Reference-Based Restriction-Site-Associated DNA Sequencing Data Are Useful for Species Delineation in a Recently Diverged Asexually Reproducing Species Complex (Parmeliaceae, Ascomycota)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Sampling
2.2. DNA Extraction and RAD Library Preparation
2.3. RADseq Assembly
2.4. Phylogenomic Analyses
2.5. Analysis of Population Structure
3. Results and Discussion
3.1. Assembly of RAD Sequencing
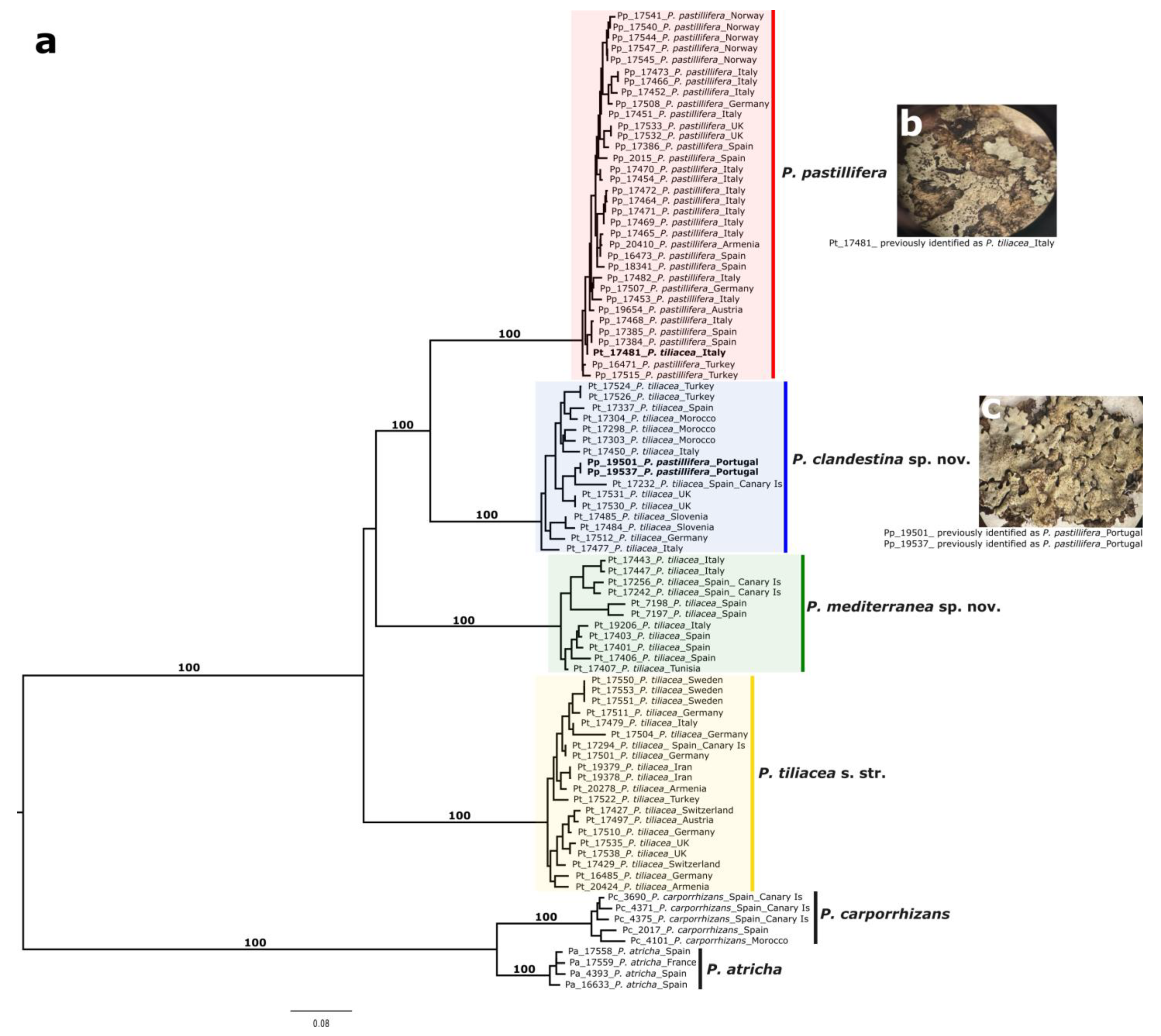
3.2. Phylogenomic Analyses
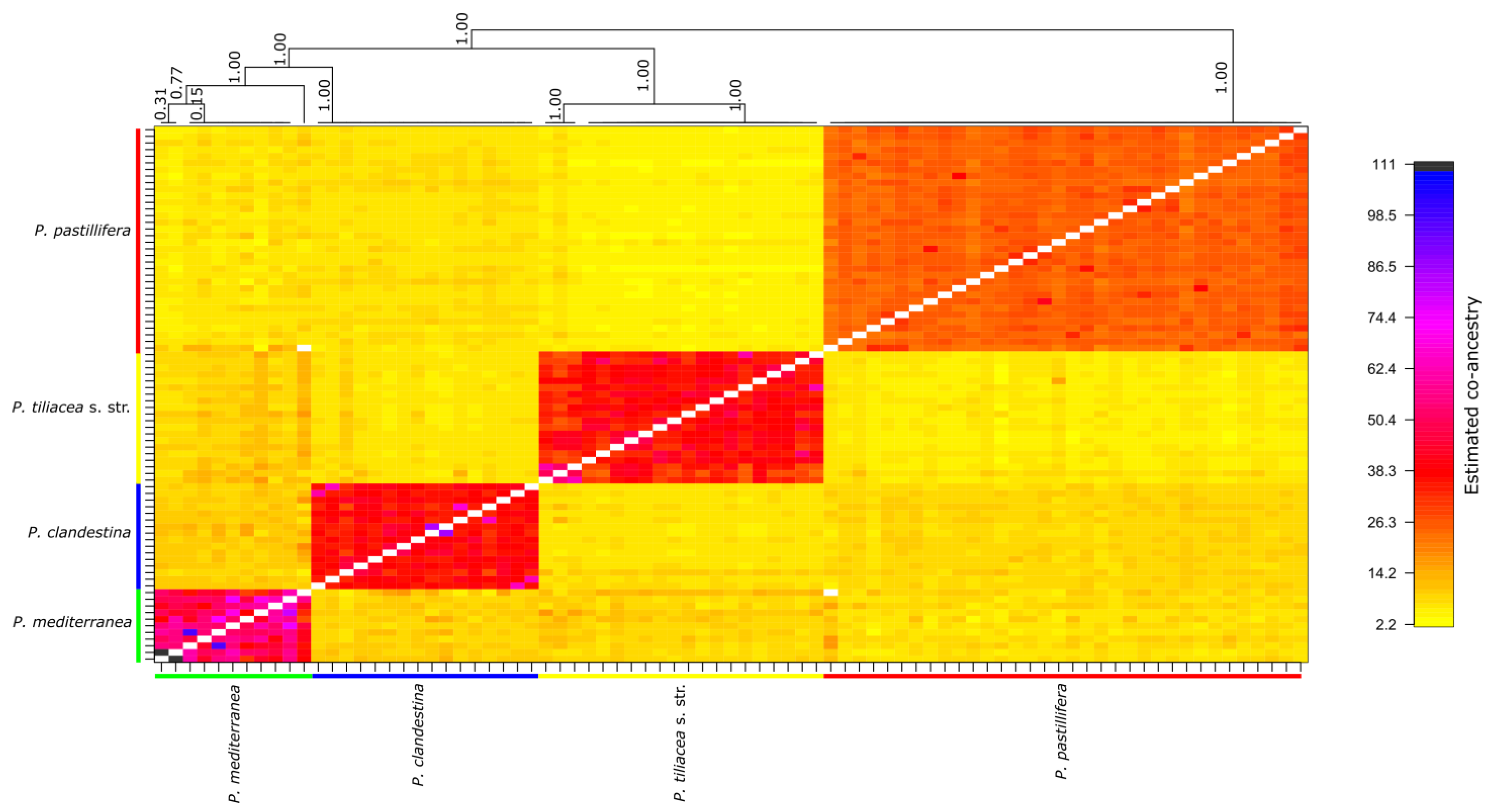
3.3. Analysis of Population Structure
3.4. Taxonomy
- MycoBank: MB850743
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lücking, R.; Leavitt, S.D.; Hawksworth, D.L. Species in lichen-forming fungi: Balancing between conceptual and practical considerations, and between phenotype and phylogenomics. Fungal Divers. 2021, 109, 99–154. [Google Scholar] [CrossRef]
- Crespo, A.; Lumbsch, H.T. Cryptic species in lichen-forming fungi. IMA Fungus 2010, 1, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Crespo, A.; Pérez-Ortega, S. Cryptic species and species pairs in lichens: A discussion on the relationship between molecular phylogenies and morphological characters. An. Del Jardín Botánico De Madr. 2009, 66, 71–81. [Google Scholar] [CrossRef]
- Grube, M.; Kroken, S. Molecular approaches and the concept of species and species complexes in lichenized fungi. Mycol. Res. 2000, 104, 1284–1294. [Google Scholar] [CrossRef]
- Leavitt, S.D.; Moreau, C.S.; Lumbsch, H.T. The Dynamic Discipline of Species Delimitation: Progress Toward Effectively Recognizing Species Boundaries in Natural Populations. In Recent Advances in Lichenology; Upreti, D.K., Divakar, P.K., Shukla, V., Bajpai, R., Eds.; Springer: Delhi, India, 2015; pp. 11–44. [Google Scholar]
- Leavitt, S.D.; Divakar, P.K.; Crespo, A.; Lumbsch, H.T. A matter of time—Understanding the limits of the power of molecular data for delimiting species boundaries. Herzogia 2016, 29, 479–492. [Google Scholar] [CrossRef]
- Lumbsch, H.T.; Leavitt, S.D. Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Divers. 2011, 50, 59–72. [Google Scholar] [CrossRef]
