1. Introduction
The introduction of an exotic pathogen into a new area leads to the establishment of new biotic interactions, giving rise to a process of coevolution between the host and the pathogen [
1,
2]. For the pathogen, the ability to establish itself is closely linked to its ability to survive and reproduce within the host [
3], and therefore to its pathogenicity (defined as the ability of a pathogen to cause disease in a particular host [
4]) and virulence (defined as the degree of damage to the host [
4]). Genetic variability in the pathogen population is essential to ensure the successful adaptation of the pathogen to the new environment [
5].
Fusarium circinatum is an introduced pathogen currently established in Europe in Spain and Portugal infecting
Pinus radiata and
P. pinaster [
6]. This pathogen causes the Pine Pitch Canker (PPC) disease on mature trees,
P. radiata being one of the most susceptible species while
P. pinaster is moderately resistant to the disease. Typical symptoms are sunken cankers on branches and stem with abundant resin, and wilt and necrosis of needles above the infection point [
7].
Fusarium circinatum also infects seedlings in nurseries causing pre- and post-emergence damping-off [
7]. PPC is one of the most important diseases worldwide [
7,
8]. In the European Plant Protection Organization (EPPO) territory, it is a disease included in the A2 list (includes pests locally present in the EPPO region), and in the EU, it is regulated as a quarantine pest (Commission Decision 2007/433/EC and 2019/2032/EC) in order to prevent the introduction and spread of
F. circinatum in the EU.
DNA-based studies have been carried out on
Fusarium circinatum populations worldwide to analyze their structure and infer the reproduction mode and pathways for pathogen introduction and migration [
9]. First studies done in Japan and California showed low levels of genetic diversity and concluded the lack of sexual reproduction [
10,
11,
12]. A global analysis including isolates from different geographical locations [
13] revealed that populations from Mexico, Portugal, South Africa, Uruguay, and the USA were more diverse than those from Spain, Chile, France, and Japan. MLGs identified in the USA were spread worldwide which suggests a common source for introductions. Regarding sexual reproduction, occurrence was rejected for populations from the USA and Spain, but not from Chile and South Africa. However, in a recent study in South Africa population [
14], ongoing sexual reproduction was rejected, indicating that the highly diverse genetic diversity detected was probably a consequence of multiple different introductions.
Genetic diversity analysis revealed that Spanish population is structured in two clusters, each one with a predominant haplotype (named MLG32 and MLG59), a situation compatible with two independent pathogen introductions in Spain [
13]. Genetic association analysis showed linkage disequilibrium and clonality in the population, supporting the lack of sexual reproduction in the
F. circinatum Spanish population. Both mating types, MAT-1 and MAT-2, were found, although one of the two clusters (with predominant haplotype MLG32) only contained isolates of mating type MAT-2 located in the northeastern region (País Vasco). A further study with isolates representing the main haplotypes in each of the two clusters [
15] showed that variability on life history traits such as disease severity, spore germination and sporulation do not compromise the pathogen evolutionary potential. This question is particularly important for introduced pathogens, where a reduced genetic variation may exist [
1] because of the small population size when established in a new environment.
There are different mechanisms potentially associated with significant changes in genetic variability of
F. circinatum. Heterokaryons are originated through: (1) hyphal anastomosis of isolates of the same vegetative compatibility group (VCG) [
16] and (2) selfing, which results from fusion of haploid cells produced by the same individual [
16,
17]. For
Fusarium circinatum the sexual state has not been reported in nature [
9], and VCGs have been determined in some populations. In southeastern USA, where PPC disease was first described in 1946, a total of 45 VCGs were found [
11], while in California, where disease was reported 40 years later, eight VCGs were identified [
18]. In northeastern Spain, VCGs were determined in isolates from País Vasco [
19] revealing the existence of only two vegetative compatibility groups (VCGs) of the same mating type. No other study to determine VCGs in a broader Spanish population including other regions but País Vasco has been carried out.
