A Mad7 System for Genetic Engineering of Filamentous Fungi
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. PCR and Assembly of Plasmids by USER Cloning
2.3. Transformation and Strain Validation by Diagnostic PCR
2.4. Two-Phase Protocol
2.5. Fluorescence Photography
3. Results
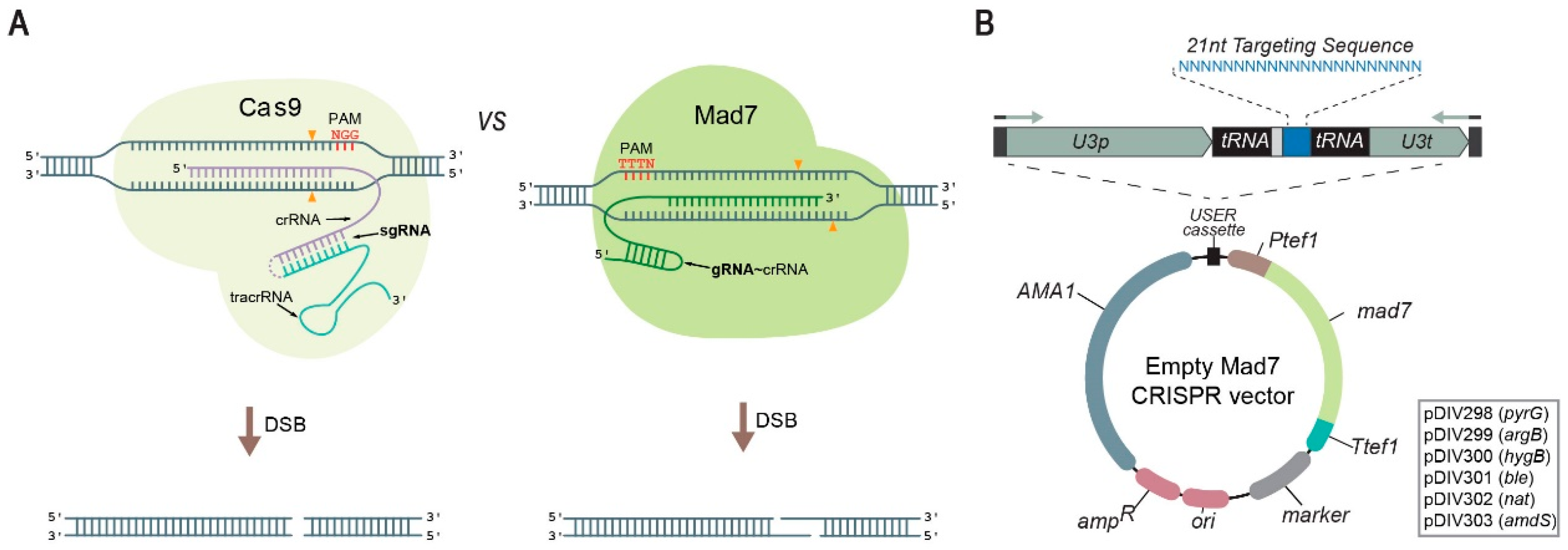
3.1. A Versatile Mad7 System to Facilitate CRISPR-Based Gene Editing
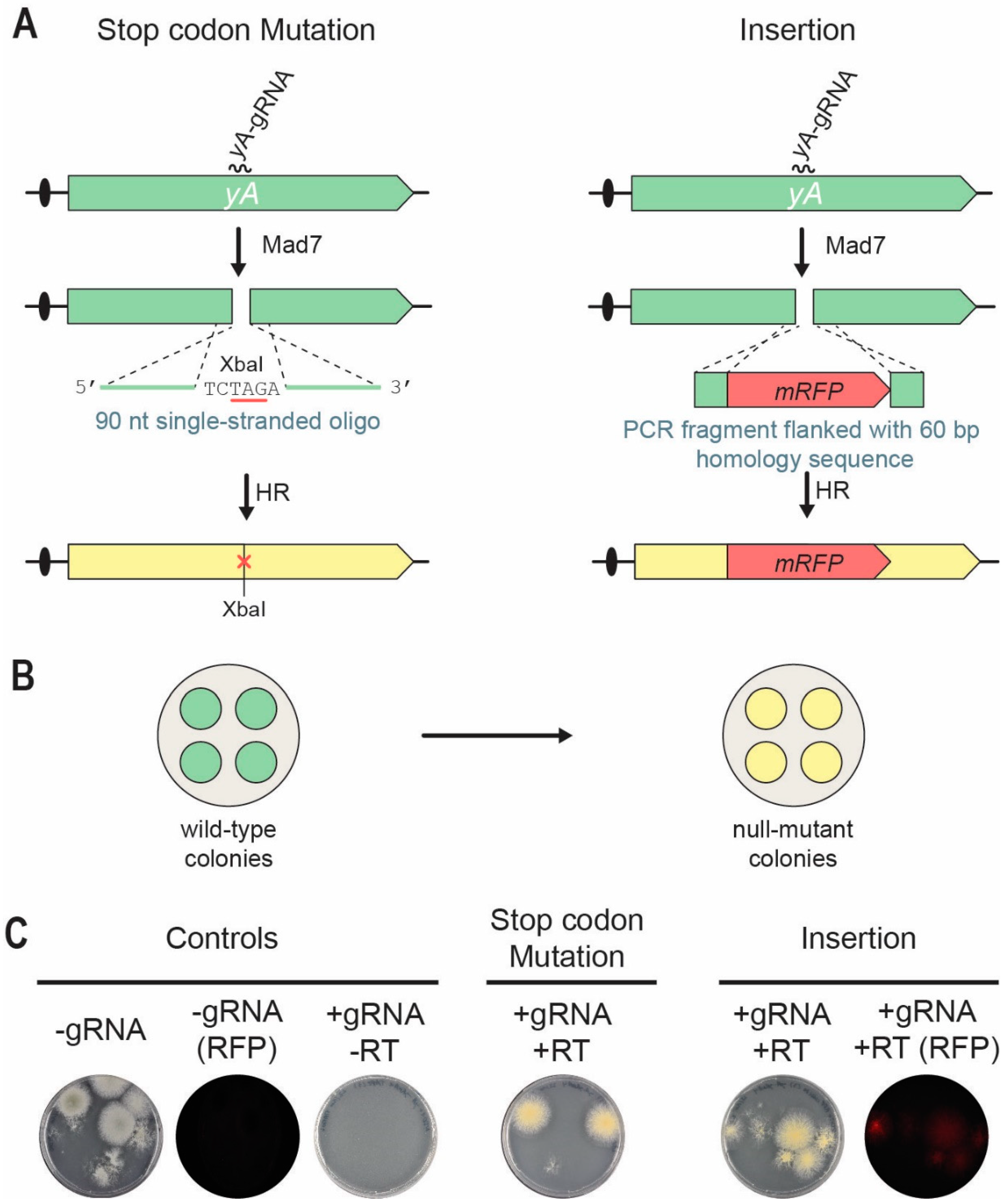
3.2. Mad7 Efficiently Mediates Template-Directed Mutagenesis and Gene Disruption in A. nidulans
3.3. Mad7 Can Be Used for Marker-Free Gene Deletion in NHEJ-Proficient A. oryzae and A. campestris
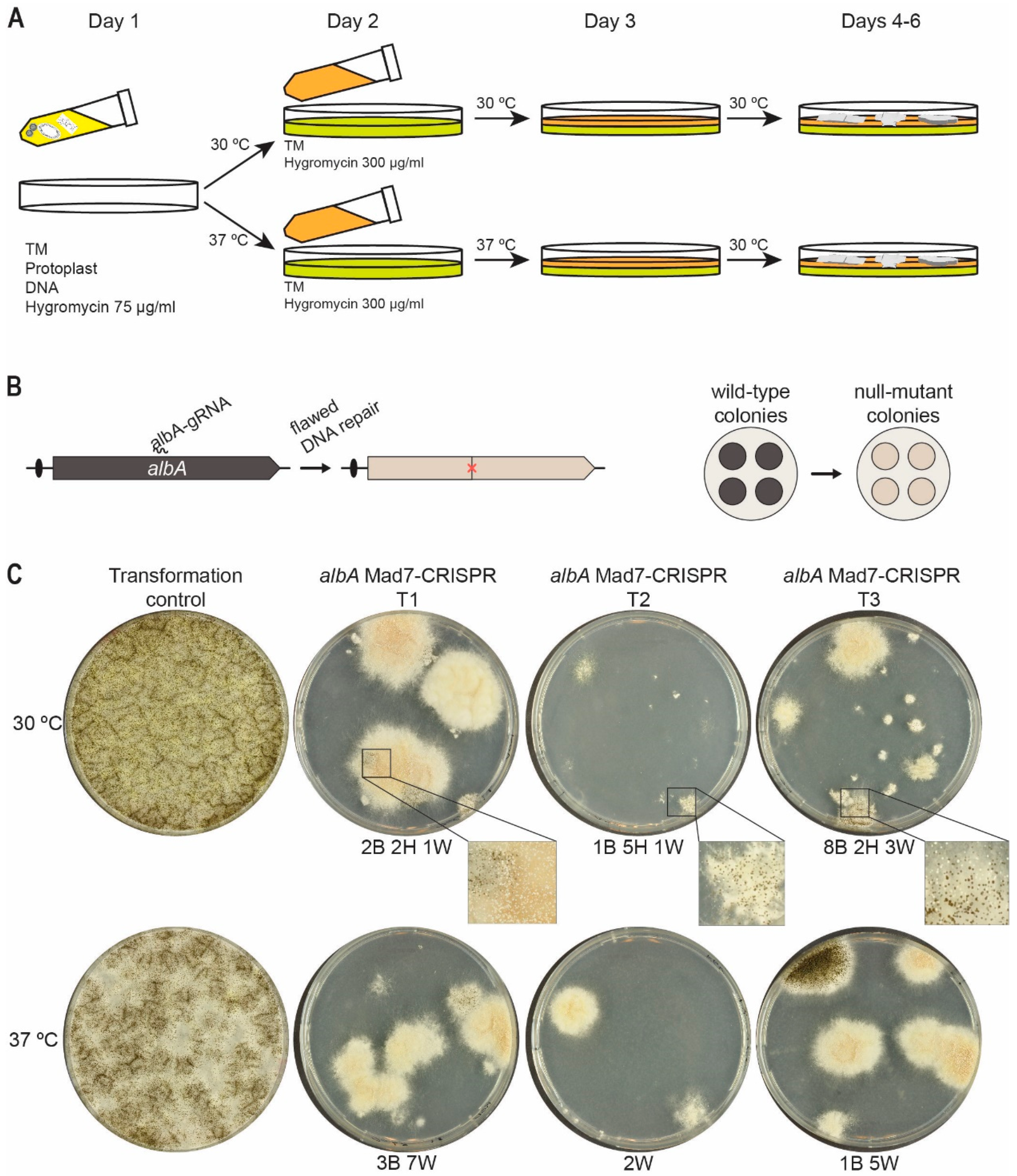
3.4. Site-Directed Mad7-Induced Gene Mutation in A. niger
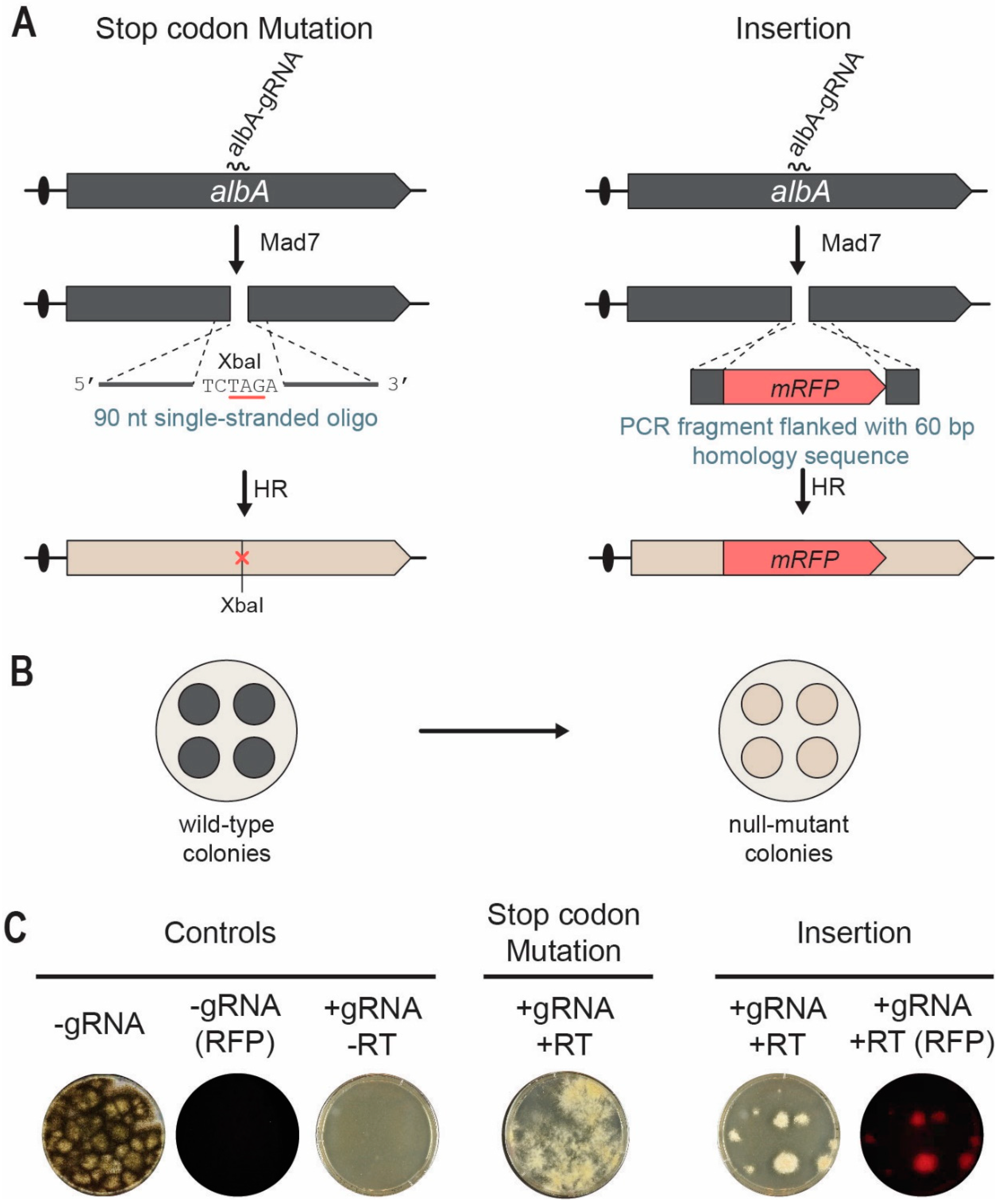
3.5. Mad7 Efficiently Mediates Template-Directed Mutagenesis and Gene Disruption in A. niger
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meyer, V.; Basenko, E.Y.; Benz, J.P.; Braus, G.H.; Caddick, M.X.; Csukai, M.; De Vries, R.P.; Endy, D.; Frisvad, J.C.; Gunde-Cimerman, N.; et al. Growing a circular economy with fungal biotechnology: A white paper. Fungal Biol. Biotechnol. 