- Vondrak, J.; Riha, P.; Arup, U.; Søchting, U. The taxonomy of the Caloplaca citrina group (Teloschistaceae) in the Black Sea region; with contributions to the cryptic species concept in lichenology. Lichenologist 2009, 41, 571–604. [Google Scholar] [CrossRef]
- Nuñez-Zapata, J.; Divakar, P.K.; Del-Prado, R.; Cubas, P.; Hawksworth, D.L.; Crespo, A. Conundrums in species concepts: The discovery of a new cryptic species segregated from Parmelina tiliacea (Ascomycota: Parmeliaceae). Lichenologist 2011, 43, 603–616. [Google Scholar] [CrossRef]
- Hodkinson, B.P.; Lendemer, J.C. Molecular analyses reveal semi-cryptic species in Xanthoparmelia tasmanica. Bibl. Lichenol. 2011, 106, 108–119. [Google Scholar]
- Coca, L.F.; Lücking, R.; Moncada, B. Two new, sympatric and semi-cryptic species of Sulzbacheromyces (Lichenized Basidiomycota, Lepidostromatales) from the Chocó biogeographic region in Colombia. Bryologist 2018, 121, 297–305. [Google Scholar] [CrossRef]
- Altermann, S.; Leavitt, S.D.; Goward, T.; Nelsen, M.P.; Lumbsch, H.T. How do you solve a problem like Letharia? A new look at cryptic species in lichen-forming fungi using Bayesian clustering and SNPs from multilocus sequence data. PLoS ONE 2014, 9, e97556. [Google Scholar] [CrossRef]
- Lücking, R.; Dal-Forno, M.; Sikaroodi, M.; Gillevet, P.M.; Bungartz, F.; Moncada, B.; Yynez-Ayabaca, A.; Chaves, J.L.; Coca, L.F.; Lawrey, J.D. A single macrolichen constitutes hundreds of unrecognized species. Proc. Natl. Acad. Sci. USA 2014, 111, 11091–11096. [Google Scholar] [CrossRef]
- Ankita, H.; Jiang, S.H.; Lücking, R.; Liu, H.J.; Wei, X.L.; Xavier-Leite, A.B.; Portilla, C.V.; Ren, Q.; Wei, J.C. Twelve new species reveal cryptic diversification in foliicolous lichens of Strigula s.lat. (Strigulales, Ascomycota). J. Fungi 2022, 8, 2. [Google Scholar]
- Cornejo, C.; Scheidegger, C. Multi-gene phylogeny of the genus Lobaria: Evidence of species-pair and allopatric cryptic speciation in East Asia. Am. J. Bot. 2015, 102, 2058–2073. [Google Scholar] [CrossRef] [PubMed]
