Biosynthesis of Fatty Acid Derivatives by Recombinant Yarrowia lipolytica Containing MsexD2 and MsexD3 Desaturase Genes from Manduca sexta
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media Composition, and Culture Conditions
2.2. Plasmid and Strain Construction
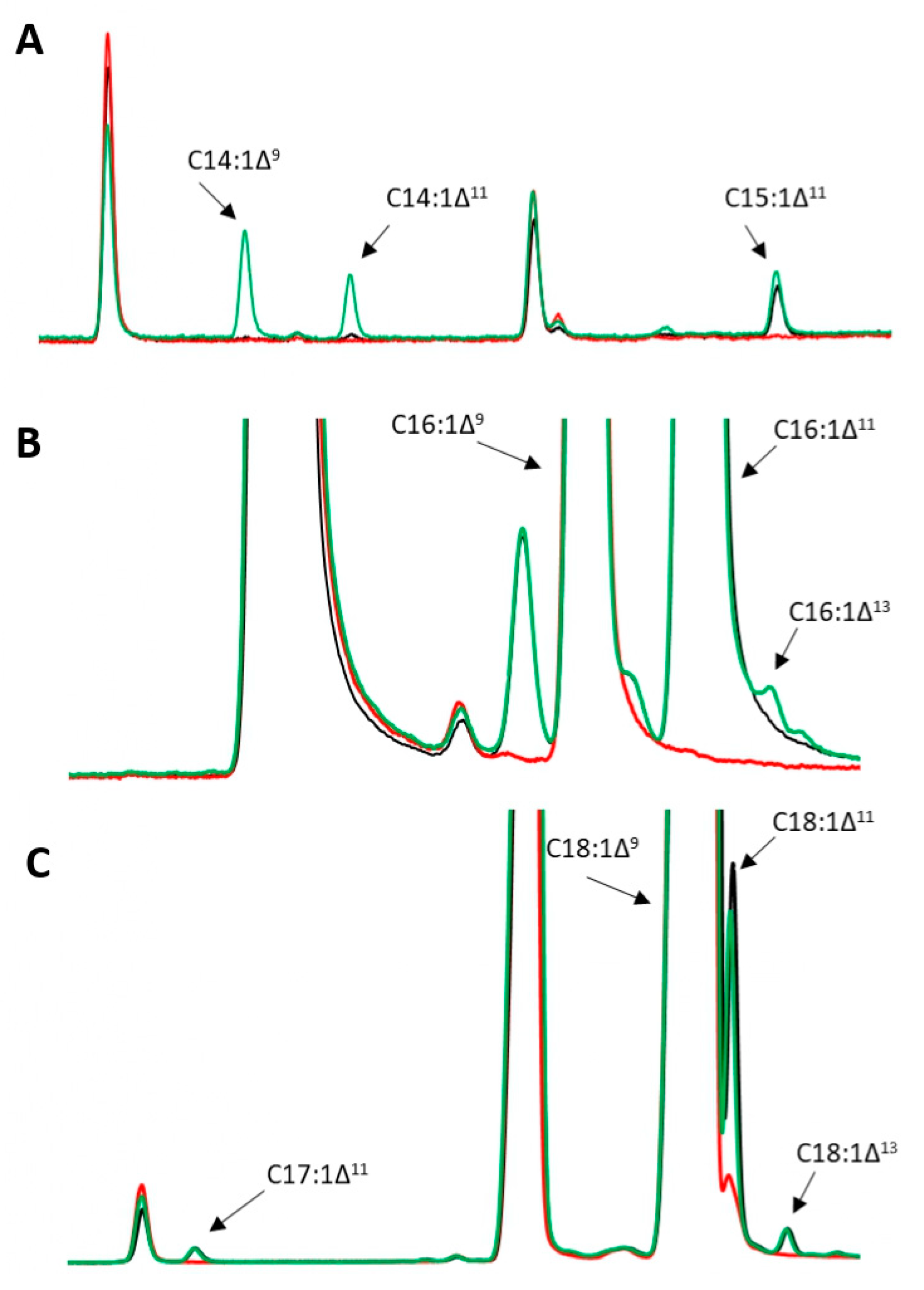
2.3. Analytical Methods
3. Results
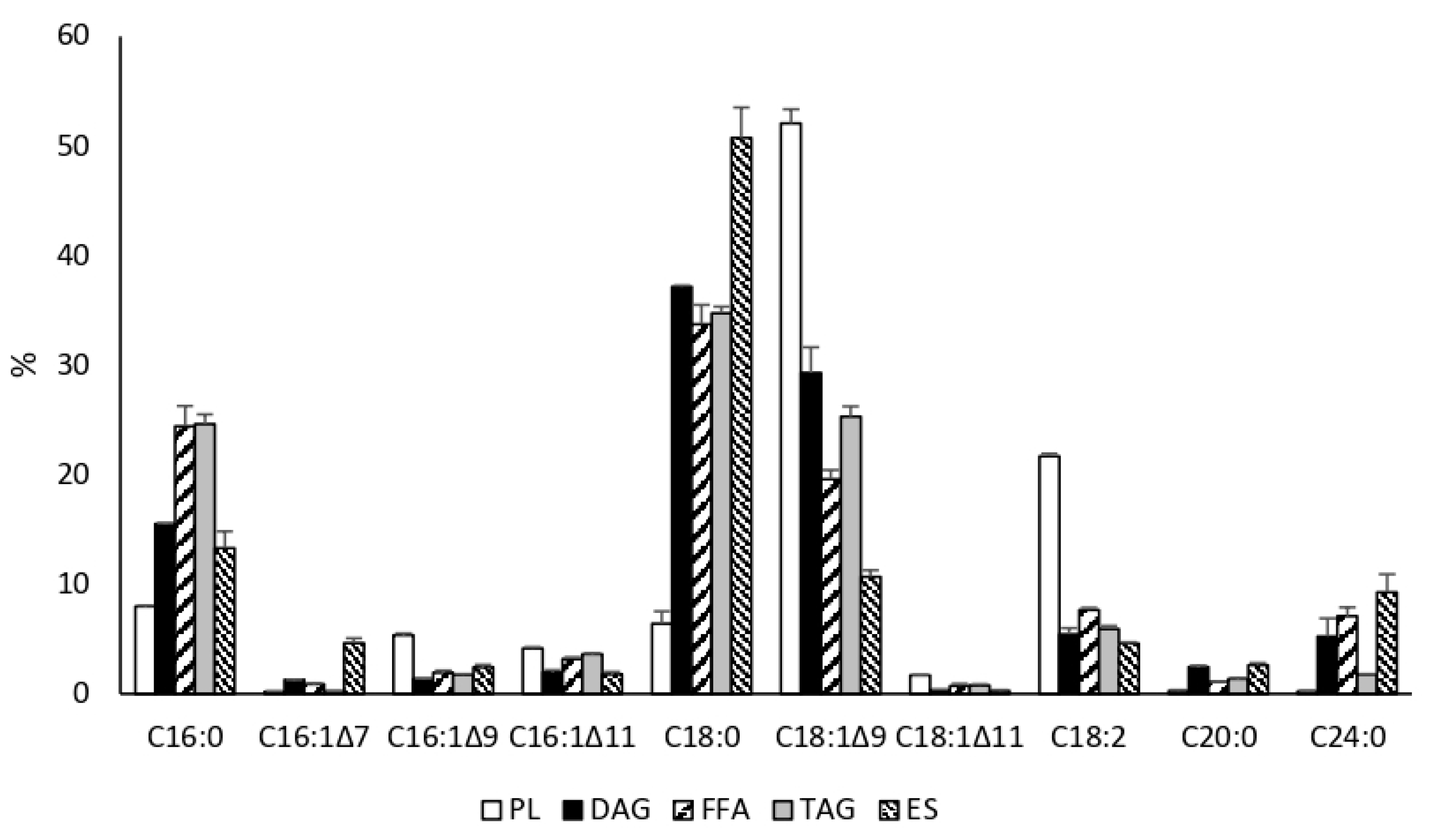
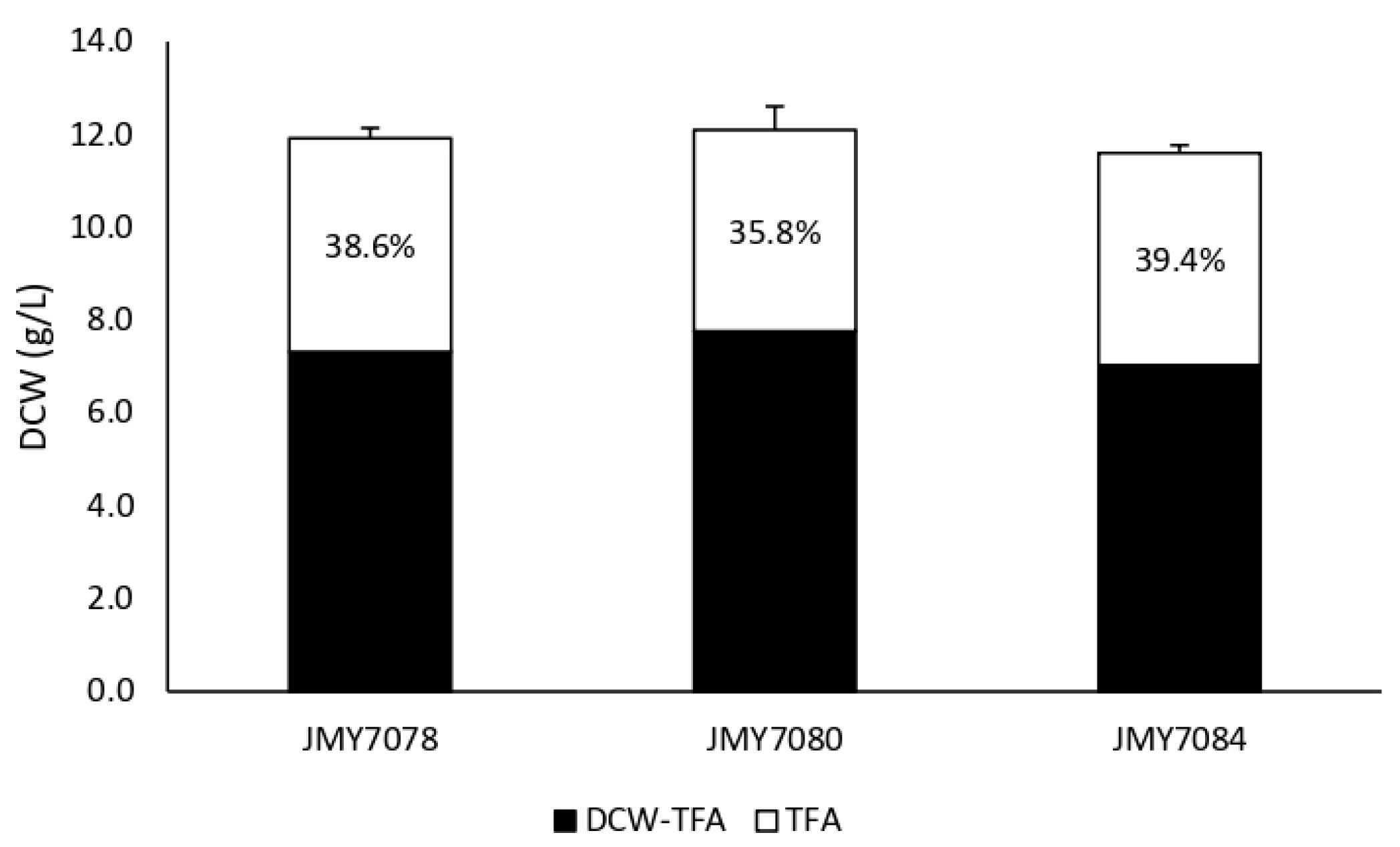
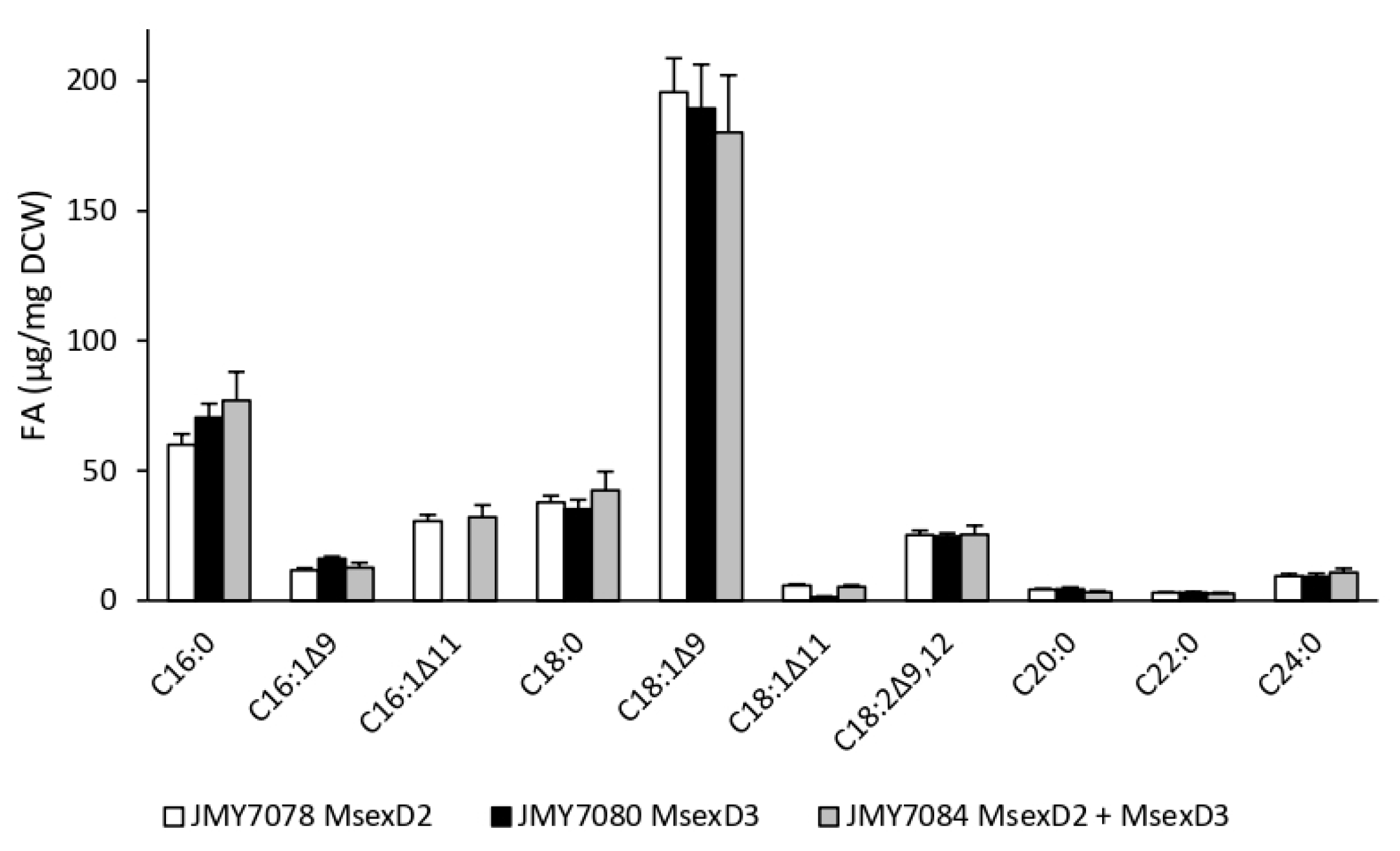
3.1. Growth and Production of Fatty Acid Derivatives in Y. lipolytica Strains Expressing MsexD2 and MsexD3 FADs
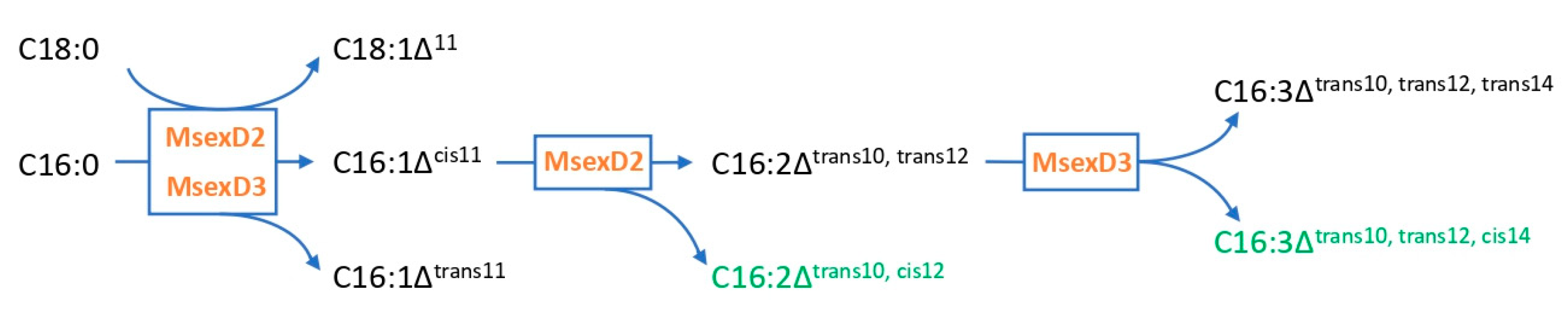
3.2. Metabolic Engineering of Y. lipolytica for Effective Production of FA Derivatives
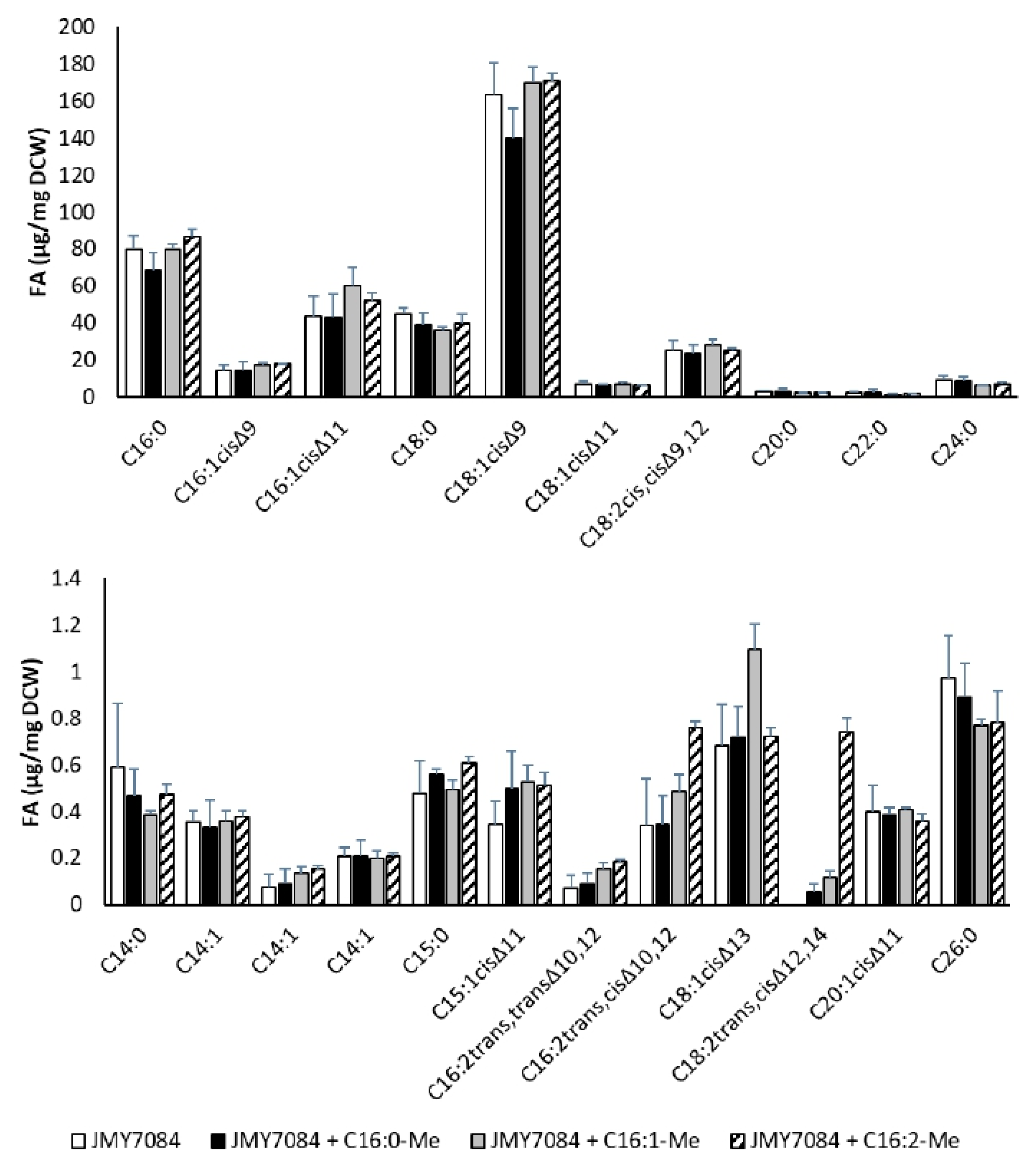
3.3. Supplementation of Media with Fatty Acid Methyl Esters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ledesma-Amaro, R. Microbial Oils: A Customizable Feedstock through Metabolic Engineering. Eur. J. Lipid Sci. Technol. 2015, 117, 141–144. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Nicaud, J.-M. Yarrowia Lipolytica as a Biotechnological Chassis to Produce Usual and Unusual Fatty Acids. Prog. Lipid Res. 2016, 61, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Kamisaka, Y.; Kimura, K.; Uemura, H.; Yamaoka, M. Overexpression of the Active Diacylglycerol Acyltransferase Variant Transforms Saccharomyces Cerevisiae into an Oleaginous Yeast. Appl. Microbiol. Biotechnol. 