Regarding selfing mechanism, it is differentiated haploid from diploid selfing [
17] with different consequences on the genetic population structure [
17]. Diploid selfing is the fusion of haploid cells originated from meiosis in the same individual (unlike outcrossing, where haploid cells are from different individuals). In heterotallic fungi such as
Fusarium circinatum, haploid selfing is prevented [
17] since the haploid cells are originated by mitosis and do not carry different alleles for the mating types that are required for the fusion of the cells. Furthermore, there is growing evidence that cryptic sex occurs in fungi for which sexual reproduction has not been reported [
17,
20,
21]. In the case of
Fusarium circinatum, the sexual stage has not been reported in the field [
9], but outcrossing was successfully conducted under laboratory conditions [
18,
22,
23], producing perithecia with viable ascospores that indicates that both mating types are functional and sexual reproduction is possible. These three types of reproduction, haploid and diploid selfing, and outcrossing, in which meiosis and recombination are involved, are considered different modes of sexual reproduction occurring in fungi [
17].
Population genetics studies provide insight into the evolutionary process and changes in gene frequencies that shape pathogen population structure [
24,
25]. Introduction of a pathogen into a new area leads to new host-pathogen interactions usually without a coevolutionary history, which have impact on their distribution and abundance and possible changes in genotype frequencies [