- Baloch, E.; Grube, M. Pronounced genetic diversity in tropical epiphyllous lichen fungi. Mol. Ecol. 2009, 18, 2185–2197. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Huescar, E.; Gonzalez-Burgos, E.; Kirika, P.M.; Boustie, J.; Ferron, S.; Gomez-Serranillos, M.P.; Lumbsch, H.T.; Divakar, P.K. A New Cryptic Lineage in Parmeliaceae (Ascomycota) with Pharmacological Properties. J. Fungi 2022, 8, 826. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, S.D.; Fankhauser, J.D.; Leavitt, D.H.; Porter, L.D.; Johnson, L.A.; St Clair, L.L. Complex patterns of speciation in cosmopolitan “rock posy” lichens—Discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Mol. Phylogenetics Evol. 2011, 59, 587–602. [Google Scholar] [CrossRef]
- Otálora, M.A.G.; Martínez, I.; Aragón, G.; Molina, M.C. Phylogeography and divergence date estimates of a lichen species complex with a disjunct distribution pattern. Am. J. Bot. 2010, 97, 216–223. [Google Scholar] [CrossRef]
- Jorna, J.; Linde, J.B.; Searle, P.C.; Jackson, A.C.; Nielsen, M.-E.; Nate, M.S.; Saxton, N.A.; Grewe, F.; Herrera-Campos, M.d.l.A.; Spjut, R.W.; et al. Species boundaries in the messy middle—A genome-scale validation of species delimitation in a recently diverged lineage of coastal fog desert lichen fungi. Ecol. Evol. 2021, 11, 18615–18632. [Google Scholar] [CrossRef]
- Pino-Bodas, R.; Rosa Burgaz, A.; Martin, M.P.; Lumbsch, H.T. Phenotypical plasticity and homoplasy complicate species delimitation in the Cladonia gracilis group (Cladoniaceae, Ascomycota). Org. Divers. Evol. 2011, 11, 343–355. [Google Scholar] [CrossRef]
- Steinová, J.; Stenroos, S.; Grube, M.; Škaloud, P. Genetic diversity and species delimitation of the zeorin-containing red-fruited Cladonia species (lichenized Ascomycota) assessed with ITS rDNA and ß-tubulin data. Lichenologist 2013, 45, 665–684. [Google Scholar] [CrossRef]
- Velmala, S.; Myllys, L.; Halonen, P.; Goward, T.; Ahti, T. Molecular data show that Bryoria fremontii and B. Tortuosa (Parmeliaceae) Are Conspecific. Lichenologist 2009, 41, 231–242. [Google Scholar] [CrossRef]
- Kotelko, R.; Piercey-Normore, M.D. Cladonia pyxidata and C. pocillum; genetic evidence to regard them as conspecific. Mycologia 2010, 102, 534–545. [Google Scholar] [CrossRef]
- Grewe, F.; Huang, J.P.; Leavitt, S.D.; Lumbsch, H.T. Reference-based RADseq resolves robust relationships among closely related species of lichen-forming fungi using metagenomic DNA. Sci. Rep. 2017, 7, 9884. [Google Scholar] [CrossRef]