2013, 97, 7345–7355. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Amaro, R.; Santos, M.A.; Jiménez, A.; Revuelta, J.L. Strain Design of Ashbya Gossypii for Single-Cell Oil Production. Appl. Environ. Microbiol. 2014, 80, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, M.; Boekhout, T.; Neuvéglise, C.; Gaillardin, C.; van Dijck, P.W.M.; Wyss, M. Yarrowia Lipolytica: Safety Assessment of an Oleaginous Yeast with a Great Industrial Potential. Crit. Rev. Microbiol. 2014, 40, 187–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Rong, C.; Chen, H.; Song, Y.; Zhang, H.; Chen, W. De Novo Synthesis of Trans-10, Cis-12 Conjugated Linoleic Acid in Oleaginous Yeast Yarrowia Lipolytica. Microb. Cell Factories 2012, 11, 51. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, H.; Li, M.; Gu, Z.; Song, Y.; Ratledge, C.; Chen, Y.Q.; Zhang, H.; Chen, W. Genetic Engineering of Yarrowia Lipolytica for Enhanced Production of Trans-10, Cis-12 Conjugated Linoleic Acid. Microb. Cell Factories 2013, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Imatoukene, N.; Verbeke, J.; Beopoulos, A.; Idrissi Taghki, A.; Thomasset, B.; Sarde, C.-O.; Nonus, M.; Nicaud, J.