1,
2]. Moreover, pathogens must evolve constantly to adapt to changes in their environment and survive [
1,
3].
Fusarium circinatum is extended in Spain to the Atlantic coast [
26] where most
Pinus radiata plantations are grown. It is the only exotic species of pine planted in Spain and cohabits in this area with
P. pinaster, a native Mediterranean species. Moreover, some new environmental situations have been identified after its introduction [
15], such as the differential genetic resistance that pine species have in Spain (
P. radiata more susceptible than
P. pinaster [
27]) that exert different pressure to the pathogen, or the management measures taken to mitigate the damage.
The objective of this work is to quantify the genetic diversity of Spanish Fusarium circinatum populations infecting pine stands and to determine how this variation is distributed in a temporal and spatial scale. The aim is to understand the biological significance of the variation detected and to make inferences about the evolutionary processes that may influence the population structure.
4. Discussion
In this work, we analyzed the genetic variation of Spanish
Fusarium circinatum population sampled during 2018–2021 years to study changes over time since first outbreak as an exotic pathogen. Results indicate that temporal variation in genetic variability is low and decreased since 2004–2011 (
Table 3). At that time, a survey was done as consequence of a severe outbreak of PPC disease in Northern regions using microsatellite (SSR) markers. Microsatellite have been the markers of choice for many population genetic studies due to their characteristics and technical advantages compared with others [
53]. A faster rate of evolution makes them more sensitive to identify recent phylogeographic and demographic events, although some caution is needed when making inferences from the results because of their hypervariability [
24,
53,
54]. A few other studies have been done but using other molecular markers [
9]. Specifically, they were RFLPs [
12], sequence-characterized markers used on the South African population [
55] and AFLP markers used on population from País Vasco [
19]. When genetic variability in the Spanish population was analyzed by regions between surveys, results indicated that variability decreases in northwestern regions while in País Vasco it is maintained (
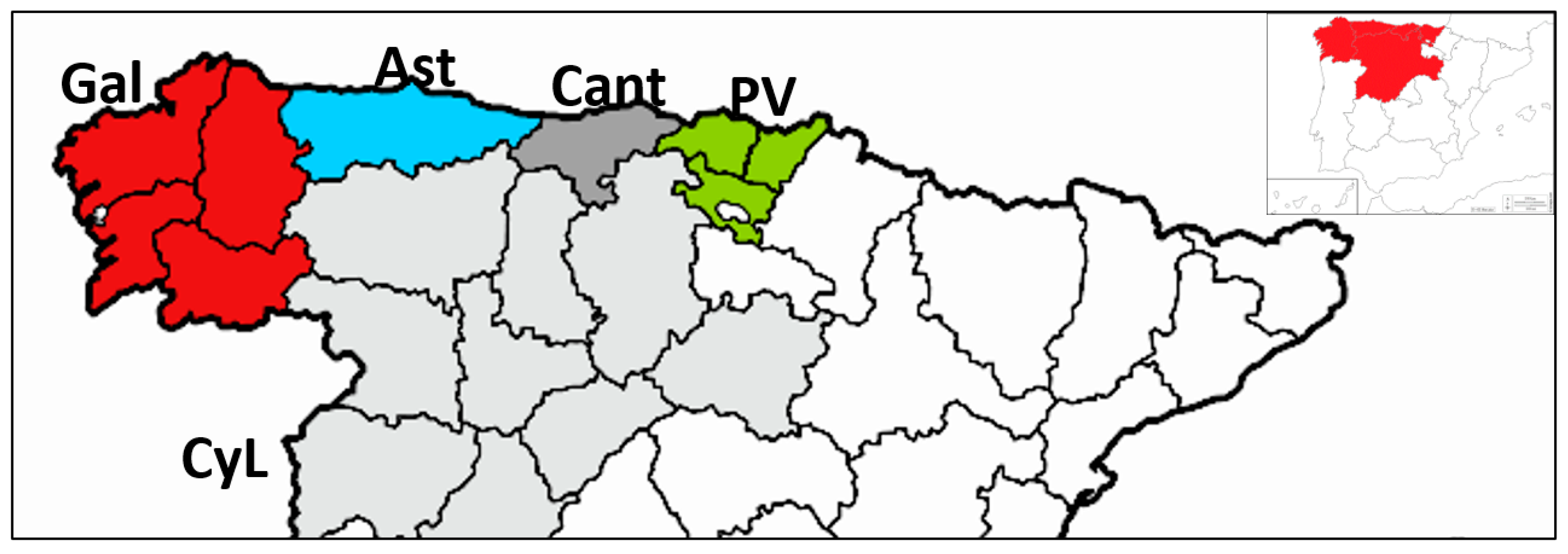
Figure 1;
Table 3). This situation with differences in variability associated with geographical locations has also been described in South African populations [
14]. Using SSR markers, they detected a highly diverse population in nurseries and in plantations located in some regions.
Results presented here show that Spanish population of
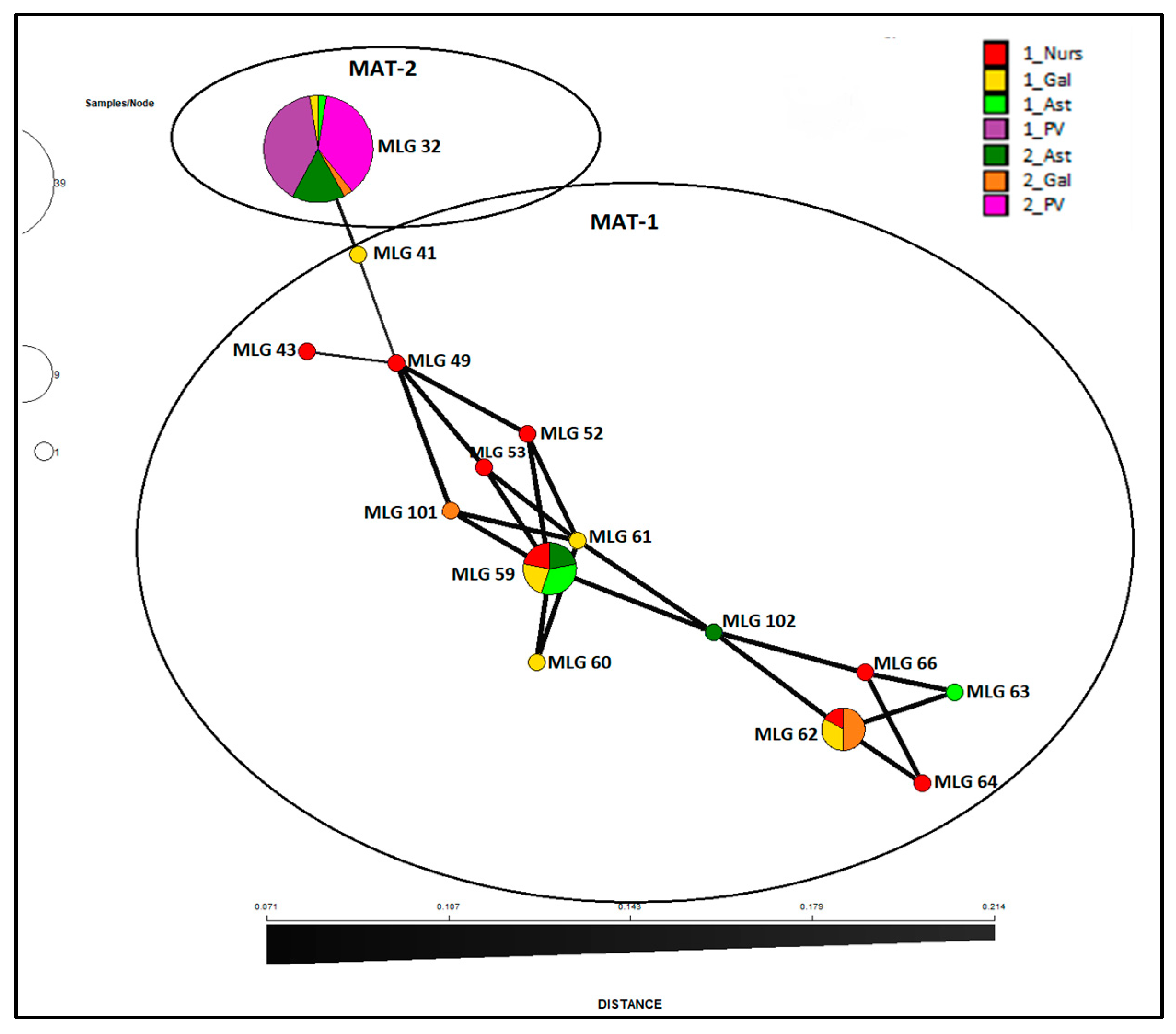
Fusarium circinatum is structured in two broad groups as visualized by the minimum spanning network analysis of MLG by surveys and regions (
