How Different Molecular Markers Estimate the Diversity of European Species of the Ganoderma Genus
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jargalmalaa, S.; Eimes, J.A.; Park, M.S.; Oh, S.-Y.; Lim, Y.W. Taxonomic evaluation of selected Ganoderma species and database sequence validation. PeerJ 2017, 5, e3596. [Google Scholar] [CrossRef] [PubMed]
- Moncalvo, J.-M.; Ryvarden, L. A nomenclatural study of the Ganodermataceae Donk. Synop. Fungorum 1997, 11, 1–114. [Google Scholar]
- Ryvarden, L.; Melo, I. Poroid Fungi of Europe, 2nd ed.; Fungiflora: Oslo, Norway, 2017. [Google Scholar]
- Læessøe, T.; Petersen, J.H. Fungi of Temperate Europe; Princeton University Press: Princeton, NJ, USA, 2019; Volume 2. [Google Scholar]
- Papp, V. Global diversity of the genus Ganoderma: Taxonomic uncertainties and challenges. In Advances in Macrofungi: Diversity, Ecology and Biotechnology; Sridhar, K.R., Deshmukh, S.K., Eds.; CRC Press Taylor & Francis Group: London, UK, 2019; pp. 10–30. [Google Scholar]
- Rivoire, B. Polypores de France et d’Europe; Mycopolydev: Orliénas, France, 2020. [Google Scholar]
- Bernicchia, A.; Gorjón, S.P. Polypores of the Mediterranean Region; Romar: Segrate, Italy, 2020. [Google Scholar]
- Yang, H.-D.; Ding, Y.; Wen, T.C.; Hapuarachichi, K.K.; Wei, D.-P. Ganoderma ovisporum sp. nov. (Polyporales, Polyporaceae) from Southwest China. Biodivers. Data J. 2022, 10, e80034. [Google Scholar] [CrossRef]
- Moncalvo, J.M.; Wang, H.H.; Hseu, R.S. Gene phylogeny of the Ganoderma lucidum complex based on ribosomal DNA sequences: Comparison with traditional taxonomic characters. Mycol. Res. 1995, 99, 1489–1499. [Google Scholar] [CrossRef]
- Moncalvo, J.M.; Buchanan, P.K. Molecular evidence for long distance dispersal across the Southern Hemisphere in the Ganoderma applanatum–australe species complex (Basidiomycota). Mycol. Res. 2008, 112, 425–436. [Google Scholar] [CrossRef]
