LtGAPR1 Is a Novel Secreted Effector from Lasiodiplodia theobromae That Interacts with NbPsQ2 to Negatively Regulate Infection. Journal of Fungi. 2023; 9(2):188. https://doi.org/10.3390/jof9020188
Chicago/Turabian StyleHuang, Caiping, Junbo Peng, Wei Zhang, Thilini Chethana, Xuncheng Wang, Hui Wang, and Jiye Yan. 2023. "LtGAPR1 Is a Novel Secreted Effector from Lasiodiplodia theobromae That Interacts with NbPsQ2 to Negatively Regulate Infection" Journal of Fungi 9, no. 2: 188. https://doi.org/10.3390/jof9020188
APA StyleHuang, C., Peng, J., Zhang, W., Chethana, T., Wang, X., Wang, H., & Yan, J. (2023). LtGAPR1 Is a Novel Secreted Effector from Lasiodiplodia theobromae That Interacts with NbPsQ2 to Negatively Regulate Infection. Journal of Fungi, 9(2), 188. https://doi.org/10.3390/jof9020188