-M. A Metabolic Engineering Strategy for Producing Conjugated Linoleic Acids Using the Oleaginous Yeast Yarrowia Lipolytica. Appl. Microbiol. Biotechnol. 2017, 101, 4605–4616. [Google Scholar] [CrossRef] [PubMed]
- Dyer, J.M.; Chapital, D.C.; Kuan, J.-C.W.; Mullen, R.T.; Turner, C.; McKeon, T.A.; Pepperman, A.B. Molecular Analysis of a Bifunctional Fatty Acid Conjugase/Desaturase from Tung. Implications for the Evolution of Plant Fatty Acid Diversity. Plant Physiol. 2002, 130, 2027–2038. [Google Scholar] [CrossRef]
- Rawat, R.; Yu, X.-H.; Sweet, M.; Shanklin, J. Conjugated Fatty Acid Synthesis: Residues 111 and 115 influence product partitioning of momordica charantia conjugase. J. Biol. Chem. 2012, 287, 16230–16237. [Google Scholar] [CrossRef]
- Buček, A.; Matoušková, P.; Vogel, H.; Šebesta, P.; Jahn, U.; Weißflog, J.; Svatoš, A.; Pichová, I. Evolution of Moth Sex Pheromone Composition by a Single Amino Acid Substitution in a Fatty Acid Desaturase. Proc. Natl. Acad. Sci. USA 2015, 112, 12586–12591. [Google Scholar] [CrossRef]
- Buček, A.; Vazdar, M.; Tupec, M.; Svatoš, A.; Pichová, I. Desaturase Specificity Is Controlled by the Physicochemical Properties of a Single Amino Acid Residue in the Substrate Binding Tunnel. Comput. Struct. Biotechnol. J. 2020, 18, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.F.; Russell, D. Molecular Cloning: A Laboratory Manual (3-Volume Set); Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 1, ISBN 978-0-87969-577-4. [Google Scholar]
- Holdsworth, J.E.; Veenhuis, M.; Ratledge, C. Enzyme Activities in Oleaginous Yeasts Accumulating and Utilizing Exogenous or Endogenous Lipids. J. Gen. Microbiol. 1988, 134, 2907–2915. [Google Scholar] [CrossRef] [PubMed]
- Lazar, Z.; Rossignol, T.; Verbeke, J.; Crutz-Le Coq, A.-M.; Nicaud, J.-M.; Robak, M. Optimized Invertase Expression and Secretion Cassette for Improving Yarrowia Lipolytica Growth on Sucrose for Industrial Applications. J. Ind. Microbiol. Biotechnol. 2013, 40, 1273–1283. [Google Scholar] [CrossRef]
- Dulermo, R.; Brunel, F.; Dulermo, T.; Ledesma-Amaro, R.; Vion, J.; Trassaert, M.; Thomas, S.; Nicaud, J.-M.; Leplat, C. Using a Vector Pool Containing Variable-Strength Promoters to Optimize Protein Production in Yarrowia Lipolytica. Microb. Cell Factories 2017, 16, 31. [Google Scholar] [CrossRef]
- Barth, G.; Gaillardin, C. Physiology and Genetics of the Dimorphic Fungus. FEMS Microbiol. Rev. 1997, 19, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Lazar, Z.; Dulermo, T.; Neuvéglise, C.; Crutz-Le Coq, A.-M.; Nicaud, J.