- Grewe, F.; Lagostina, E.; Wu, H.; Printzen, C.; Lumbsch, H.T. Population genomic analyses of RAD sequences resolves the phylogenetic relationship of the lichen-forming fungal species Usnea antarctica and Usnea aurantiacoatra. Mycokeys 2018, 43, 91–113. [Google Scholar] [CrossRef]
- Otero, A.; Barcenas Peña, A.; Lumbsch, H.T.; Grewe, F. Reference-based RADseq unravels the evolutionary history of polar species in the ‘Crux Lichenogorum’ genus Usnea (Parmeliaceae, Ascomycota). J. Fungi 2023, 9, 99. [Google Scholar] [CrossRef]
- Widhelm, T.J.; Rao, A.; Grewe, F.; Lumbsch, H.T. High-throughput sequnecing confirms the boundary between traditionally considered species pairs in a group of lichenized fungi (Peltigeraceae, Pseudocyphellaria). Bot. J. Linn. Soc. 2023, 201, 471–482. [Google Scholar] [CrossRef]
- Alonso-García, M.; Grewe, F.; Payette, S.; Villarreal, A.J.C. Population genomics of a reindeer lichen species from North American lichen woodlands. Am. J. Bot. 2021, 108, 159–171. [Google Scholar] [CrossRef]
- Thell, A.; Crespo, A.; Divakar, P.K.; Kärnefelt, I.; Leavitt, S.D.; Lumbsch, H.T.; Seaward, M.R.D. A review of the lichen family Parmeliaceae—History, phylogeny and current taxonomy. Nord. J. Bot. 2012, 30, 641–664. [Google Scholar] [CrossRef]
- Núñez-Zapata, J.; Alors, D.; Cubas, P.; Divakar, P.K.; Leavitt, S.D.; Lumbsch, H.T.; Crespo, A. Understanding disjunct distribution patterns in lichen forming fungi—Insights from the genus Parmelina (Parmeliaceae, Ascomycota). Bot. J. Linn. Soc. 2017, 184, 238–253. [Google Scholar] [CrossRef]
- Crespo, A.; Kauff, F.; Divakar, P.K.; Amo, G.; Arguello, A.; Blanco, O.; Cubas, P.; del Prado, R.; Elix, J.A.; Esslinger, T.L.; et al. Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon 2010, 59, 1735–1753. [Google Scholar] [CrossRef]
- Arguello, A.; Del Prado, R.; Cubas, P.; Crespo, A. Parmelina quercina (Parmeliaceae, Lecanorales) includes four phylogenetically supported morphospecies. Biol. J. Linn. Soc. 2007, 91, 455–467. [Google Scholar] [CrossRef]
- Crespo, A.; Ferencova, Z.; Pérez-Ortega, S.; Argüello, A.; Elix, J.A.; Divakar, P.K. Austroparmelina, a new Australasian lineage in parmelioid lichens (Parmeliaceae, Ascomycota): A multigene and morphological approach. Syst. Biodivers. 2010, 8, 209–221. [Google Scholar] [CrossRef]
