Metabarcoding and Metabolome Analyses Reveal Mechanisms of Leymus chinensis Growth Promotion by Fairy Ring of Leucocalocybe mongolica
Abstract
:1. Introduction
2. Materials and Methods
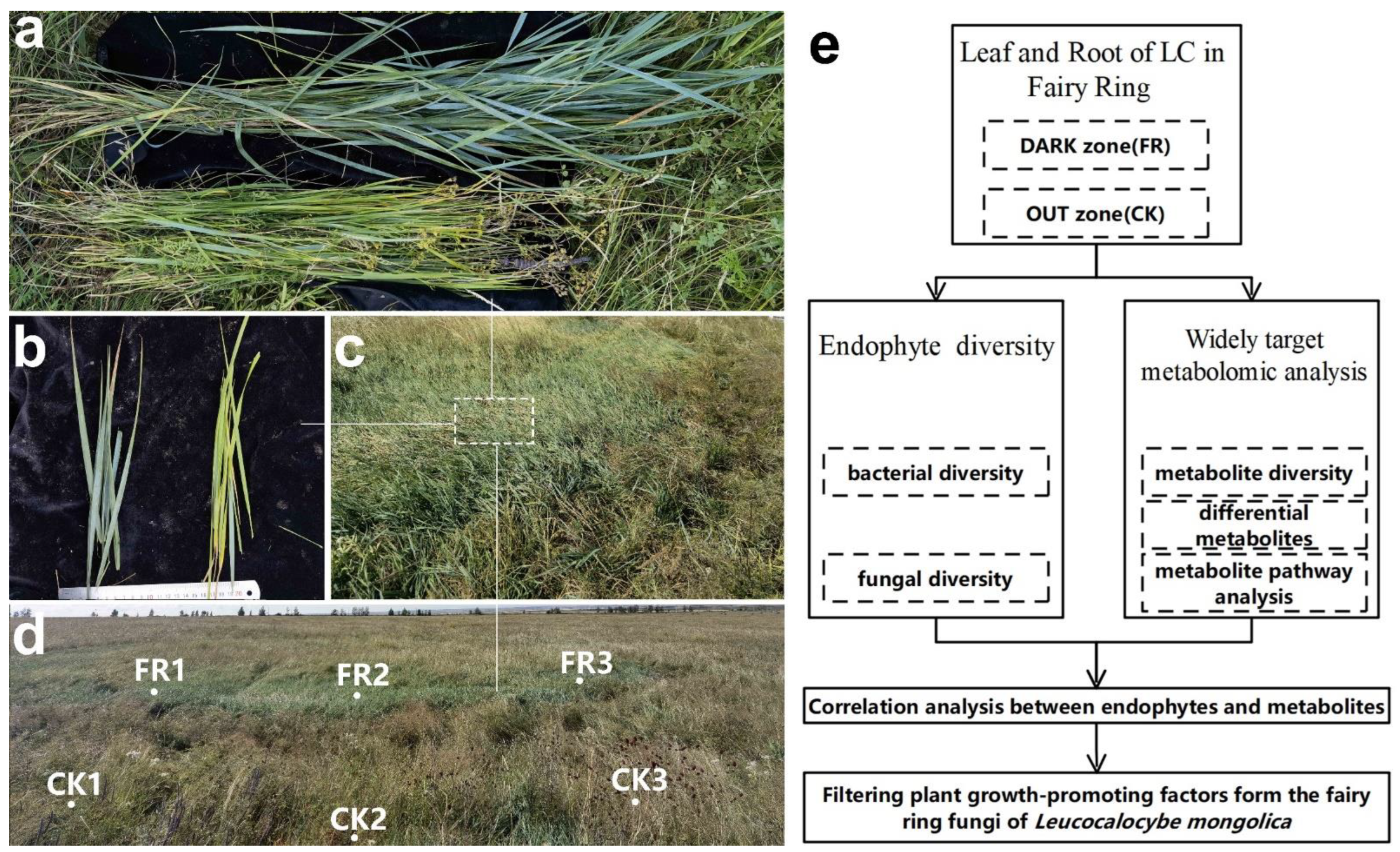
2.1. Materials
2.2. Methods
2.2.1. Metabarcoding Survey
2.2.2. Widely Targeted Metabolome Analysis
2.2.3. Correlation Analysis
3. Results
3.1. Microbial Diversity
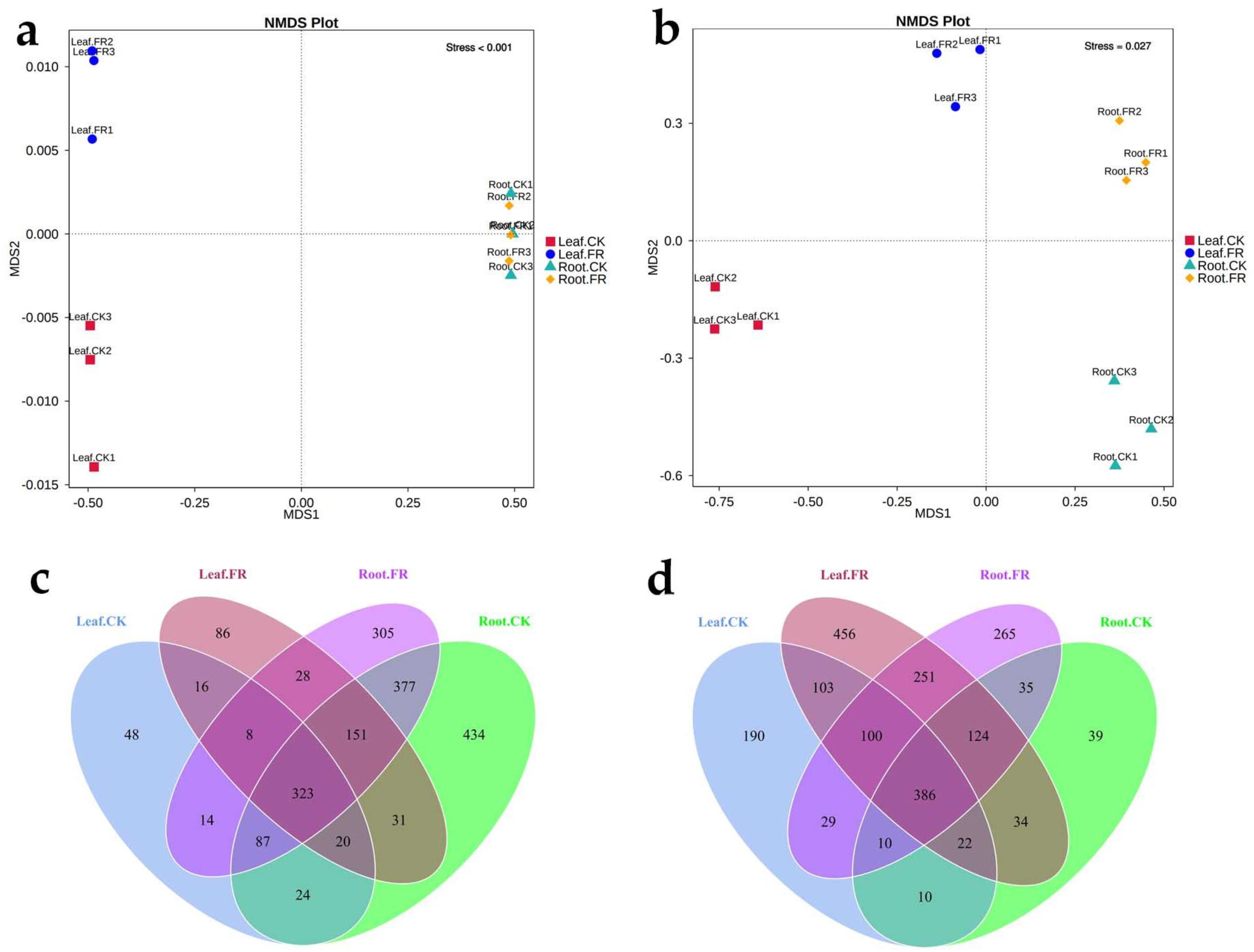
3.1.1. Metabarcoding Sequencing
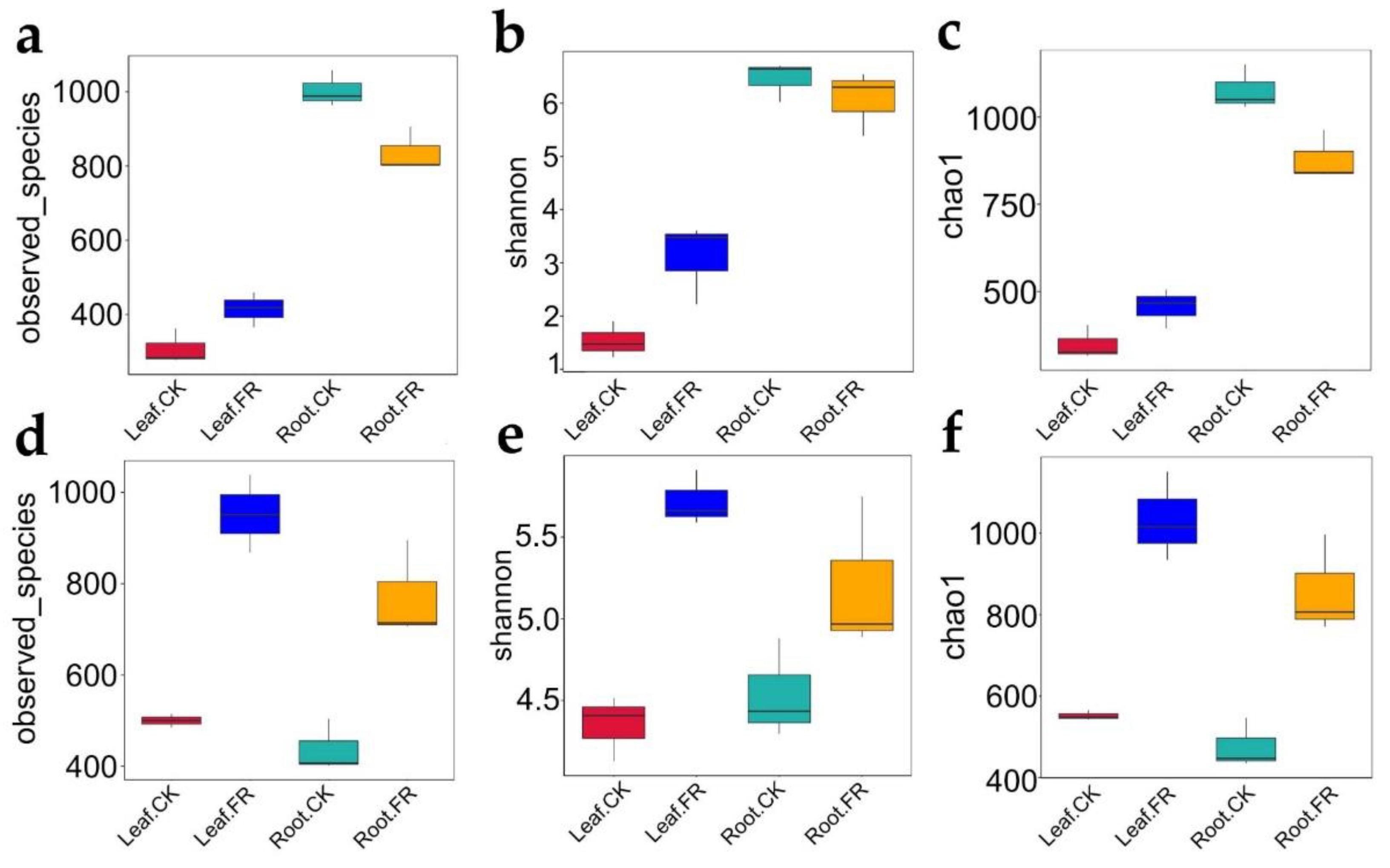
3.1.2. Microbial Alpha Diversity
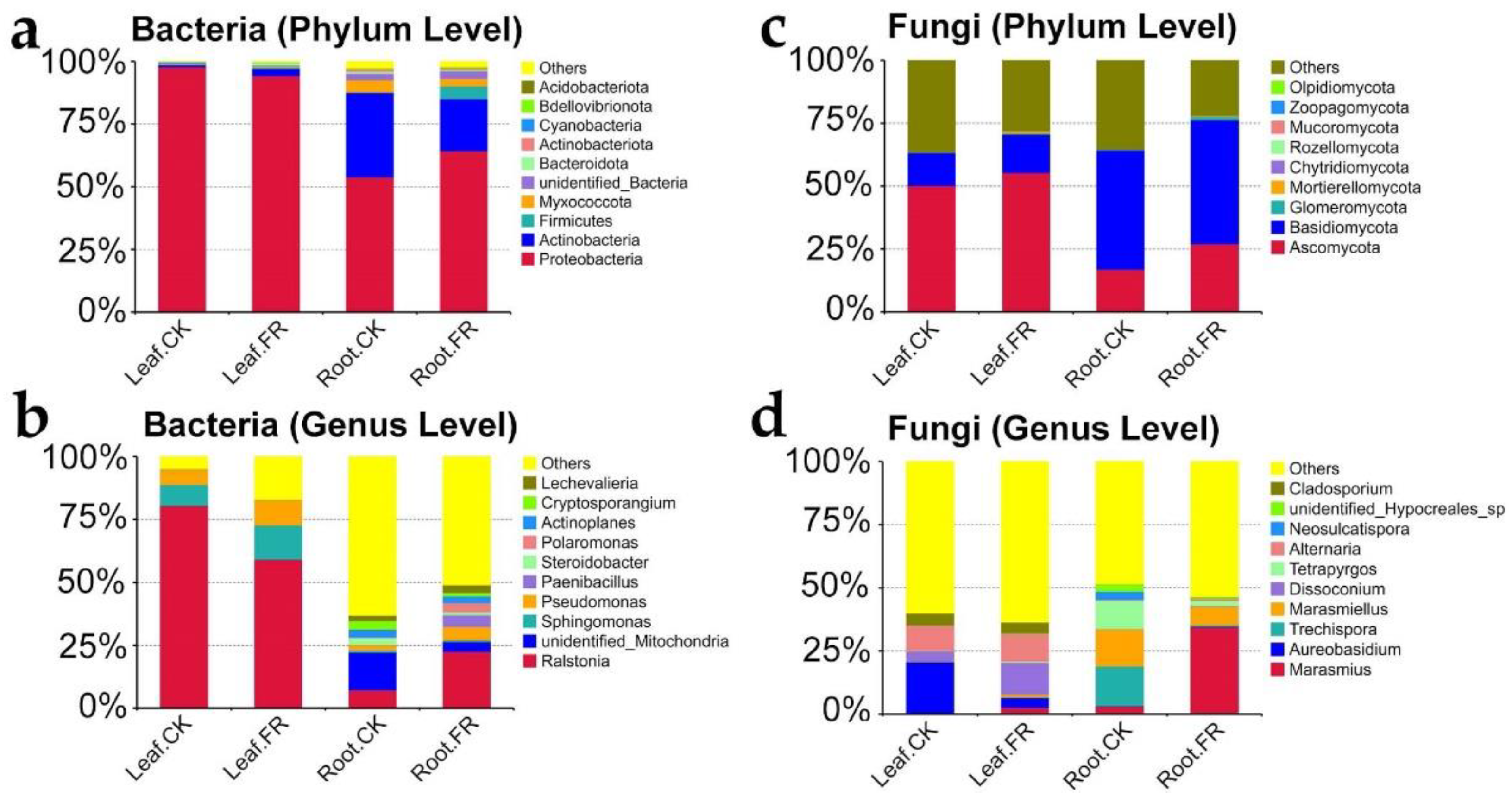
3.1.3. Key Microbial Taxon
3.2. Metabolome Analysis
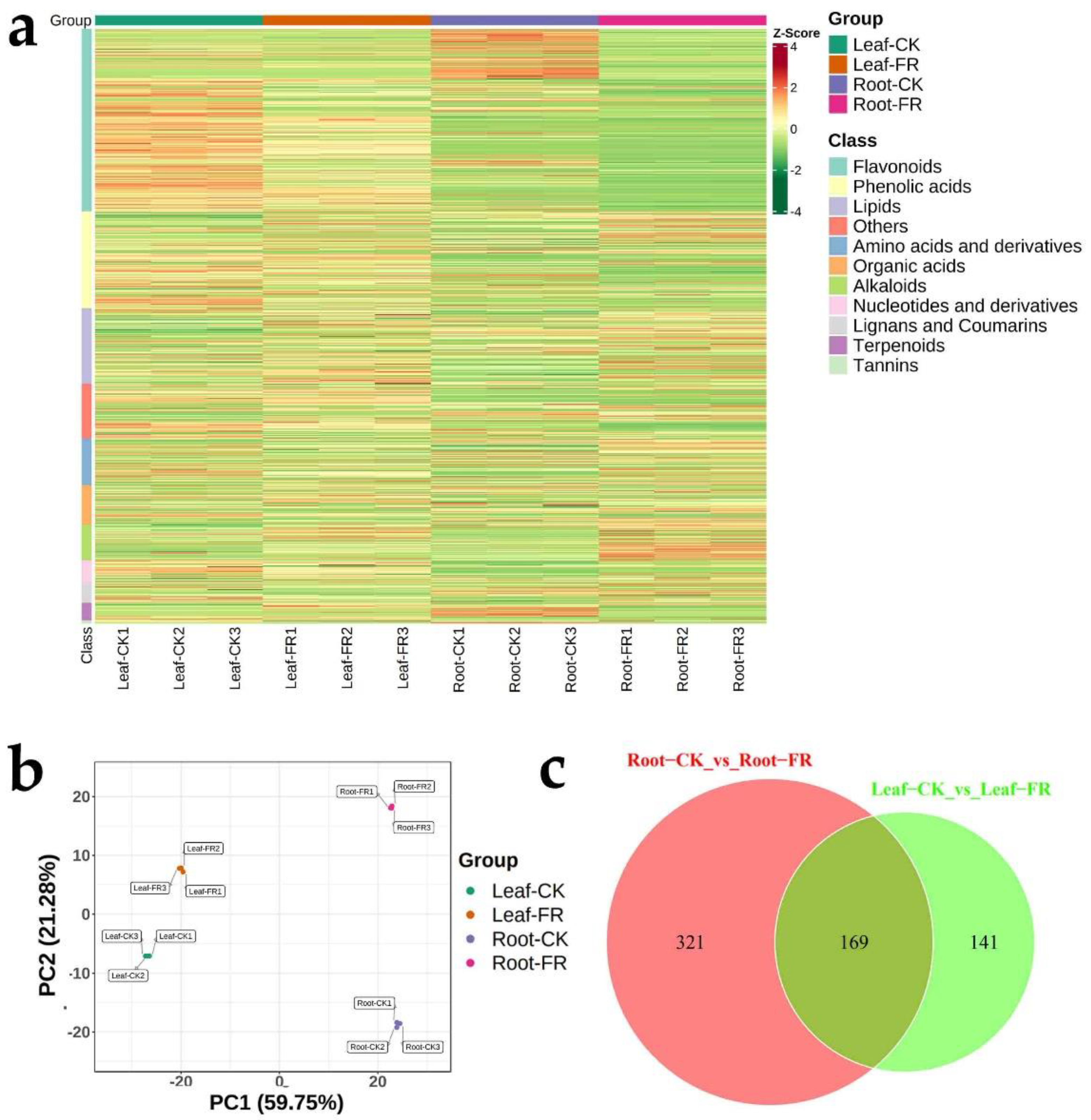
3.2.1. Metabolite Composition
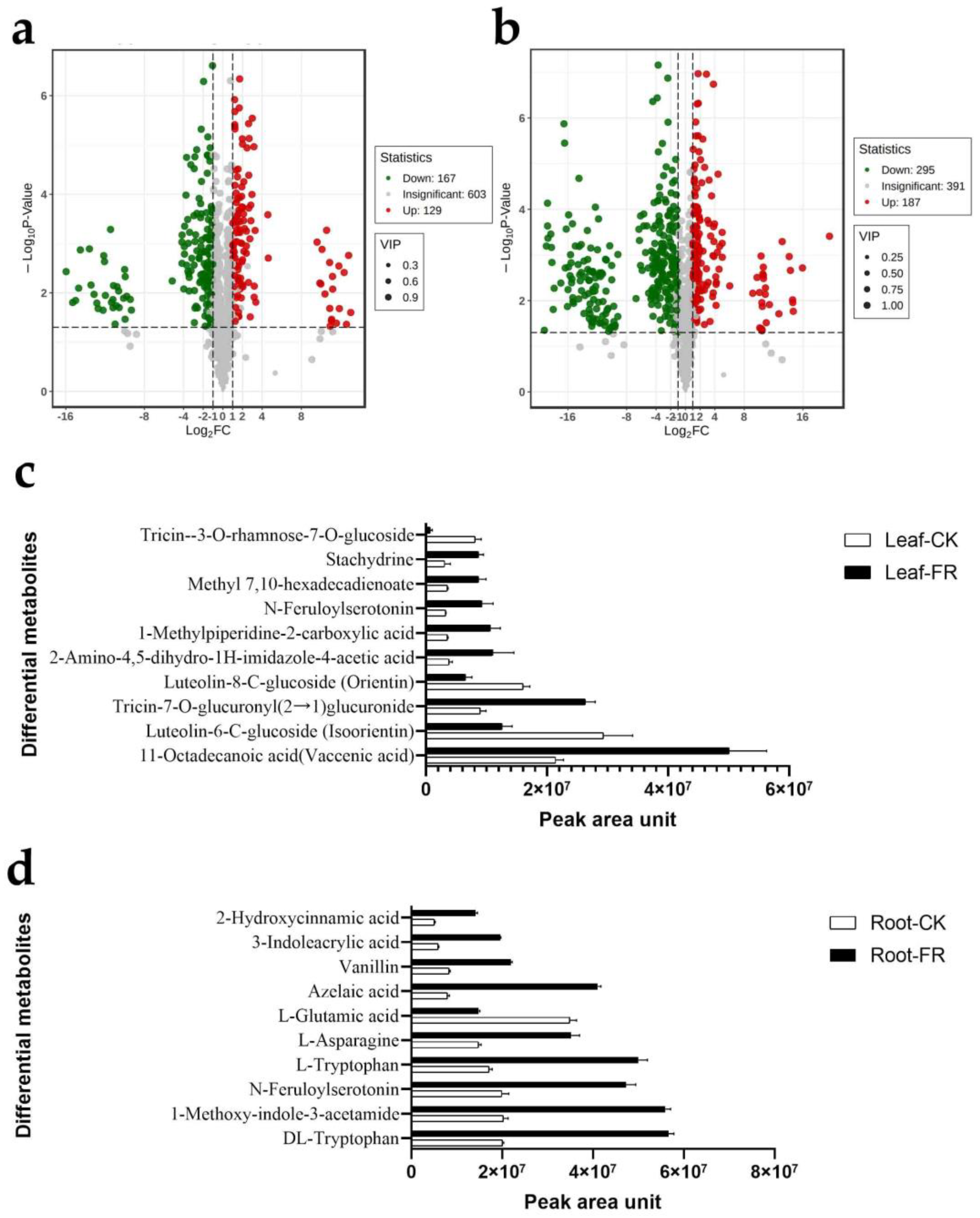
3.2.2. Differential Metabolites
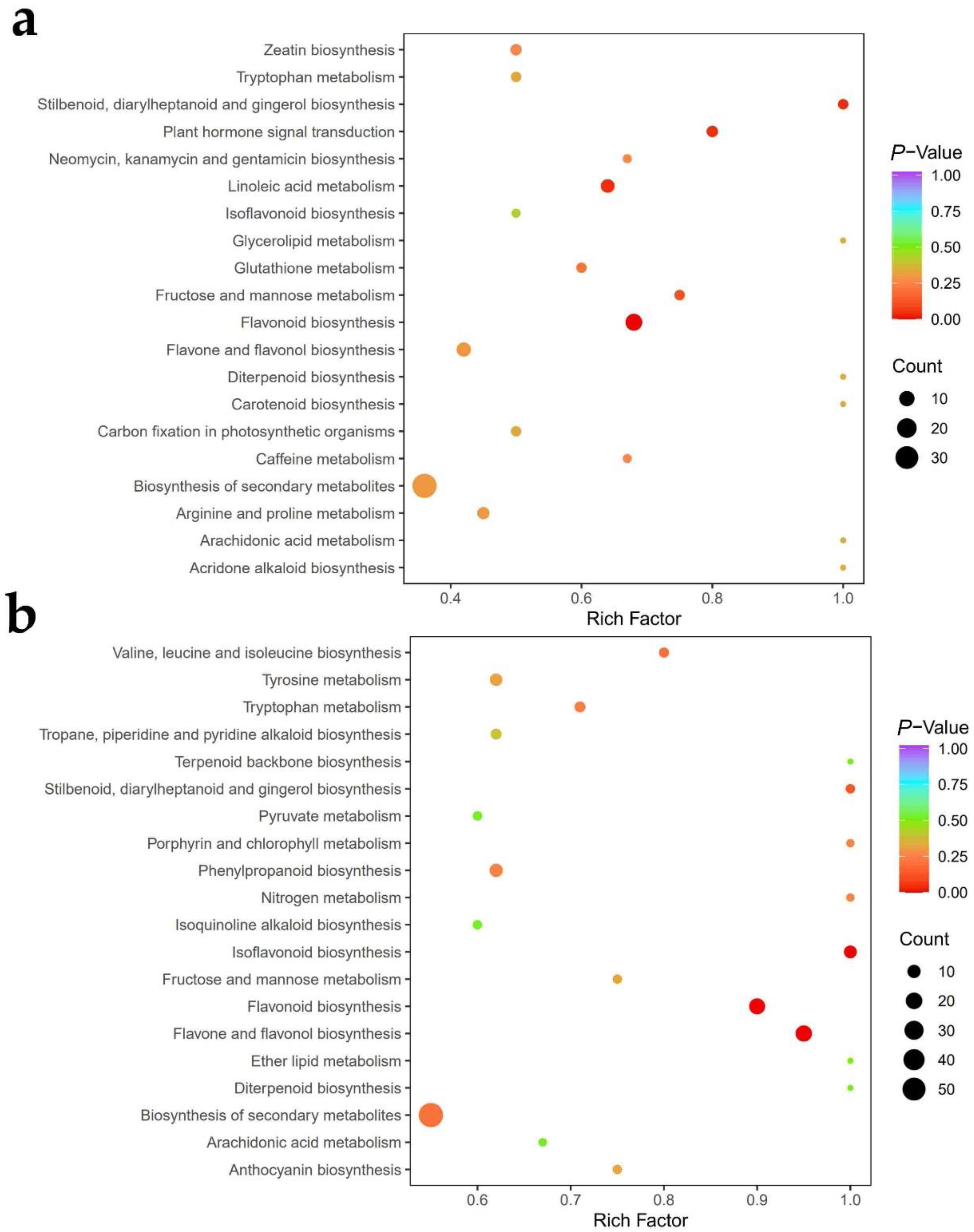
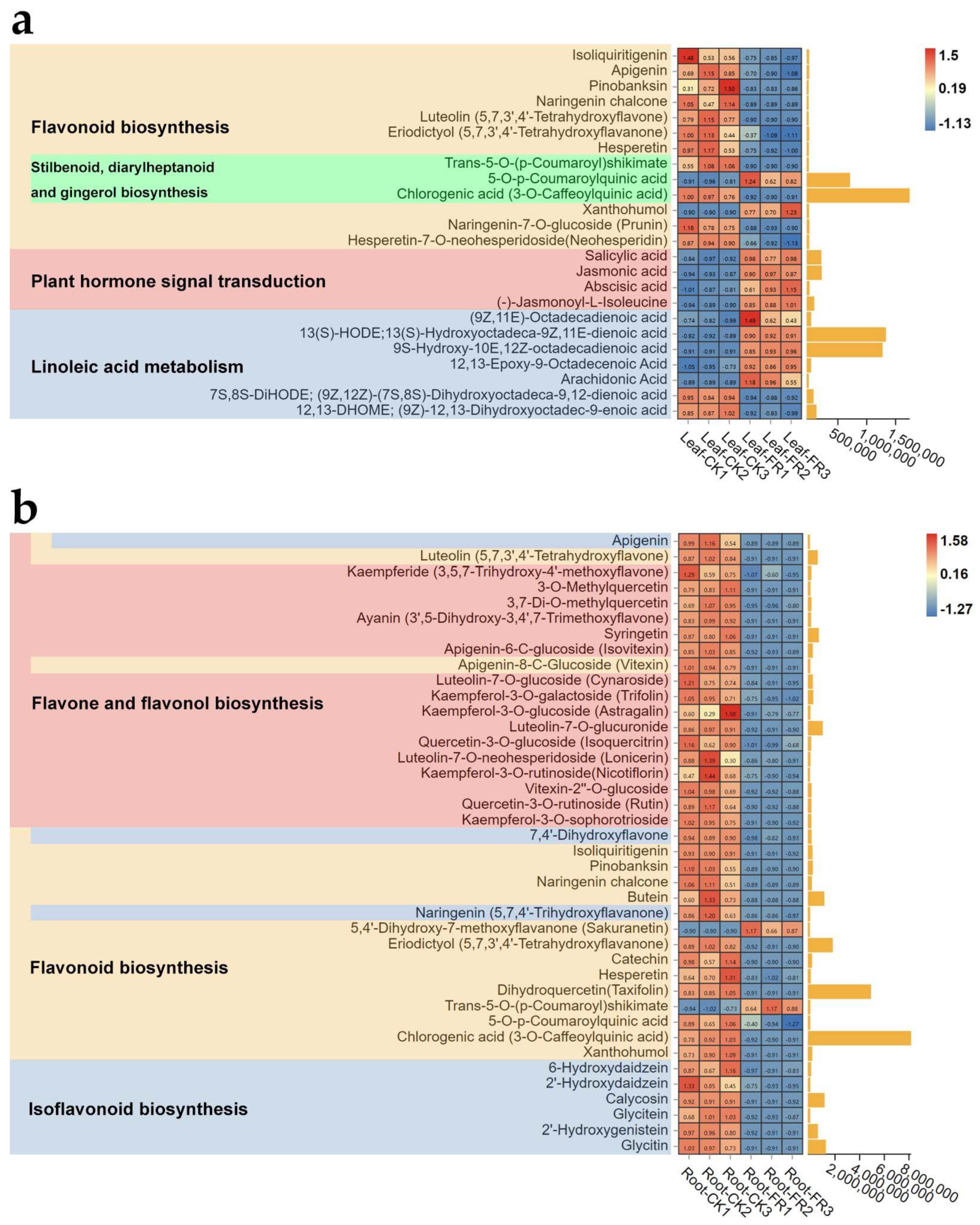
3.2.3. Metabolite Pathway Analysis
3.3. Correlation Analysis
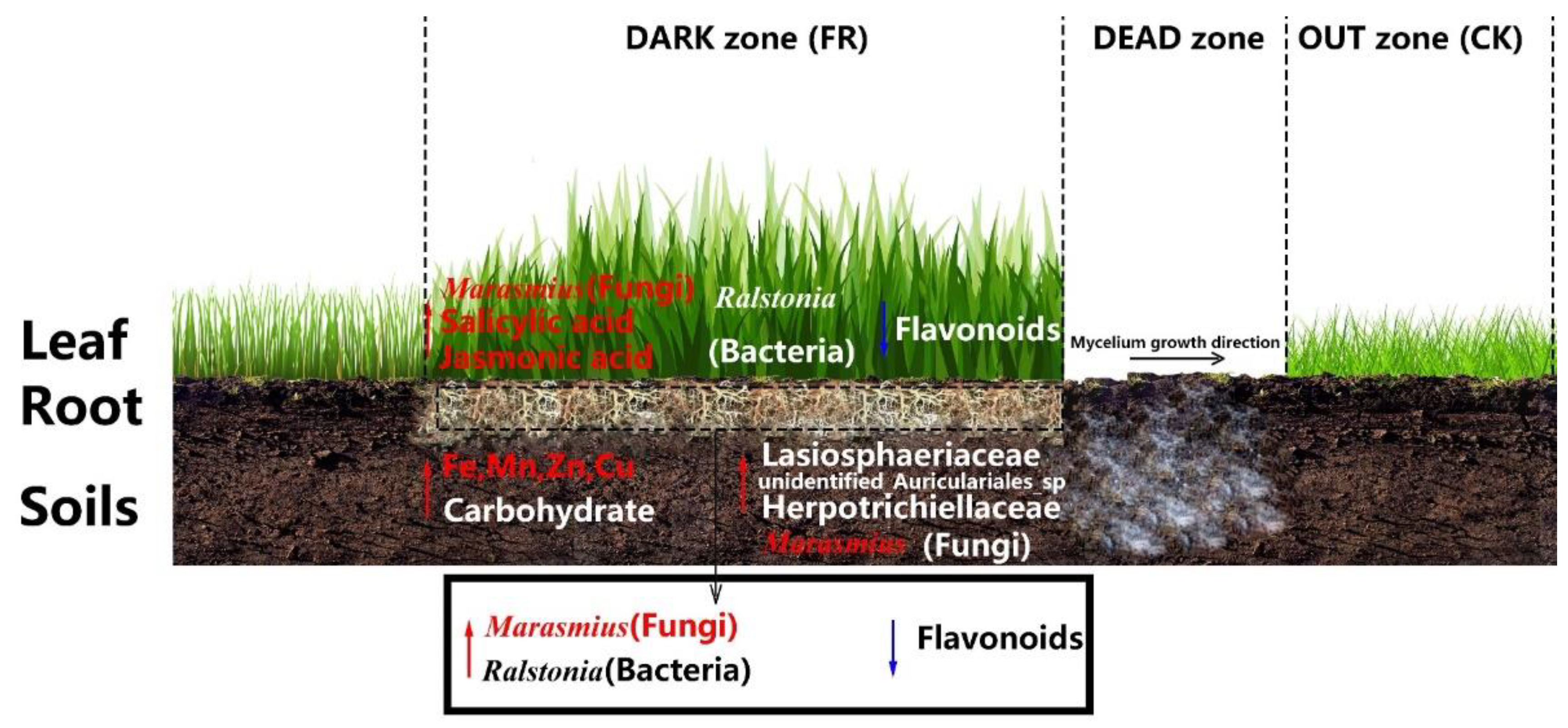
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, Q.; Ling, N.; Chen, H.; Duan, Y.; Wang, S.; Shen, Q.; Vandenkoornhuyse, P. Long-Term Chemical-Only Fertilization Induces a Diversity Decline and Deep Selection on the Soil Bacteria. mSystems 2020, 5, e00337-20. [Google Scholar] [CrossRef]