-M. Hexokinase—A Limiting Factor in Lipid Production from Fructose in Yarrowia Lipolytica. Metab. Eng. 2014, 26, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Le Dall, M.-T.; Nicaud, J.-M.; Gaillardin, C. Multiple-Copy Integration in the Yeast Yarrowia Lipolytica. Curr. Genet. 1994, 26, 38–44. [Google Scholar] [CrossRef]
- Querol, A.; Barrio, E.; Ramón, D. A Comparative Study of Different Methods of Yeast Strain Characterization. Syst. Appl. Microbiol. 1992, 15, 439–446. [Google Scholar] [CrossRef]
- Beopoulos, A.; Mrozova, Z.; Thevenieau, F.; Le Dall, M.-T.; Hapala, I.; Papanikolaou, S.; Chardot, T.; Nicaud, J.-M. Control of Lipid Accumulation in the Yeast Yarrowia Lipolytica. Appl. Environ. Microbiol. 2008, 74, 7779–7789. [Google Scholar] [CrossRef]
- Lazar, Z.; Walczak, E.; Robak, M. Simultaneous Production of Citric Acid and Invertase by Yarrowia Lipolytica SUC+ Transformants. Bioresour. Technol. 2011, 102, 6982–6989. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Sloane Stanley, G. A Simple Method for the Isolation and Purification of Total Lipides from Animal Tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Gajdoš, P.; Nicaud, J.-M.; Rossignol, T.; Čertík, M. Single Cell Oil Production on Molasses by Yarrowia Lipolytica Strains Overexpressing DGA2 in Multicopy. Appl. Microbiol. Biotechnol. 2015, 99, 8065–8074. [Google Scholar] [CrossRef] [PubMed]