- Dobson, F.S.; Hawksworth, D.L. Parmelia pastillifera (Harm.) Schub. & Klem. and P. tiliacea (Hoffm.) Ach. in the British Isles. Lichenologist 1976, 8, 47–59. [Google Scholar]
- Nuñez-Zapata, J.; Cubas, P.; Hawksworth, D.L.; Crespo, A. Biogeography and Genetic Structure in Populations of a Widespread Lichen (Parmelina tiliacea, Parmeliaceae, Ascomycota). PLoS ONE 2015, 10, e0126981. [Google Scholar] [CrossRef]
- Hörandl, E.; Stuessy, T.F. Paraphyletic groups as natural units of biological classification. Taxon 2010, 59, 1641–1653. [Google Scholar] [CrossRef]
- Nuñez-Zapata, J.A. Variabilidad Genética, Especies Crípticas y Filogenia Molecular en Hongos Liquenizados Del Género Parmelina (Parmeliaceae, Ascomycota); Universidad Complutense de Madrid: Madrid, Spain, 2013. [Google Scholar]
- Kim, W.; Liu, R.; Woo, S.; Kang, K.B.; Park, H.; Yu, Y.H.; Ha, H.-H.; Oh, S.-Y.; Yang, J.H.; Kim, H.; et al. Linking a gene cluster to atranorin, a major cortical substance of lichens, through genetic dereplication and heterologous expression. Am. Soc. Microbiol. 2021, 12, 10–1128. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Malinsky, M.; Trucchi, E.; Lawson, D.; Falush, D. RADpainter and fineRADstructure: Population inference from RADseq data. Mol. Biol. Evol. 2018, 35, 1284–1290. [Google Scholar] [CrossRef]
- Stamakasis, A. RAxML Version 8: A Tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Eaton, D.A.; Overcast, I. Ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 2020, 36, 2592–2594. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Jombart, T.; Balloux, F.; Dray, S. Adephylo: New tools for investigating the phylogenetic signal in biological traits. Bioinformatics 2010, 26, 1907–1909. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Josh, L. GST and its relatives do not measure differentiation. Mol. Ecol. 2008, 17, 4015–4026. [Google Scholar]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: Fst and related measures. Mol. Ecol. Resour. 2011, 11, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P.W. A standardized genetic differentiation measure. Evolution 2005, 59, 1633–1638. [Google Scholar]
