Trichoderma harzianum Cellulase Gene thph2 Affects Trichoderma Root Colonization and Induces Resistance to Southern Leaf Blight in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Fungal Strains and Culture
2.3. Constructing the Vectors
2.4. Transformation by ATMT and Screening of Transformants
2.5. Southern Blotting Analysis
2.6. Determination of Trichoderma Growth and Reproductive Ability
2.7. Plate Confrontation Assay of Trichoderma and the Pathogen
2.8. Inoculum Preparation
2.9. Infection Assay
2.10. Quantification of Trichoderma Root Colonization
2.11. Analysis of Gene Expression Using RT-qPCR
2.12. Data Analysis
3. Results
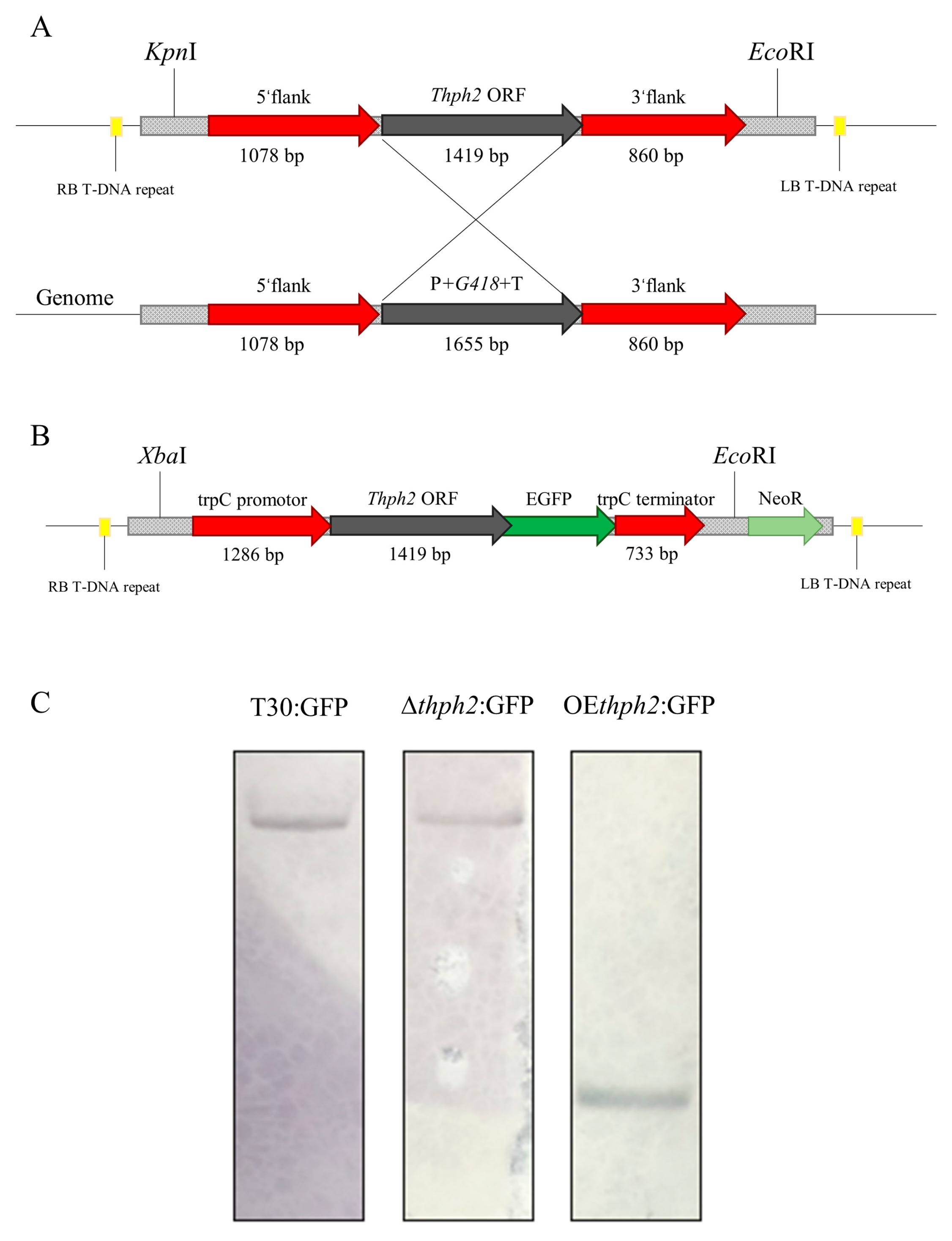
3.1. Construction and Identification of thph2-Deletion and -Overexpression Transformants
3.2. Expression of thph2 in Transformants
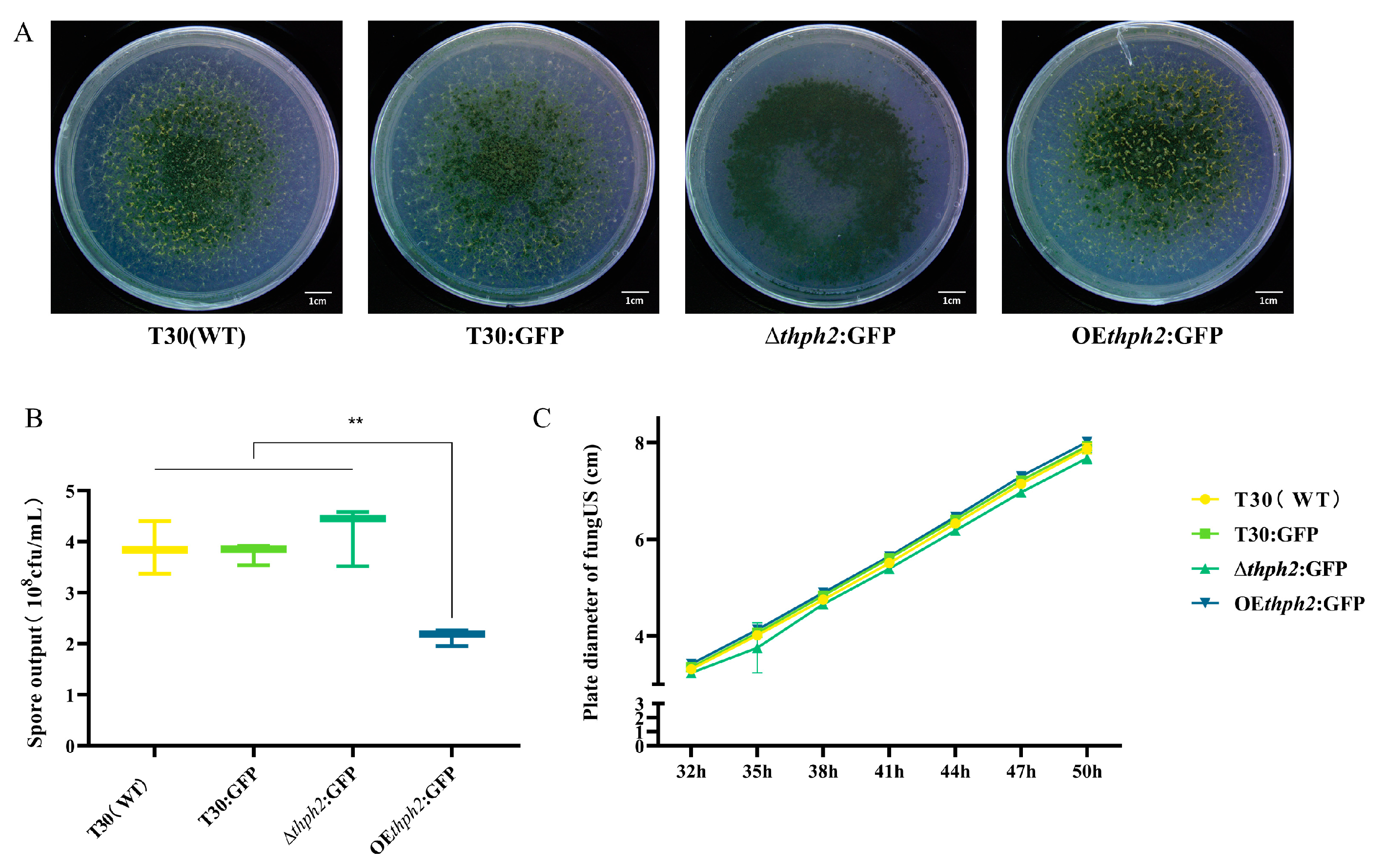
3.3. Evaluation of Growth, Reproduction, and Phenotype of thph2 Mutants
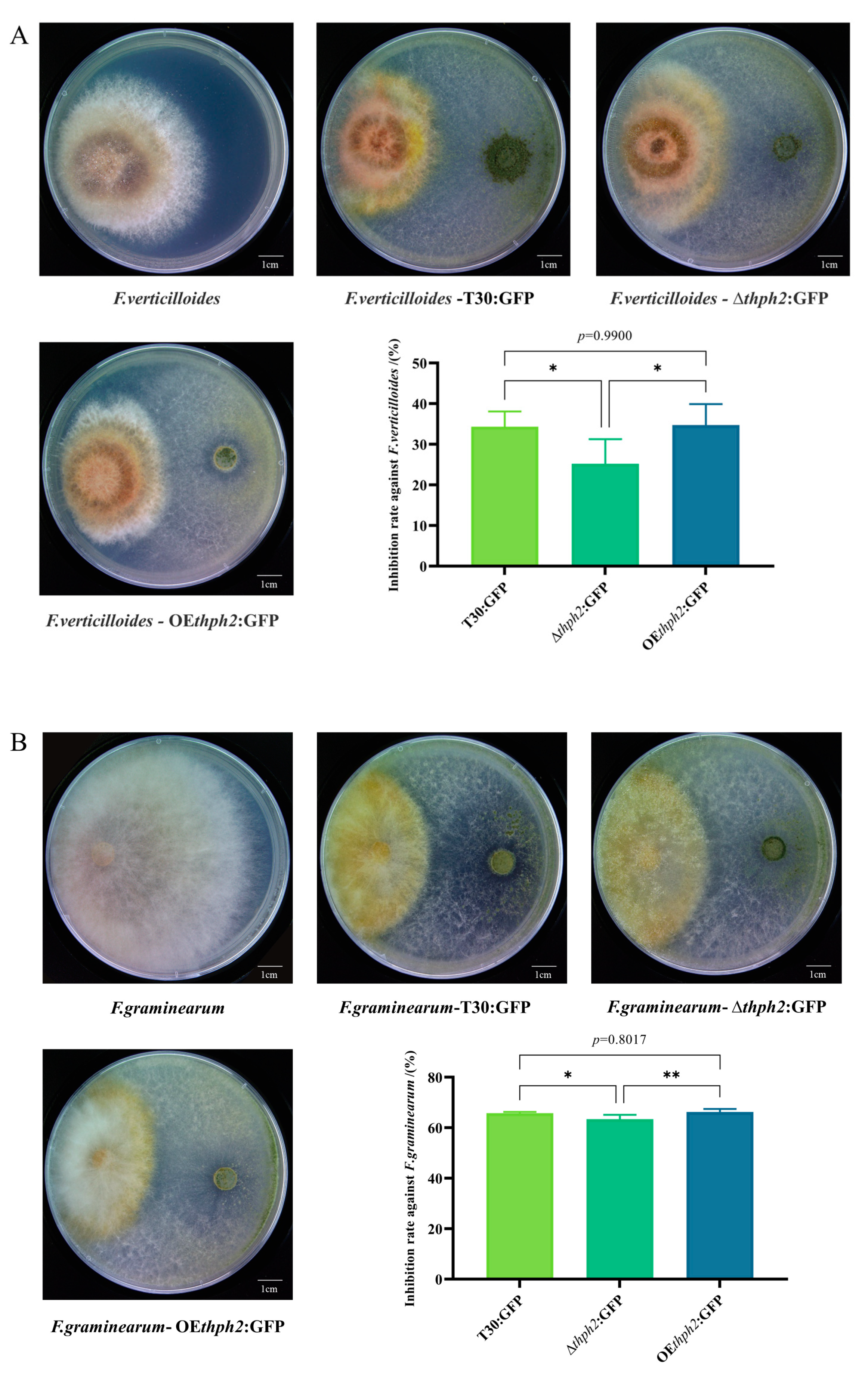
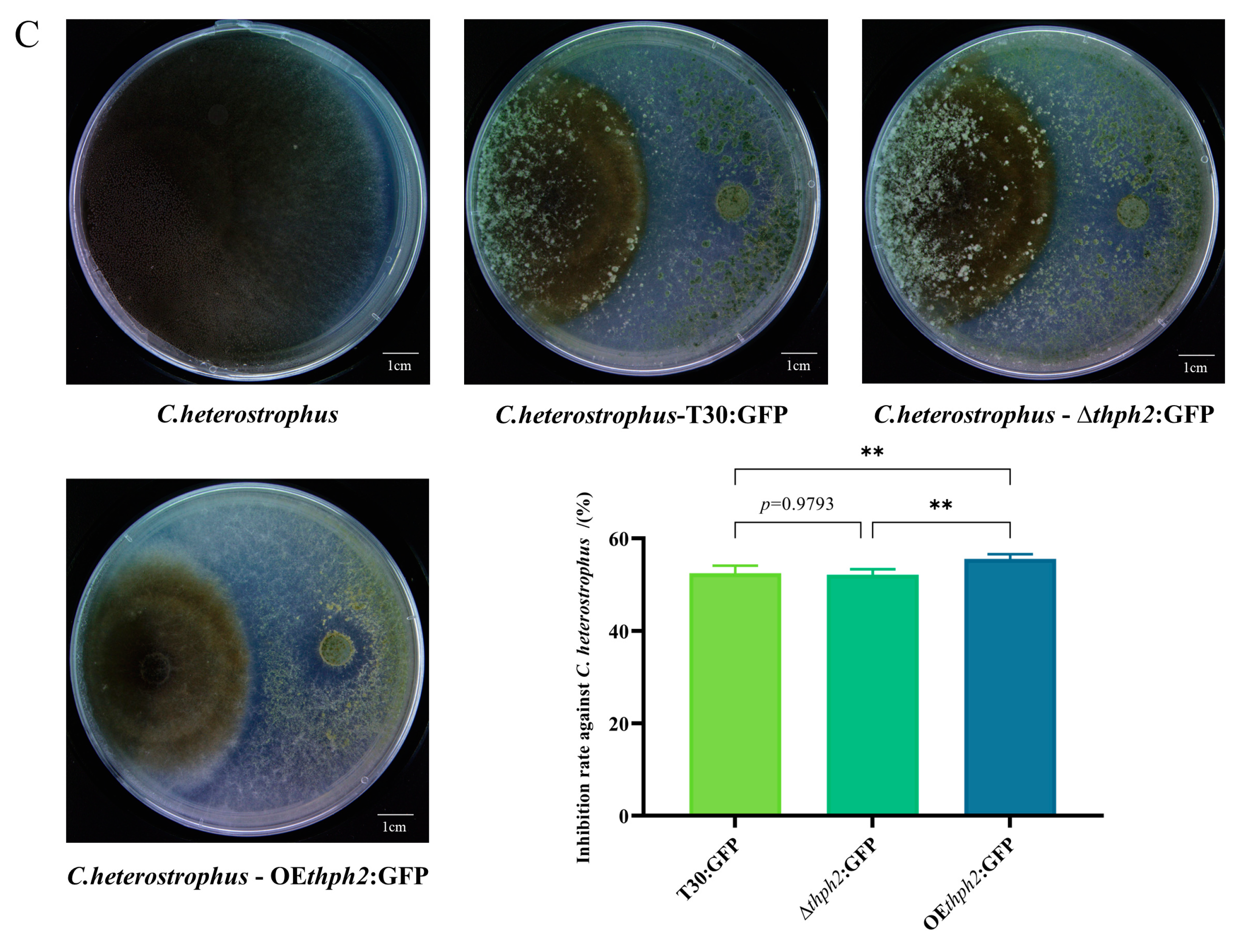
3.4. Evaluation of Trichoderma Strain Antagonism
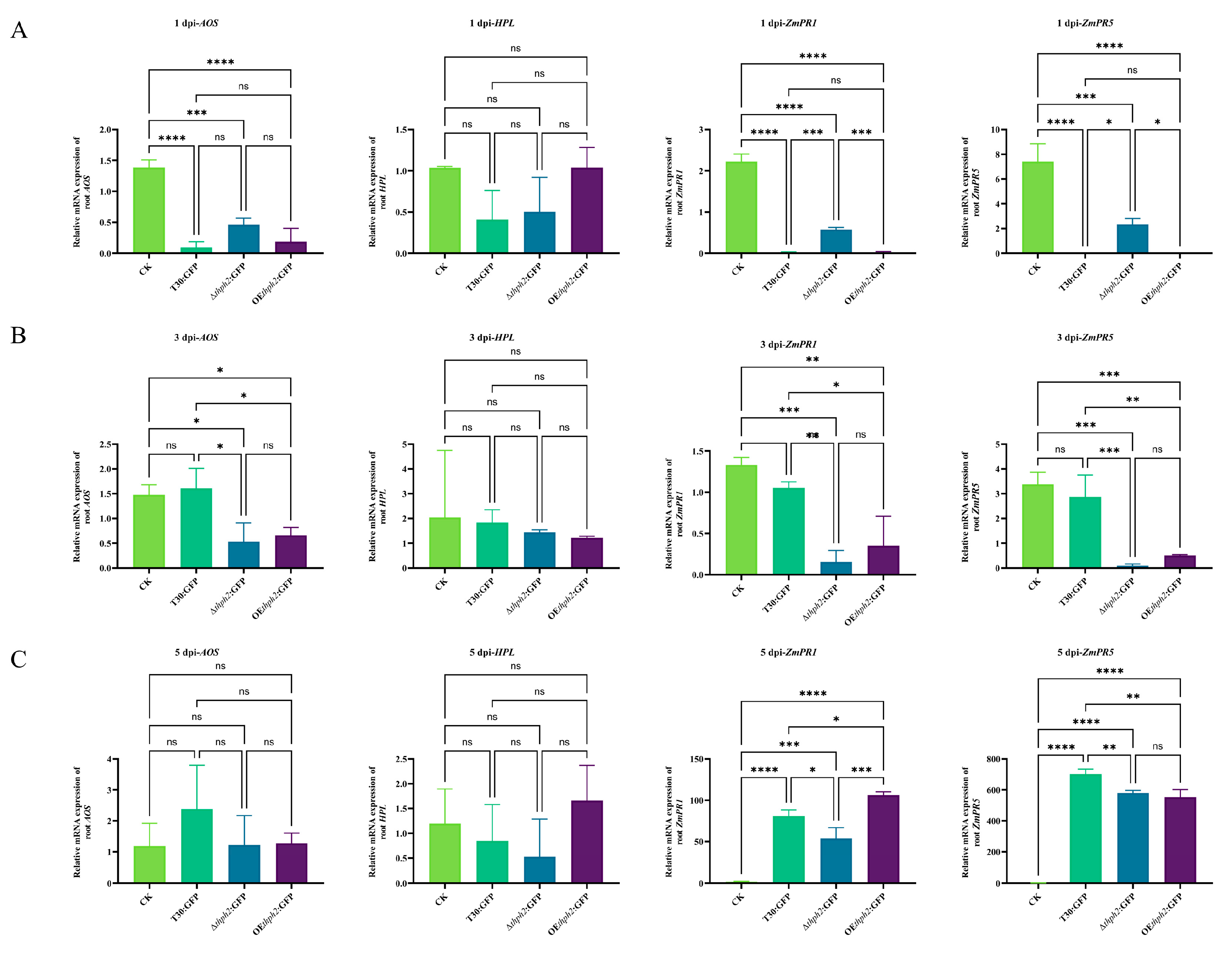
3.5. The Analysis of the Effects of thph2 on Trcihoderma Root Colonization and Host Defense Response Induction
3.6. Thph2-Induced Resistance against C. heterostrophus in Maize, Mainly through ISR
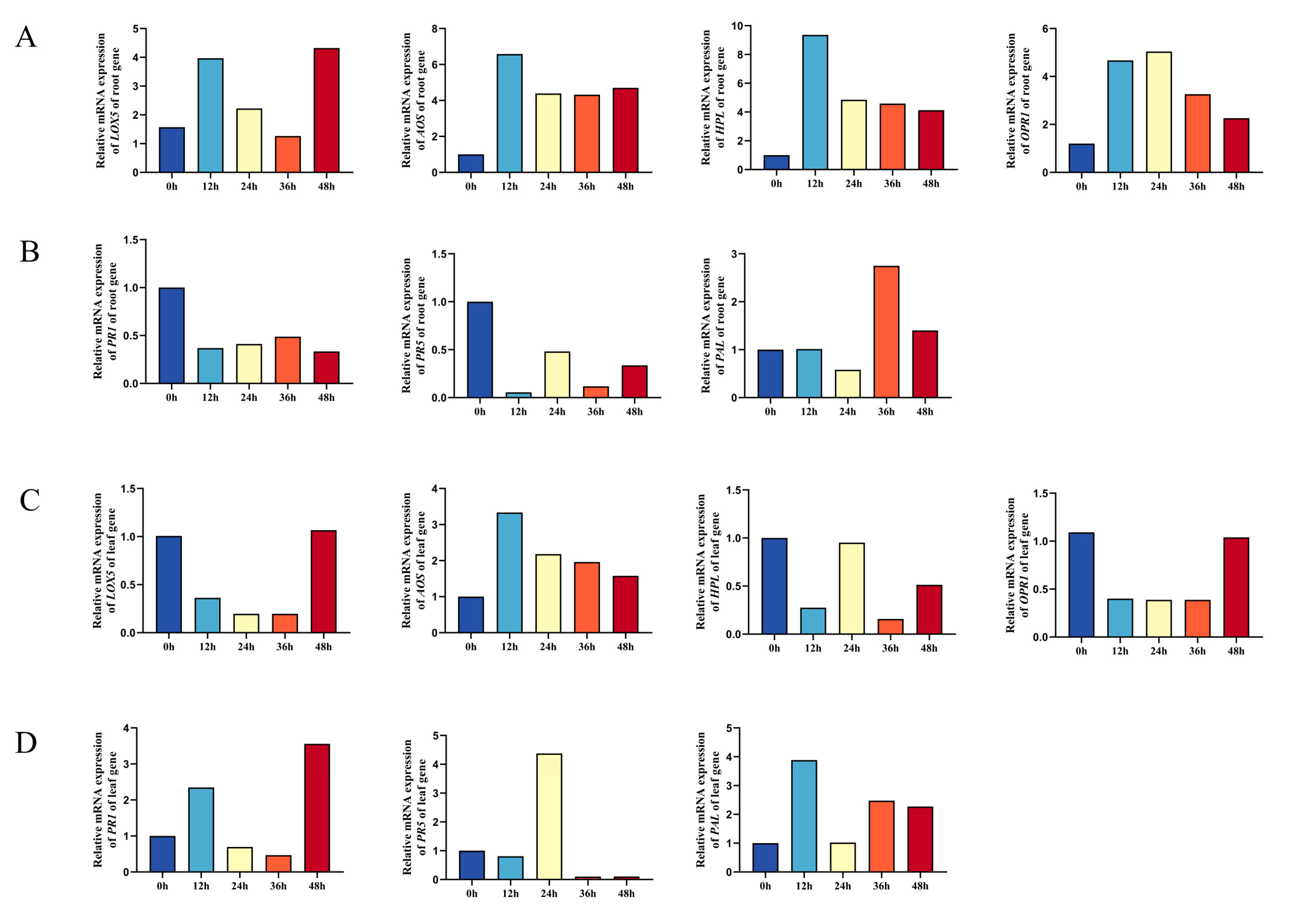
3.7. Thph2 Protein Induces Defense Response Gene Expression in Maize