- Christopherson, S.W.; Glass, R.L. Preparation of Milk Fat Methyl Esters by Alcoholysis in an Essentially Nonalcoholic Solution1. J. Dairy Sci. 1969, 52, 1289–1290. [Google Scholar] [CrossRef]
- Matoušková, P.; Pichová, I.; Svatoš, A. Functional Characterization of a Desaturase from the Tobacco Hornworm Moth (Manduca Sexta) with Bifunctional Z11- and 10,12-Desaturase Activity. Insect Biochem. Mol. Biol. 2007, 37, 601–610. [Google Scholar] [CrossRef]
- Rigouin, C.; Croux, C.; Borsenberger, V.; Ben Khaled, M.; Chardot, T.; Marty, A.; Bordes, F. Increasing Medium Chain Fatty Acids Production in Yarrowia Lipolytica by Metabolic Engineering. Microb. Cell Factories 2018, 17, 142. [Google Scholar] [CrossRef]
- Rigouin, C.; Gueroult, M.; Croux, C.; Dubois, G.; Borsenberger, V.; Barbe, S.; Marty, A.; Daboussi, F.; André, I.; Bordes, F. Production of Medium Chain Fatty Acids by Yarrowia Lipolytica: Combining Molecular Design and TALEN to Engineer the Fatty Acid Synthase. ACS Synth. Biol. 2017, 6, 1870–1879. [Google Scholar] [CrossRef]


| Strain (Host Strain) | Plasmid/Genotype | References |
|---|---|---|
| Escherichia coli | ||
| JME1046 | JMP62-pTEF-URA3ex | Lazar et al. 2013 [15] |
| JME2563 | JMP62-pTEF-LEU2ex | Dulermo et al. 2017 [16] |