- Winter, D.J. MMOD: An R library for the calculation of population differentiation statistics. Mol. Ecol. Resour. 2012, 12, 1158–1160. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]


| Nei’s Gst | |||
|---|---|---|---|
| P. clandestina | P. mediterranea | P. tiliacea s. str. | |
| P. mediterranea | 0.7389792 | ||
| P. tiliacea s. str. | 0.7892928 | 0.7798357 | |
| P. pastillifera | 0.7938066 | 0.8331741 | 0.8652509 |
| Hedrick’s G’st | |||
| P. clandestina | P. mediterranea | P. tiliacea s. str. | |
| P. mediterranea | 0.9105523 | ||
| P. tiliacea s. str. | 0.9452315 | 0.9314394 | |
| P. pastillifera | 0.9291603 | 0.9452874 | 0.9654279 |
| AMOVA Components of Covariance | % |
|---|---|
| Variations between samples | 94.906133 |
| Variations within samples | 5.093867 |
| Total variations | 100 |
| Phi-samples-total = 0.9490613 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barcenas-Peña, A.; Divakar, P.K.; Crespo, A.; Nuñez-Zapata, J.; Lumbsch, H.T.; Grewe, F. Reference-Based Restriction-Site-Associated DNA Sequencing Data Are Useful for Species Delineation in a Recently Diverged Asexually Reproducing Species Complex (Parmeliaceae, Ascomycota). J. Fungi 2023, 9, 1180. https://doi.org/10.3390/jof9121180
Barcenas-Peña A, Divakar PK, Crespo A, Nuñez-Zapata J, Lumbsch HT, Grewe F. Reference-Based Restriction-Site-Associated DNA Sequencing Data Are Useful for Species Delineation in a Recently Diverged Asexually Reproducing Species Complex (Parmeliaceae, Ascomycota). Journal of Fungi. 2023; 9(12):1180. https://doi.org/10.3390/jof9121180
Chicago/Turabian StyleBarcenas-Peña, Alejandrina, Pradeep K. Divakar, Ana Crespo, Jano Nuñez-Zapata, H. Thorsten Lumbsch, and Felix Grewe. 2023. "Reference-Based Restriction-Site-Associated DNA Sequencing Data Are Useful for Species Delineation in a Recently Diverged Asexually Reproducing Species Complex (Parmeliaceae, Ascomycota)" Journal of Fungi 9, no. 12: 1180. https://doi.org/10.3390/jof9121180
APA StyleBarcenas-Peña, A., Divakar, P. K., Crespo, A., Nuñez-Zapata, J., Lumbsch, H. T., & Grewe, F. (2023). Reference-Based Restriction-Site-Associated DNA Sequencing Data Are Useful for Species Delineation in a Recently Diverged Asexually Reproducing Species Complex (Parmeliaceae, Ascomycota). Journal of Fungi, 9(12), 1180. https://doi.org/10.3390/jof9121180