2020, 7, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Torres, F.A.E.; Zaccarim, B.R.; de Lencastre Novaes, L.C.; Jozala, A.F.; Santos, C.A.; Teixeira, M.F.S.; Ebinuma, V.C.S. Natural colorants from filamentous fungi. Appl. Microbiol. Biotechnol. 2016, 100, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Berka, R.M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S.E.; Chapman, J.; Chertkov, O.; Coutinho, P.M.; Cullen, D.; et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 2008, 26, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Vesth, T.C.; Nybo, J.L.; Theobald, S.; Frisvad, J.C.; Larsen, T.O.; Nielsen, K.F.; Hoof, J.B.; Brandl, J.; Salamov, A.; Riley, R.; et al. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat. Genet. 2018, 50, 1688–1695. [Google Scholar] [CrossRef]
- Choque, E.; Klopp, C.; Valiere, S.; Raynal, J.; Mathieu, F. Whole-genome sequencing of Aspergillus tubingensis G131 and overview of its secondary metabolism potential. BMC Genom. 2018, 19, 200. [Google Scholar] [CrossRef]
- Schuster, M.; Kahmann, R. CRISPR-Cas9 Genome Editing Approaches in Filamentous Fungi and Oomycetes. Fungal Genet. Biol. 2019, 130, 43–53. [Google Scholar] [CrossRef]