- Guglielmo, F.; Gonthier, P.; Garbelotto, M.; Nicolotti, G. Optimization of sampling procedures for DNA-based diagnosis of wood decay fungi in standing trees. Lett. Appl. Microbiol. 2010, 51, 90–97. [Google Scholar] [CrossRef]
- Beck, T.; Gáperová, S.; Gáper, J.; Náplavová, K.; Šebesta, M.; Kisková, J.; Pristaš, P. Genetic (non)-homogeneity of the bracket fungi of the genus Ganoderma (Basidiomycota) in Central Europe. Mycosphere 2020, 11, 225–238. [Google Scholar] [CrossRef]
- Fryssouli, V.; Zervakis, G.I.; Polemis, E.; Typas, M.A. A global meta-analysis of ITS rDNA sequences from material belonging to the genus Ganoderma (Basidiomycota, Polyporales) including new data from selected taxa. MycoKeys 2020, 75, 71–143. [Google Scholar] [CrossRef]
- He, M.Q.; Zhao, R.L.; Liu, D.M.; Denchev, T.; Begerow, D.; Yurkov, A.; Kemler, M.; Millanes, A.; Wedin, M.; McTaggart, A.R.; et al. Species diversity of Basidiomycota. Fungal Divers. 2022, 114, 281–325. [Google Scholar] [CrossRef]
- Papp, V.; Barina, Z.; Finy, P.; Dima, B.; Redhead, S.A. Nomenclature of the Beeswax bracket (Ganoderma pfeifferi), a European wood decay fungus with medicinal properties. Taxon 2022, 71, 1299–1304. [Google Scholar] [CrossRef]
- Catarbia, M.; Girometta, C.E.; Baiguera, R.M.; Buratti, S.; Babbini, S.; Bernicchia, A.; Savino, E. Lignicolous fungi collected in Northern Italy: Identification and morphological description of isolates. Diversity 2022, 14, 413. [Google Scholar]
- Kotlaba, F.; Pouzar, Z. Ecology of the Lacquered bracket—Ganoderma resinaceum—and its distribution in Bohemia. Mykol. Listy. 2009, 107, 14–19. (In Czech) [Google Scholar]
- Naplavova, K.; Beck, T.; Pristas, P.; Gaperova, S.; Sebesta, M.; Piknova, M.; Gaper, J. Molecular data reveal unrecognized diversity in the European Ganoderma resinaceum. Forests 2020, 11, 850. [Google Scholar] [CrossRef]