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma Species-Opportunistic, Avirulent Plant Symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- Hermosa, R.; Viterbo, A.; Chet, I.; Monte, E. Plant-Beneficial Effects of Trichoderma and of Its Genes. Microbiology 2012, 158, 17–25. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Fan, L.; Fu, K.; Yu, C.; Wang, M.; Xia, H.; Sun, J.; Li, Y.; Chen, J. Cellulase from Trichoderma harzianum Interacts with Roots and Triggers Induced Systemic Resistance to Foliar Disease in Maize. Sci. Rep. 2016, 6, 35543. [Google Scholar] [CrossRef]
- Nawrocka, J.; Małolepsza, U. Diversity in Plant Systemic Resistance Induced by Trichoderma. Biol. Control 2013, 67, 149–156. [Google Scholar] [CrossRef]
- Fernández, E.; Segarra, G.; Trillas, M.I. Physiological Effects of the Induction of Resistance by Compost or Trichoderma asperellum Strain T34 against Botrytis cinerea in Tomato. Biol. Control 2014, 78, 77–85. [Google Scholar] [CrossRef]
- Chen, J. Recent progress in studies on disease resistance induced by Trichoderma. Chin. J. Biol. Control 2015, 31, 733–741. [Google Scholar] [CrossRef]
- Salas-Marina, M.A.; Isordia-Jasso, M.I.; Islas-Osuna, M.A.; Delgado-Sánchez, P.; Jiménez-Bremont, J.F.; RodrÃguez-Kessler, M.; Rosales-Saavedra, M.T.; Herrera-Estrella, A.; Casas-Flores, S. The Epl1 and Sm1 Proteins from Trichoderma atroviride and Trichoderma virens Differentially Modulate Systemic Disease Resistance against Different Life Style Pathogens in Solanum Lycopersicum. Front. Plant Sci. 2015, 6, 77. [Google Scholar] [CrossRef]