- Cui, M.; Zeng, L.; Qin, W.; Feng, J. Measures for reducing nitrate leaching in orchards:A review. Environ. Pollut. 2020, 263, 114553. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lei, Y.; Zhang, Y.; Liu, J.; Shi, X.; Jia, H.; Wang, C.; Chen, F.; Chu, Q. Rational trade-offs between yield increase and fertilizer inputs are essential for sustainable intensification: A case study in wheat-maize cropping systems in China. Sci. Total Environ. 2019, 679, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Vitousek, P.M.; Naylor, R.; Crews, T.; David, M.B.; Drinkwater, L.E.; Holland, E.; Johnes, P.J.; Katzenberger, J.; Martinelli, L.A.; Matson, P.A.; et al. Agriculture. Nutrient imbalances in agricultural development. Science 2009, 324, 1519–1520. [Google Scholar] [CrossRef] [PubMed]
- Hiltunen, M.; Ament-Velásquez, S.L.; Johannesson, H. The Assembled and Annotated Genome of the Fairy-Ring Fungus Marasmius oreades. Genome Biol. Evol. 2021, 13, evab126. [Google Scholar] [CrossRef]
- Takano, T.; Yamamoto, N.; Suzuki, T.; Dohra, H.; Choi, J.H.; Terashima, Y.; Yokoyama, K.; Kawagishi, H.; Yano, K. Genome sequence analysis of the fairy ring-forming fungus Lepista sordida and gene candidates for interaction with plants. Sci. Rep. 2019, 9, 5888. [Google Scholar] [CrossRef]
- Shantz, H.L.; Piemeisel, R.L. Fungus fairy rings in eastern Colorado and their effect on vegetation. J. Agric. Res. 1917, 11, 191–246. [Google Scholar]
- Zotti, M.; De Filippis, F.; Cesarano, G.; Ercolini, D.; Tesei, G.; Allegrezza, M.; Giannino, F.; Mazzoleni, S.; Bonanomi, G. One ring to rule them all: An ecosystem engineer fungus fosters plant and microbial diversity in a Mediterranean grassland. New Phytol. 2020, 227, 884–898. [Google Scholar] [CrossRef]
- Miller, S.L.; Gongloff, A. Fairy rings, associated fungi, and assessment of their distribution across environmental variables using GIS. Fungal Ecol. 2021, 50, 101040. [Google Scholar] [CrossRef]
- Duan, M.; Bau, T. Grassland fairy rings of Leucocalocybe mongolica represent the center of a rich soil microbial community. Braz. J. Microbiol. 2021, 52, 1357–1369. [Google Scholar] [CrossRef]