- Wasser, S.P.; Zmitrovich, I.V.; Diduch, M.Y.; Spirin, W.A.; Malysheva, V.F. Morphological Traits of Ganoderma lucidum Complex. Highlighting G. tsugae var. jannieae: The Current Generalization; A.R.A. Gantner Verlag: Rugell, Lichtenstein, 2006. [Google Scholar]
- Krisai-Greilhuber, I.; Flechtmann, S.; Friebes, G.; Koller, G.; Kresitschnig, P.; Stoik, O. Notable fungal species from Austria. Osterr. Z. Für Pilzkd. 2017, 26, 269–281. (In German) [Google Scholar]
- Vlasák, J.P. Collection of Dr. Josef Vlasák, Hluboká nad Vltavou, Czech Republic, Edition 18. II. 2015. Available online: http://mykoweb.prf.jcu.cz/polypores/ (accessed on 10 July 2023).
- Zhou, L.-W.; Cao, Y.; Wu, S.-H.; Vlasák, J.; Li, D.-W.; Li, M.-J.; Dai, Y.-C. Global diversity of the Ganoderma lucidum complex (Ganodermataceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry 2015, 114, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, S.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Aoki, T.; Ariyawansa, H.A.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, S.M.; et al. Fungal taxonomy and sequence-based nomenclature. Nat. Microbiol. 2021, 6, 540–548. [Google Scholar] [CrossRef]
- Tekpinar, A.D.; Kalmer, A. Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwig. 2019, 109, 187–224. [Google Scholar] [CrossRef]
- Zhang, N.; Luo, J.; Bhattacharya, D. Advances in Fungal Phylogenomics and Their Impact on Fungal Systematics. Adv. Genet. 2017, 100, 309–328. [Google Scholar]
- Cooper, J.; Kirk, P. CABI Bioscience Database, Landscape Research, Index Fungorum Database. Available online: http://www.speciesfungorum.org/Names/Names.asp (accessed on 12 July 2023).
- IPNI—The International Plant Names Index. Available online: http://www.ipni.org (accessed on 12 July 2023).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kotlaba, F.; Pouzar, Z. Ecology of the polypore Ganoderma adspersum in Bohemia. Mykol. Listy. 2009, 109, 11–15. (In Czech) [Google Scholar]