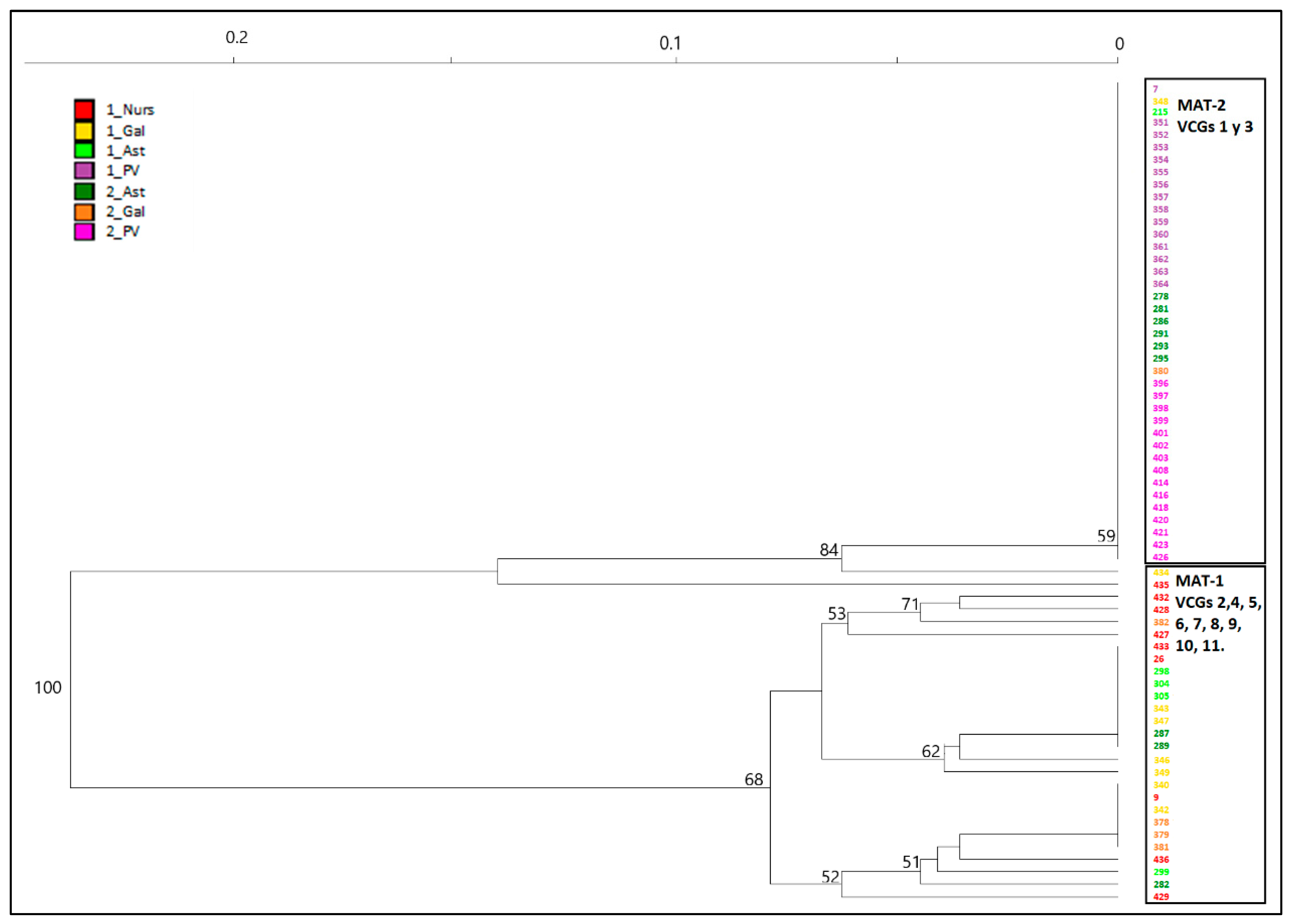
Figure 4) as well as the NJ dendrogram including all the isolates (
Figure 3). This structure was already described by Berbegal et al. [
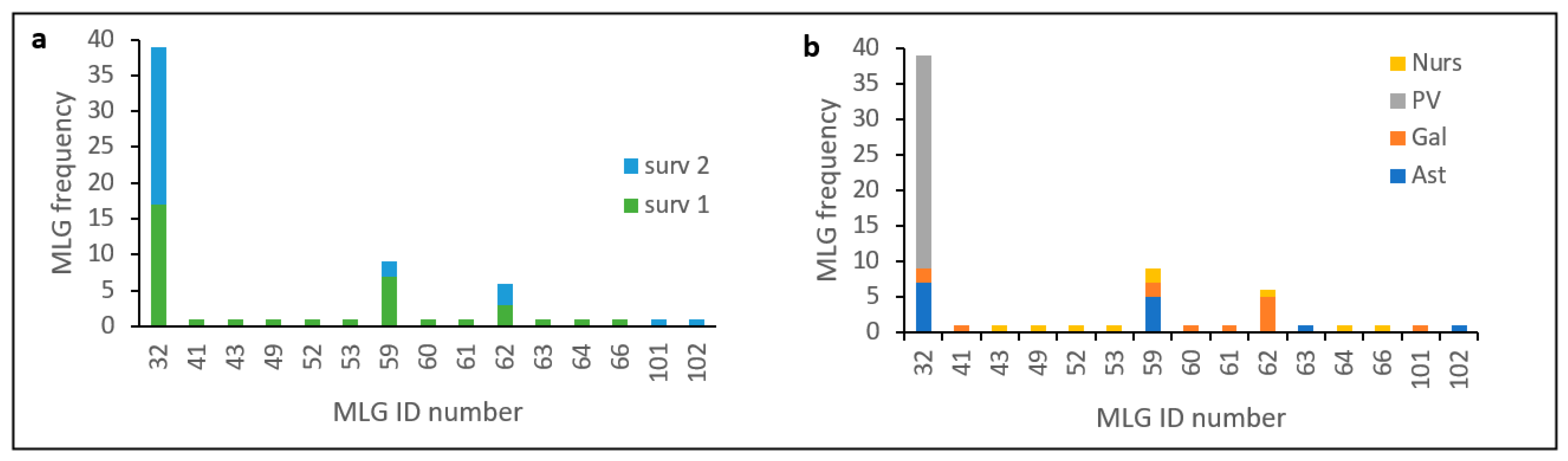
13] in an analysis that included a greater number of isolates and samples from other Spanish affected regions (Cantabria and Castilla y León), and evidenced the occurrence of at least two introductions of the pathogen in Spain. One of these clusters includes all isolates of haplotype MLG32, and MAT-2 distributed mainly in the País Vasco but also found in the Northwestern regions of Galicia and Asturias. Frequency distribution of this haplotype (
Figure 2) and minimum spanning network analysis (
Figure 4) shows that MLG32 presence has increased in the NW regions over time between the two surveys done (10 years apart on average), and continues to be the only one present in País Vasco.
This situation confirms that MLG32 is a well-adapted haplotype as it was shown in previous studies [
15] where some pathogenic life history traits were compared among isolates. Comparison demonstrated that one isolate from País Vasco representative of haplotype MLG32 was the most virulent of those studied. This isolate caused more severe disease on
Pinus radiata at 25 and 30 °C compared with the second most common haplotype (MLG59). Spore germination was also higher for the most common haplotype, and produced more spores at 20 and 25 °C.