- Choi, J.H.; Ohnishi, T.; Yamakawa, Y.; Takeda, S.; Sekiguchi, S.; Maruyama, W.; Yamashita, K.; Suzuki, T.; Morita, A.; Ikka, T.; et al. The source of “fairy rings”: 2-azahypoxanthine and its metabolite found in a novel purine metabolic pathway in plants. Angew. Chem. Int. Ed. Engl. 2014, 53, 1552–1555. [Google Scholar] [CrossRef] [PubMed]
- Zotti, M.; Bonanomi, G.; Mancinelli, G.; Barquero, M.; De Filippis, F.; Giannino, F.; Mazzoleni, S.; González-Andrés, F. Riding the wave: Response of bacterial and fungal microbiota associated with the spread of the fairy ring fungus Calocybe gambosa. Appl. Soil Ecol. 2021, 163, 103963. [Google Scholar] [CrossRef]
- Duan, M.; Bau, T. Initial sample processing can influence the soil microbial metabarcoding surveys, revealed by Leucocalocybe mongolica fairy ring ecosystem. Biotechnol. Biotechnol. Equip. 2021, 35, 1427–1438. [Google Scholar] [CrossRef]
- Duan, M.; Bao, H.; Bau, T. Analyses of transcriptomes and the first complete genome of Leucocalocybe mongolica provide new insights into phylogenetic relationships and conservation. Sci. Rep. 2021, 11, 1–12. [Google Scholar]