| JME2607 | JMP62-8UAS-pTEF-RedStae2-LEU2ex | Dulermo et al. 2017 [16] |
| JME3048 | JMP62-8UAS-pTEF-URA3ex | Dulermo et al. 2017 [16] |
| JME 4145 | JMP1046-MsexD2 | This work |
| JME 4147 | JMP1046-MsexD3 | This work |
| JME 4299 | JMP3048-MsexD2 | This work |
| JME 4301 | JMP2607-MsexD3 | This work |
| Yarrowia lipolytica | ||
| W29 | MATA, wild type | Barth and Gaillardin 1997 [17] |
| Po1d | MATA leu2–270 ura3–302 xpr2–322 + pXPR2-SUC2 | Barth and Gaillardin 1997 [17] |
| JMY6699 | Po1d, pTEF-MsexD2-URA3ex, LEU2 | This work |
| JMY6700 | Po1d, pTEF-MsexD3-URA3ex, LEU2 | This work |
| JMY3501 | W29 ura3–302 leu2–270 xpr2–322 Δpox1–6 Δtgl4 + pXPR2-SUC2 + pTEF-DGA2-LEU2ex + pTEF-GPD1-URA3ex | Lazar et al. 2014 [18] |
| JMY3820 | W29 ura3–302 leu2–270 xpr2–322 Δpox1–6 Δtgl4 + pXPR2-SUC2 + pTEF-DGA2 + pTEF-GPD1 | Lazar et al. 2014 [18] |
| JMY7078 | JMY3820, 8UAS-pTEF-MsexD2-URA3ex, LEU2 | This work |
| JMY7080 | JMY3820, 8UAS-pTEF-MsexD3-LEU2ex, URA3 | This work |
| JMY7084 | JMY3820, 8UAS-pTEF-MsexD2-URA3ex, 8UAS-pTEF-MsexD3-LEU2ex | This work |
| Strain + Addition | DCW (g/L) | TFA/DCW (%) |
|---|---|---|
| JMY7084 | 11.6 ± 0.6 | 40.1 ± 6.7 |
| JMY7084 + C16:0-Me | 12.4 ± 0.3 | 35.5 ± 6.4 |
| JMY7084 + C16:1-Me | 12.4 ± 0.5 | 41.6 ± 2.4 |