Exclusive presence of MLG32 in País Vasco during these 10 years between surveys indicates
Fusarium circinatum is inbreeding or goes under asexual reproduction, at least in this region. Only two VCGs were found in País Vasco, which contributes to a limited exchange of genetic information and maintenance of individual identity [
56]. Temporal increase of VCG 1 (from 60 to 97%) respect to VCG 3 (
Table 8) suggests that haplotype MLG32 and particularly VCG 1 has more fitness that other isolates and is reproducing asexually with no recombination. Other results also suggest that País Vasco is isolated genetically and there is not gene flow with northwestern regions. Mantel test showed that this region is subdivided and differentiated from the northwestern regions with no evidence of migration among populations (
Table 8). Pairwise population comparisons of genetic diversity also indicated that subpopulation from País Vasco is differentiated from Galicia and Asturias.
The presence of a well-adapted haplotype together with evidence for clonality in País Vasco supports that MLG32 may have existed for some time and that this pathogen established first in this region, a hypothesis already suggested by Berbegal et al. [
13]. Symptoms of PPC disease were initially observed in a nursery from País Vasco in 1990s [
57], and some of these isolates were subsequently analyzed together with isolates collected during the first survey. Results confirmed that population from País Vasco at those years was already constituted by only haplotype MLG32. Iturritxa et al. [
19] also reported a genetically homogeneous population using AFLPs markers in a sampling done in 2004 in País Vasco. This haplotype was later identified in other northwestern Spanish regions.
The maintenance of the population genetics in two clusters during these 10 years between surveys supports the occurrence of clonality or inbreeding and the lack of sexual reproduction. AMOVA results on the whole population show that genetic variation is explained mostly by regions, but not by time, supporting the idea of clonal organisms. However, this structure does not appear when AMOVA is performed on the clone-corrected dataset. This discrepancy in the results is probably due to the small size of the population that results from correction and point out the importance that the clones have in the AMOVA analysis.
The second cluster in which population of
Fusarium circinatum is structured (
Figure 3) is more diverse in terms of haplotypes and VCGs and includes most of the isolates of MAT-1 from NW regions (Gal and Ast in
Figure 1). Here, genetic diversity has decreased between surveys (
Table 3) suggesting that population is inbreeding. In addition, other results support lack of random mating. These are the standardized index of association (r
d) (
Table 4) and P
sex values associated with some of the three more frequent haplotypes within regions and between surveys (
Table 5). Lack of significance between migrant proportions of Asturias and Galicia also corroborates inbreeding populations in these regions.
In NW regions with a greater number of MLGs and VCGs, the occurrence and identification of heterokaryons is more likely as has been the case. Two isolates from the first survey, one from Galicia and other from a nursery in Asturias, have two microsatellite alleles per locus. These heterokaryons may have originated by anastomoses of hyphae or diploid selfing (processes not necessarily exclusive [
16]). Detection of two new haplotypes in the second survey, one from Galicia and other from Asturias, also suggests that these processes of hyphal fusion and/or recombination are taking place. New introductions or movement of the pathogen with plant material is not likely occurring since 2007, when the entrance and movement of material was regulated in the EU territory to prevent pathogen spread. Lack of presence of other haplotypes in País Vasco is indirect evidence of the efficiency of those regulations.
Despite the genetic uniformity found in the population of
Fusarium circinatum in País Vasco, evidence of genetic changes was found when more intensive samplings were carried out. In an intensive sampling done in 2016 in a
P. radiata plantation with symptoms of PPC, one new haplotype was detected by SSR marker analysis [
58]. Only 1 isolate out of the 69 obtained from cankers showed polymorphism for FCM-25 loci, showing 199 bp length instead of 204 bp.
In conclusion, results presented here about the temporal and spatial variation of population structure of Spanish Fusarium circinatum are explained by the asexual reproduction, but also selfing at least to a lesser extent that leads to new haplotypes. The existence of haplotype MLG32 maintained on time and widely distributed across northwestern regions suggest its well adaptation to the environment and the host. Results showed that País Vasco is a region that after many years of the first disease outbreak in Spain remains clearly differentiated from other Northwestern populations. This fact was supported with no evidence of migration among regions. The low genetic diversity observed and lack of new haplotypes in País Vasco indirectly evidences that regulation within the EU territory to prevent pathogen spread is efficient.