- Djonović, S.; Vargas, W.A.; Kolomiets, M.V.; Horndeski, M.; Wiest, A.; Kenerley, C.M. A Proteinaceous Elicitor Sm1 from the Beneficial Fungus Trichoderma virens Is Required for Induced Systemic Resistance in Maize. Plant Physiol. 2007, 145, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Gaderer, R.; Lamdan, N.L.; Frischmann, A.; Sulyok, M.; Krska, R.; Horwitz, B.A.; Seidl-Seiboth, V. Sm2, a Paralog of the Trichoderma Cerato-Platanin Elicitor Sm1, Is Also Highly Important for Plant Protection Conferred by the Fungal-Root Interaction of Trichoderma with Maize. BMC Microbiol. 2015, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Morán-Diez, E.; Hermosa, R.; Ambrosino, P.; Cardoza, R.E.; Gutiérrez, S.; Lorito, M.; Monte, E. The ThPG1 Endopolygalacturonase Is Required for the Trichoderma harzianum-Plant Beneficial Interaction. MPMI 2009, 22, 1021–1031. [Google Scholar] [CrossRef]
- Bhat, M.K. Cellulases and Related Enzymes in Biotechnology. Biotechnol. Adv. 2000, 18, 355–383. [Google Scholar] [CrossRef]
- Boisset, C.; Fraschini, C.; Schülein, M.; Henrissat, B.; Chanzy, H. Imaging the Enzymatic Digestion of Bacterial Cellulose Ribbons Reveals the Endo Character of the Cellobiohydrolase Cel6A from Humicola Insolens and Its Mode of Synergy with Cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 2000, 66, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Tsukagoshi, N.; Udaka, S. Nucleotide Sequence of the Cellobiohydrolase Gene from Trichoderma viride. Nucl. Acids Res. 1990, 18, 5559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, B.; Xia, L. High Efficient Expression of Cellobiase Gene from Aspergillus Niger in the Cells of Trichoderma reesei. Bioresour. Technol. 