- Ouedraogo, J.P.; Tsang, A. CRISPR_Cas Systems for fungal research. Fungal Biol. Rev. 2020, 34, 189–201. [Google Scholar] [CrossRef]
- Leisen, T.; Werner, J.; Pattar, P.; Safari, N.; Ymeri, E.; Sommer, F.; Schroda, M.; Suárez, I.; Collado, I.G.; Scheuring, D.; et al. Multiple knockout mutants reveal a high redundancy of phytotoxic compounds contributing to necrotrophic pathogenesis of Botrytis cinerea. PLOS Pathog. 2022, 18, e1010367. [Google Scholar] [CrossRef]
- Huang, J.; Rowe, D.; Subedi, P.; Zhang, W.; Suelter, T.; Valent, B.; Cook, D.E. CRISPR-Cas12a induced DNA double-strand breaks are repaired by multiple pathways with different mutation profiles in Magnaporthe oryzae. Nat. Commun. 2022, 13, 7168. [Google Scholar] [CrossRef]
- Wang, Q.; Coleman, J.J. Progress and challenges: Development and implementation of CRISPR/Cas9 technology in filamentous fungi. Comput. Struct. Biotechnol. J. 2019, 17, 761–769. [Google Scholar] [CrossRef]
- Xue, C.; Greene, E.C. DNA Repair pathway choices in CRISPR-Cas9-mediated genome editing. Trends Genet. 2021, 37, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, T.S.; Baudrier, L.; Billon, P.; Ciccia, A. CRISPR-based genome editing through the lens of DNA repair. Mol. Cell 2022, 82, 348–388. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Cook, D.E. The contribution of DNA repair pathways to genome editing and evolution in filamentous pathogens. FEMS Microbiol. Rev. 2022, 1–21, fuac035. [Google Scholar] [CrossRef] [PubMed]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Nødvig, C.S.; Hoof, J.B.; Kogle, M.E.; Jarczynska, Z.D.; Lehmbeck, J.; Klitgaard, D.K.; Mortensen, U.H. Efficient Oligo nucleotide mediated crispr-cas9 gene editing in Aspergilli. Fungal Genet. Biol. 2018, 115, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, Y.; Suzuki, K.; Ishii, C.; Inoue, H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 2004, 101, 12248–12253. [Google Scholar] [CrossRef]
- Matsu-ura, T.; Baek, M.; Kwon, J.; Hong, C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2015, 2, 1–7. [Google Scholar] [CrossRef]
- Pohl, C.; Kiel, J.A.; Driessen, A.J.; Bovenberg, R.A.; Nygård, Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 2016, 5, 754–764. [Google Scholar] [CrossRef]
- Rojek, J.; Basavaraju, Y.; Nallapareddy, S.; Baumgartner, R.; Schoffelen, S.; Pedersen, L. Mad7: An IP friendly CRISPR enzyme. Authorea Prepr. 2021, 1–7. [Google Scholar]