| JMY7084 + C16:2-Me | 12.5 ± 0.3 | 41.9 ± 0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hambalko, J.; Gajdoš, P.; Nicaud, J.-M.; Ledesma-Amaro, R.; Tupec, M.; Pichová, I.; Čertík, M. Biosynthesis of Fatty Acid Derivatives by Recombinant Yarrowia lipolytica Containing MsexD2 and MsexD3 Desaturase Genes from Manduca sexta. J. Fungi 2023, 9, 114. https://doi.org/10.3390/jof9010114
Hambalko J, Gajdoš P, Nicaud J-M, Ledesma-Amaro R, Tupec M, Pichová I, Čertík M. Biosynthesis of Fatty Acid Derivatives by Recombinant Yarrowia lipolytica Containing MsexD2 and MsexD3 Desaturase Genes from Manduca sexta. Journal of Fungi. 2023; 9(1):114. https://doi.org/10.3390/jof9010114
Chicago/Turabian StyleHambalko, Jaroslav, Peter Gajdoš, Jean-Marc Nicaud, Rodrigo Ledesma-Amaro, Michal Tupec, Iva Pichová, and Milan Čertík. 2023. "Biosynthesis of Fatty Acid Derivatives by Recombinant Yarrowia lipolytica Containing MsexD2 and MsexD3 Desaturase Genes from Manduca sexta" Journal of Fungi 9, no. 1: 114. https://doi.org/10.3390/jof9010114
APA StyleHambalko, J., Gajdoš, P., Nicaud, J.-M., Ledesma-Amaro, R., Tupec, M., Pichová, I., & Čertík, M. (2023). Biosynthesis of Fatty Acid Derivatives by Recombinant Yarrowia lipolytica Containing MsexD2 and MsexD3 Desaturase Genes from Manduca sexta. Journal of Fungi, 9(1), 114. https://doi.org/10.3390/jof9010114