2011, 102, 4568–4572. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-Microbe Interactions: Shaping the Evolution of the Plant Immune Response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Katiyar-Agarwal, S.; Jin, H. Role of Small RNAs in Host-Microbe Interactions. Annu. Rev. Phytopathol. 2010, 48, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Rubartelli, A.; Lotze, M.T. Inside, Outside, Upside down: Damage-Associated Molecular-Pattern Molecules (DAMPs) and Redox. Trends. Immunol. 2007, 28, 429–436. [Google Scholar] [CrossRef]
- Lorito, M.; Woo, S.L.; Harman, G.E.; Monte, E. Translational Research On Trichoderma: From ’Omics to the Field. Annu. Rev. Phytopathol. 2010, 48, 395–417. [Google Scholar] [CrossRef]
- Wu, S.; Shan, L.; He, P. Microbial Signature-Triggered Plant Defense Responses and Early Signaling Mechanisms. Plant. Sci. 2014, 228, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U. Systemic Acquired Resistance. Plant Signal. Behav. 2006, 1, 179–184. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Ton, J.; Van Pelt, J.A.; Van Loon, L.C.; Pieterse, C.M.J. Differential Effectiveness of Salicylate-Dependent and Jasmonate/Ethylene-Dependent Induced Resistance in Arabidopsis. MPMI 2002, 15, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Nogueira-Lopez, G.; Greenwood, D.R.; Middleditch, M.; Winefield, C.; Eaton, C.; Steyaert, J.M.; Mendoza-Mendoza, A. The