- Liu, Z.; Schiel, J.A.; Maksimova, E.; Strezoska, Ž.; Zhao, G.; Anderson, E.M.; Wu, Y.; Warren, J.; Bartels, A.; Van Brabant Smith, A.; et al. ErCas12a CRISPR-MAD7 for model generation in human cells, mice, and rats. CRISPR J. 2020, 3, 97–108. [Google Scholar] [CrossRef]
- Jarczynska, Z.D.; Rendsvig, J.K.H.; Pagels, N.; Viana, V.R.; Ndvig, C.S.; Kirchner, F.H.; Strucko, T.; Nielsen, M.L.; Mortensen, U.H. DIVERSIFY: A fungal multispecies gene expression platform. ACS Synth. Biol. 2021, 10, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of dna molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Jarczynska, Z.D.; Garcia Vanegas, K.; Deichmann, M.; Nørskov Jensen, C.; Scheeper, M.J.; Futyma, M.E.; Strucko, T.; Jares Contesini, F.; Sparholt Jørgensen, T.; Blæsbjerg Hoof, J.; et al. A versatile in vivo dna assembly toolbox for fungal strain engineering. ACS Synth. Biol. 2022, 11, 3251–3263. [Google Scholar] [CrossRef] [PubMed]
- Jochumsen, N.; Muzzi-Erichsen, G. Genome Editing by Guided Endonuclease and Single-Stranded Oligonucleotide. Patent EP3874051A1, 2019. [Google Scholar]
- Horizon. Available online: https://horizondiscovery.com/en/gene-editing/gene-editing-reagents/mad7 (accessed on 24 November 2022).