- Pristas, P.; Gaperova, S.; Gaper, J.; Judova, J. Genetic variability in Fomes fomentarius reconfirmed by translation elongation factor 1-α DNA sequences and 25S LSU rRNA sequences. Biologia 2013, 68, 816–820. [Google Scholar] [CrossRef]
- Korhonen, A.; Seelan, J.S.S.; Miettinen, O. Cryptic species diversity in polypores: The Skeletocutis nivea species complex. MycoKeys 2018, 36, 45–82. [Google Scholar] [CrossRef]
- Vasaitis, R.; Menkis, A.; Lim, Y.W.; Seok, S.; Tomsovsky, M.; Jankovsky, L.; Lygis, V.; Slippers, B.; Stenlid, J. Genetic variation and relationships in Laetiporus sulphureus s. lat., as determined by ITS rDNA sequences and in vitro growth rate. Mycol. Res. 2009, 113, 326–336. [Google Scholar] [CrossRef]
- Tomsovsky, M.; Vampola, P.; Sedlak, P.; Byrtusova, Z.; Jankovsky, L. Delimitation of central and northern European species of the Phellinus igniarius group (Basidiomycota, Hymenochaetales) based on analysis of ITS and translation elongation factor 1 alpha DNA sequences. Mycol. Prog. 2010, 9, 431–445. [Google Scholar] [CrossRef]
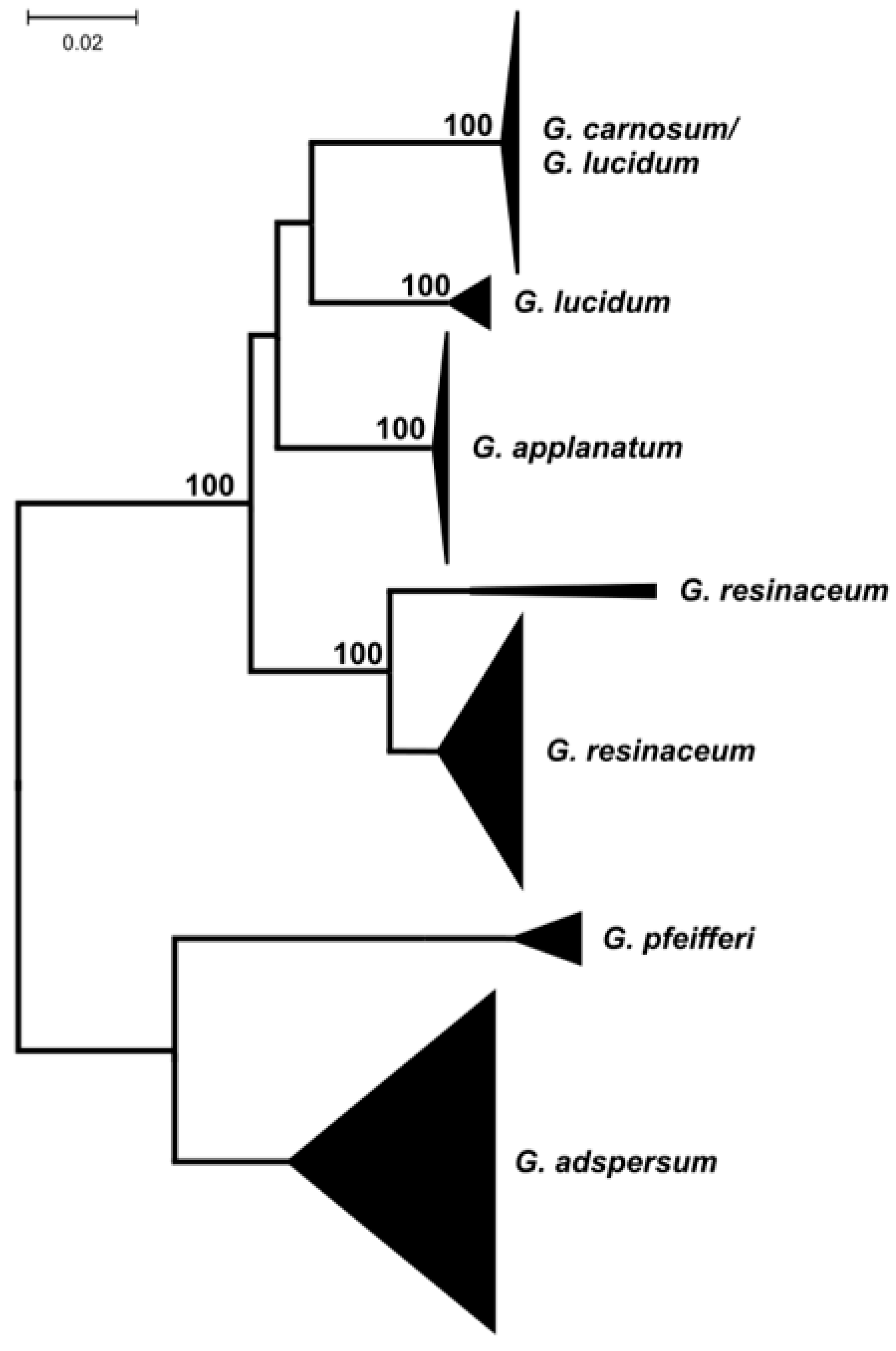
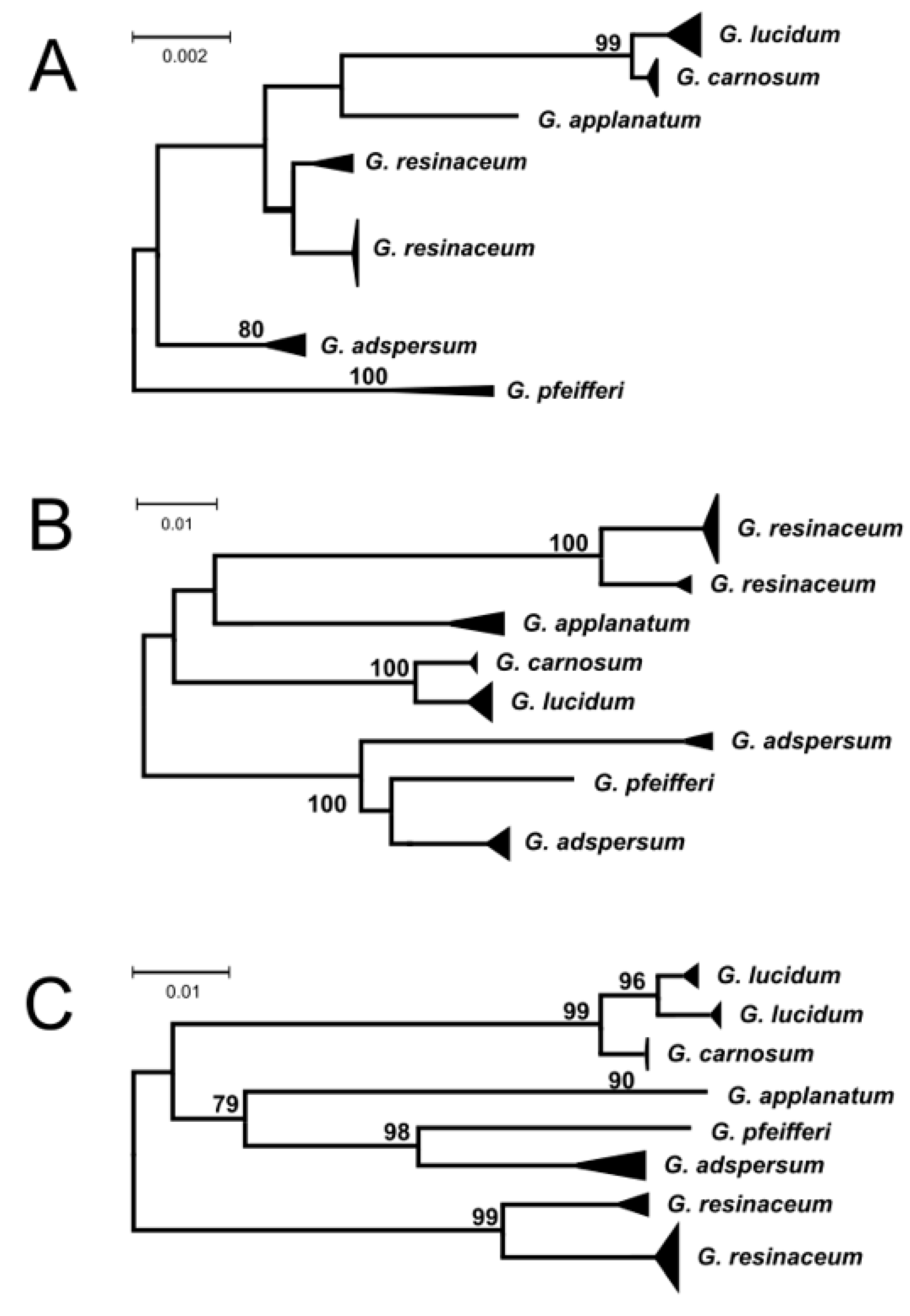
| Species | Marker | |||||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | tef1-α | Rpb2 | |||||
| No. Sequences | Diversity | No. Sequences | Diversity | No. Sequences | Diversity | No. Sequences | Diversity | |
| G. adspersum | 85 | 0.012 | 7 | 0.000 | 9 | 0.000 | 16 | 0.033 |
| G. applanatum | 57 | 0.000 | 1 | na | 1 | na | 7 | 0.009 |
| G. carnosum | 19 | 0.006 | 12 | 0.000 | 11 | 0.000 | 5 | 0.001 |
| G. lucidum | 65 | 0.025 | 13 | 0.000 | 13 | 0.006 | 12 | 0.003 |
| G. pfeifferi | 13 | 0.002 | 3 | 0.002 | 1 | na | 1 | na |
| G. resinaceum | 70 | 0.008 | 26 | 0.000 | 29 | 0.013 | 27 | 0.010 |
| average | 0.009 | 0.000 | 0.005 | 0.011 | ||||
| Species | Marker | G. adspersum | G. applanatum | G. carnosum | G. lucidum | G. pfeifferi | G. resinaceum |
|---|---|---|---|---|---|---|---|
| G. adspersum | ITS | - | |||||
| LSU | - | ||||||
| tef1-α | - | ||||||
| Rpb2 | - | marker | average | ||||