Apoplastic Secretome of Trichoderma virens during Interaction with Maize Roots Shows an Inhibition of Plant Defence and Scavenging Oxidative Stress Secreted Proteins. Front. Plant Sci. 2018, 9, 409. [Google Scholar] [CrossRef]
- Karuppiah, V.; Sun, J.; Li, T.; Vallikkannu, M.; Chen, J. Co-Cultivation of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841 Causes Differential Gene Expression and Improvement in the Wheat Growth and Biocontrol Activity. Front. Microbiol. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; del-Val, E.; Larsen, J. Ecological Functions of Trichoderma spp. and Their Secondary Metabolites in the Rhizosphere: Interactions with Plants. FEMS Microbiol. Ecol. 2016, 92, fiw036. [Google Scholar] [CrossRef] [PubMed]
- Fu, K.; Fan, L.; Li, Y.; Gao, S.; Chen, J. Tmac1, a Transcription Factor Which Regulated High Affinity Copper Transport in Trichoderma reesei. Microbiol. Res. 2012, 167, 536–543. [Google Scholar] [CrossRef]
- de Groot, M.J.; Bundock, P.; Hooykaas, P.J.; Beijersbergen, A.G. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Southern, E. Southern Blotting. Nat. Protoc. 2006, 1, 518–525. [Google Scholar] [CrossRef]
- Latif, S.; Gottschamel, J.; Syed, T.; Younus, I.; Gull, K.; Sameeullah, M.; Batool, N.; Lössl, A.G.; Mariz, F.; Müller, M.; et al. Inducible Expression of Human Papillomavirus-16 L1 Capsomeres in the Plastomes of Nicotiana Tabacum: Transplastomic Plants Develop Normal Flowers and Pollen. Biotechnol. Appl. Biochem. 2021, 69, 596–611. [Google Scholar] [CrossRef]