- Nielsen, J.B.; Nielsen, M.L.; Mortensen, U.H. Transient disruption of non-homologous end-joining facilitates targeted genome manipulations in the filamentous fungus Aspergillus nidulans. Fungal Genet. Biol. 2008, 45, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1–3. [Google Scholar] [CrossRef]
- Varga, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Aspergillus Section Candidi based on molecular, morphological and physiological data. Stud. Mycol. 2007, 59, 75–88. [Google Scholar] [CrossRef]
- Nour-Eldin, H.H.; Geu-Flores, F.; Halkier, B.A. USER cloning and USER fusion: The ideal cloning techniques for small and big laboratories. In Plant Secondary Metabolism Engineering, Methods in Molecular Biology, 1st ed.; Fett-Neto, A.G., Ed.; Springer: Berlim, Germany, 2010; Volume 1, pp. 185–200. [Google Scholar]
- Vanegas, K.G.; Jarczynska, Z.D.; Strucko, T.; Mortensen, U.H. Cpf1 Enables fast and efficient genome editing in Aspergilli. Fungal Biol. Biotechnol. 2019, 6, 1–10. [Google Scholar] [CrossRef]
- Kitamoto, N.; Matsui, J.; Kawai, Y.; Kato, A.; Yoshino, S.; Ohmiya, K.; Tsukagoshi, N. Utilization of the TEF1-α gene (TEF1) promoter for expression of polygalacturonase genes, PgaA and PgaB, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 1998, 50, 85–92. [Google Scholar] [CrossRef]
- Tamano, K.; Bruno, K.S.; Karagiosis, S.A.; Culley, D.E.; Deng, S.; Collett, J.R.; Umemura, M.; Koike, H.; Baker, S.E.; Machida, M. Increased production of fatty acids and triglycerides in Aspergillus oryzae by enhancing expressions of fatty acid synthesis-related genes. Appl. Microbiol. Biotechnol. 2013, 97, 269–281. [Google Scholar] [CrossRef]
- Jöchl, C.; Rederstorff, M.; Hertel, J.; Stadler, P.F.; Hofacker, I.I.; Schrettl, M.; Haas, H.; Hüttenhofer, A. Small NcRNA Transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008, 36, 2677–2689. [Google Scholar] [CrossRef]
- Price, M.A.; Cruz, R.; Bryson, J.; Escalettes, F.; Rosser, S.J. Expanding and understanding the CRISPR toolbox for Bacillus subtilis with MAD7 and dMAD7. Biotechnol. Bioeng. 2020, 117, 1805–1816. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, E.B.; Timberlake, W.E. Molecular characterization of the Aspergillus nidulans yA locus. Genetics 1989, 121, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Yelton, M.M.; Timberlake, W.E.; Hondel, C.A. A cosmid for selecting genes by complementation in Aspergillus nidulans: Selection of the developmentally regulated yA locus. Proc. Natl. Acad. Sci. USA 1985, 82, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, K.G.; Lehka, B.J.; Mortensen, U.H. SWITCH: A dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae. Microb. Cell Factories 2017, 16, 1–12. [Google Scholar] [CrossRef]
- Hoof, J.B.; Nødvig, C.S.; Mortensen, U.H. Genome Editing: CRISPR-Cas9. In Fungal Genomics: Methods and Protocols, 2nd ed.; de Vries, R.P., Tsang, A., Grigoriev, I.V., Eds.; Springer: Berlim, Germany, 2018; Volume 1775, pp. 119–132. [Google Scholar] [CrossRef]