| G. applanatum | ITS | 0.194 | - | ITS | 0.154 | ||
| LSU | 0.009 | - | LSU | 0.010 | |||
| tef1-α | 0.093 | - | tef1-α | 0.099 | |||
| Rpb2 | 0.092 | - | Rpb2 | 0.067 | |||
| G. carnosum | ITS | 0.201 | 0.083 | - | |||
| LSU | 0.014 | 0.010 | - | ||||
| tef1-α | 0.106 | 0.108 | - | ||||
| Rpb2 | 0.102 | 0.091 | - | ||||
| G. lucidum | ITS | 0.199 | 0.076 | 0.022 | - | ||
| LSU | 0.015 | 0.011 | 0.002 | - | |||
| tef1-α | 0.112 | 0.109 | 0.016 | - | |||
| Rpb2 | 0.106 | 0.086 | 0.016 | - | |||
| G. pfeifferi | ITS | 0.181 | 0.194 | 0.204 | 0.204 | - | |
| LSU | 0.011 | 0.018 | 0.016 | 0.017 | - | ||
| tef1-α | 0.054 | 0.091 | 0.108 | 0.110 | - | ||
| Rpb2 | 0.055 | 0.088 | 0.096 | 0.104 | - | ||
| G. resinaceum | ITS | 0.224 | 0.099 | 0.110 | 0.105 | 0.217 | - |
| LSU | 0.006 | 0.007 | 0.010 | 0.010 | 0.012 | - | |
| tef1-α | 0.115 | 0.114 | 0.110 | 0.119 | 0.119 | - | |
| Rpb2 | 0.128 | 0.107 | 0.106 | 0.109 | 0.126 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pristas, P.; Beck, T.; Nosalova, L.; Gaperova, S.; Gaper, J. How Different Molecular Markers Estimate the Diversity of European Species of the Ganoderma Genus. J. Fungi 2023, 9, 1023. https://doi.org/10.3390/jof9101023
Pristas P, Beck T, Nosalova L, Gaperova S, Gaper J. How Different Molecular Markers Estimate the Diversity of European Species of the Ganoderma Genus. Journal of Fungi. 2023; 9(10):1023. https://doi.org/10.3390/jof9101023
Chicago/Turabian StylePristas, Peter, Terezia Beck, Lea Nosalova, Svetlana Gaperova, and Jan Gaper. 2023. "How Different Molecular Markers Estimate the Diversity of European Species of the Ganoderma Genus" Journal of Fungi 9, no. 10: 1023. https://doi.org/10.3390/jof9101023
APA StylePristas, P., Beck, T., Nosalova, L., Gaperova, S., & Gaper, J. (2023). How Different Molecular Markers Estimate the Diversity of European Species of the Ganoderma Genus. Journal of Fungi, 9(10), 1023. https://doi.org/10.3390/jof9101023