- Hao, D.; Lang, B.; Wang, Y.; Wang, X.; Liu, T.; Chen, J. Designing Synthetic Consortia of Trichoderma Strains That Improve Antagonistic Activities against Pathogens and Cucumber Seedling Growth. Microb. Cell Fact. 2022, 21, 234. [Google Scholar] [CrossRef]
- Lawry, R. Cross-Communication between Trichoderma and Plants during Root Colonisation. Ph.D. Thesis, Lincoln University, Philadelphia, PA, USA, 2016. [Google Scholar]
- Torres, A.M.; Weeden, N.F.; Martín, A. Linkage among Isozyme, RFLP and RAPD Markers in Vicia Faba. Theoret. Appl. Genet. 1993, 85, 937–945. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.M.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.-A.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting Ready for Battle. MPMI 2006, 19, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Feussner, I.; Wasternack, C. The Lipoxygenase Pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Fokar, M.; Abd, H.; Zhang, H.; Allen, R.D.; Par, P.W. (Z)-3-Hexenol Induces Defense Genes and Downstream Metabolites in Maize. Planta 2004, 220, 900–909. [Google Scholar] [CrossRef]
- Tang, S. Study on the Application of Trichoderma in Biological Control of Maize Diseases. Agric. Technol. Equip. 2022, 3, 91–92+95. [Google Scholar]
- Li, L.; Liu, B.; Yang, K.; Zhao, Z.; Chen, K.; Li, H.; Li, S. Control Effects of Trichoderma Strains against Wheat Powdery Mildew in Field. Shandong Agric. Sci. 2021, 53, 96–100. [Google Scholar] [CrossRef]