- Guo, Y.; Contesini, F.J.; Wang, X.; Ghidinelli, S.; Tornby, D.S.; Andersen, T.E.; Mortensen, U.H.; Larsen, T.O. Biosynthesis of calipyridone a represents a fungal 2-pyridone formation without ring expansion in Aspergillus californicus. Org. Lett. 2022, 24, 804–808. [Google Scholar] [CrossRef]
- Wierson, W.A.; Simone, B.W.; WareJoncas, Z.; Mann, C.; Welker, J.M.; Kar, B.; Emch, M.J.; Friedberg, I.; Gendron, W.A.C.; Barry, M.A.; et al. Expanding the CRISPR toolbox with ErCas12a in zebrafish and human cells. CRISPR J. 2019, 2, 417–433. [Google Scholar] [CrossRef]
- Lin, Q.; Zhu, Z.; Liu, G.; Sun, C.; Lin, D.; Xue, C.; Li, S.; Zhang, D.; Gao, C.; Wang, Y.; et al. Genome editing in plants with MAD7 nuclease. J. Genet. Genom. 2021, 48, 444–451. [Google Scholar] [CrossRef]
- Chiang, Y.M.; Meyer, K.M.; Praseuth, M.; Baker, S.E.; Bruno, K.S.; Wang, C.C. Characterization of a polyketide synthase in Aspergillus niger whose product is a precursor for both dihydroxynaphthalene (DHN) melanin and naphtho-γ-pyrone. Fungal Genet. Biol. 2011, 48, 430–437. [Google Scholar] [CrossRef]
- Rendsvig, J.K.H. Filamentous Fungi for Protein Production and Identification of Superior Cell Factories—Genetic Tools and Expression Systems. Ph.D. Thesis, Technical University of Denmark, Lyngby, Denmark, 2020. [Google Scholar]
- Smith, C.; Day, P.J.; Walker, M.R. Generation of cohesive ends on PCR products by UDG-mediated excision of dU, and application for cloning into restriction digest-linearized vectors. Genome Res. 1993, 2, 328–332. [Google Scholar] [CrossRef]
- Jiang, D.; Hatahet, Z.; Melamede, R.J.; Kow, Y.W.; Wallace, S.S. Characterization of Escherichia coli Endonuclease VIII. J. Biol. Chem. 1997, 272, 32230–32239. [Google Scholar] [CrossRef]
- Melamede, R.J.; Hatahet, Z.; Kow, Y.W.; Ide, H.; Wallace, S.S. Isolation and Characterization of Endonuclease VIII from Escherichia coli. Biochemistry 1994, 33, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanegas, K.G.; Rendsvig, J.K.H.; Jarczynska, Z.D.; Cortes, M.V.d.C.B.; van Esch, A.P.; Morera-Gómez, M.; Contesini, F.J.; Mortensen, U.H. A Mad7 System for Genetic Engineering of Filamentous Fungi. J. Fungi 2023, 9, 16. https://doi.org/10.3390/jof9010016
Vanegas KG, Rendsvig JKH, Jarczynska ZD, Cortes MVdCB, van Esch AP, Morera-Gómez M, Contesini FJ, Mortensen UH. A Mad7 System for Genetic Engineering of Filamentous Fungi. Journal of Fungi. 2023; 9(1):16. https://doi.org/10.3390/jof9010016
Chicago/Turabian StyleVanegas, Katherina Garcia, Jakob Kræmmer Haar Rendsvig, Zofia Dorota Jarczynska, Marcio Vinicius de Carvalho Barros Cortes, Abel Peter van Esch, Martí Morera-Gómez, Fabiano Jares Contesini, and Uffe Hasbro Mortensen. 2023. "A Mad7 System for Genetic Engineering of Filamentous Fungi" Journal of Fungi 9, no. 1: 16. https://doi.org/10.3390/jof9010016
APA StyleVanegas, K. G., Rendsvig, J. K. H., Jarczynska, Z. D., Cortes, M. V. d. C. B., van Esch, A. P., Morera-Gómez, M., Contesini, F. J., & Mortensen, U. H. (2023). A Mad7 System for Genetic Engineering of Filamentous Fungi. Journal of Fungi, 9(1), 16. https://doi.org/10.3390/jof9010016