- Ma, G.; Liang, N.; Li, M.; Liu, M.; Chen, Y.; Lian, H. Effect of Trichoderma on Cucumber Damping-off and Physiological Characteristics. Chin. J. Biol. Control 2021, 37, 277–285. [Google Scholar] [CrossRef]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-Glucanases: Their Biological Functions and Transgenic Expression against Phytopathogenic Fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef] [PubMed]
- Grover, A. Plant Chitinases: Genetic Diversity and Physiological Roles. CRC. Crit. Rev. Plant. Sci. 2012, 31, 57–73. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Puri, N.; Sharma, P.; Gupta, N. Mannanases: Microbial Sources, Production, Properties and Potential Biotechnological Applications. Appl. Microbiol. Biotechnol. 2012, 93, 1817–1830. [Google Scholar] [CrossRef]
- Dhawan, S.; Kaur, J. Microbial Mannanases: An Overview of Production and Applications. Crit. Rev. Biotechnol. 2007, 27, 197–216. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.; Liu, F.; Liang, J.; Zhao, P.; Tsui, C.K.M.; Cai, L. Cross-Kingdom Synthetic Microbiota Supports Tomato Suppression of Fusarium Wilt Disease. Nat. Commun. 2022, 13, 7890. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Dou, K.; Wang, S.; Wu, Q.; Ni, M.; Zhang, T.; Lu, Z.; Tang, J.; Chen, J. Elicitor Hydrophobin Hyd1 Interacts with Ubiquilin1-like to Induce Maize Systemic Resistance. J. Integr. Plant Biol. 2019, 62, 509–526. [Google Scholar] [CrossRef]
- Matsui, K. Green Leaf Volatiles: Hydroperoxide Lyase Pathway of Oxylipin Metabolism. Curr. Opin. Plant. Biol. 2006, 9, 274–280. [Google Scholar] [CrossRef]
- Zhang, J.; Simmons, C.; Yalpani, N.; Crane, V.; Wilkinson, H.; Kolomiets, M. Genomic Analysis of the 12-Oxo-Phytodienoic Acid Reductase Gene Family of Zea Mays. Plant Mol. Biol. 2005, 59, 323–343. [Google Scholar] [CrossRef]
- Mei, C.; Qi, M.; Sheng, G.; Yang, Y. Inducible Overexpression of a Rice Allene Oxide Synthase Gene Increases the Endogenous Jasmonic Acid Level, PR Gene Expression, and Host Resistance to Fungal Infection. MPMI 2006, 19, 1127–1137. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lang, B.; Chen, J. Trichoderma harzianum Cellulase Gene thph2 Affects Trichoderma Root Colonization and Induces Resistance to Southern Leaf Blight in Maize. J. Fungi 2023, 9, 1168. https://doi.org/10.3390/jof9121168
Lang B, Chen J. Trichoderma harzianum Cellulase Gene thph2 Affects Trichoderma Root Colonization and Induces Resistance to Southern Leaf Blight in Maize. Journal of Fungi. 2023; 9(12):1168. https://doi.org/10.3390/jof9121168
Chicago/Turabian StyleLang, Bo, and Jie Chen. 2023. "Trichoderma harzianum Cellulase Gene thph2 Affects Trichoderma Root Colonization and Induces Resistance to Southern Leaf Blight in Maize" Journal of Fungi 9, no. 12: 1168. https://doi.org/10.3390/jof9121168
APA StyleLang, B., & Chen, J. (2023). Trichoderma harzianum Cellulase Gene thph2 Affects Trichoderma Root Colonization and Induces Resistance to Southern Leaf Blight in Maize. Journal of Fungi, 9(12), 1168. https://doi.org/10.3390/jof9121168