- Caesar-Tonthat, T.C.; Espeland, E.; Caesar, A.J.; Sainju, U.M.; Lartey, R.T.; Gaskin, J.F. Effects of Agaricus lilaceps fairy rings on soil aggregation and microbial community structure in relation to growth stimulation of western wheatgrass (Pascopyrum smithii) in Eastern Montana rangeland. Microb. Ecol. 2013, 66, 120–131. [Google Scholar] [CrossRef]
- Fidanza, M.A.; Cisar, J.L.; Kostka, S.J.; Gregos, J.S.; Schlossberg, M.J.; Franklin, M. Preliminary investigation of soil chemical and physical properties associated with type-I fairy ring symptoms in turfgrass. Hydrol. Processes 2007, 21, 2285–2290. [Google Scholar] [CrossRef]
- Gramss, G.; Voigt, K.-D.; Bergmann, H. Factors influencing water solubility and plant availability of mineral compounds in the tripartite fairy rings of Marasmius oreades (Bolt.: Fr.) Fr. J. Basic Microbiol. 2005, 45, 41–54. [Google Scholar] [CrossRef]
- Duan, M.; Lu, M.; Lu, J.; Yang, W.; Li, B.; Ma, L.; Wang, L. Soil Chemical Properties, Metabolome, and Metabarcoding Give the New Insights into the Soil Transforming Process of Fairy Ring Fungi Leucocalocybe mongolica. J. Fungi 2022, 8, 680. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019, 49, 73–82. [Google Scholar] [CrossRef]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial endophytes: Recent developments and applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qin, J.; Chen, W.; Zhou, Y.; Ren, A.; Gao, Y. Pathogen resistant advantage of endophyte-infected over endophyte-free Leymus chinensis is strengthened by pre-drought treatment. Eur. J. Plant Pathol. 2016, 144, 477–486. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, S.; Wu, X.; Syed, S.I.; Syed, I.U.S.; Huang, B.; Guan, P.; Wang, D. Grazing Affects Bacterial and Fungal Diversities and Communities in the Rhizosphere and Endosphere Compartments of Leymus chinensis through Regulating Nutrient and Ion Distribution. Microorganisms 2021, 9, 476. [Google Scholar] [CrossRef]
- Chen, J.; Deng, Y.; Yu, X.; Wu, G.; Gao, Y.; Ren, A. Epichloë Endophyte Infection Changes the Root Endosphere Microbial Community Composition of Leymus Chinensis Under Both Potted and Field Growth Conditions. Microb. Ecol. 2022, 1–13. [Google Scholar] [CrossRef]
- Kosmacz, M.; Sokołowska, E.M.; Bouzaa, S.; Skirycz, A. Towards a functional understanding of the plant metabolome. Curr. Opin. Plant Biol. 2020, 55, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, P.; Wang, F.; Li, R.; Liu, J.; Wang, Q.; Liu, W.; Wang, B.; Hu, G. Integrated Analysis of the Transcriptome and Metabolome Revealed Candidate Genes Involved in GA3-Induced Dormancy Release in Leymus chinensis Seeds. Int. J. Mol. Sci. 2021, 22, 4161. [Google Scholar] [CrossRef] [PubMed]
- Hongna, C.; Leyuan, T.; Junmei, S.; Xiaori, H.; Xianguo, C. Exogenous salicylic acid signal reveals an osmotic regulatory role in priming the seed germination of Leymus chinensis under salt-alkali stress. Environ. Exp. Bot. 2021, 188, 104498. [Google Scholar] [CrossRef]
- Chen, L.H.; Zhang, Y.B.; Yang, X.W.; Xu, W.; Wang, Y.P. Characterization and quantification of ginsenosides from the root of Panax quinquefolius L. by integrating untargeted metabolites and targeted analysis using UPLC-Triple TOF-MS coupled with UFLC-ESI-MS/MS. Food Chem. 2022, 384, 132466. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wen, L.; Chen, Z.; Zhang, Z.; Pang, X.; Deng, Z.; Liu, T.; Guo, Y. Study on metabolic variation in whole grains of four proso millet varieties reveals metabolites important for antioxidant properties and quality traits. Food Chem. 2021, 357, 129791. [Google Scholar] [CrossRef]
- Cheng, Y.-J.; Guo, W.-W.; Yi, H.-L.; Pang, X.-M.; Deng, X. An efficient protocol for genomic DNA extraction fromCitrus species. Plant Mol. Biol. Rep. 2003, 21, 177–178. [Google Scholar] [CrossRef]
- Beckers, B.; Op De Beeck, M.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Performance of 16s rDNA Primer Pairs in the Study of Rhizosphere and Endosphere Bacterial Microbiomes in Metabarcoding Studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kujawska, M.B.; Rudawska, M.; Wilgan, R.; Leski, T. Similarities and Differences among Soil Fungal Assemblages in Managed Forests and Formerly Managed Forest Reserves. Forests 2021, 12, 353. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemura, H.; Choi, J.H.; Matsuzaki, N.; Taniguchi, Y.; Wu, J.; Hirai, H.; Motohashi, R.; Asakawa, T.; Ikeuchi, K.; Inai, M.; et al. A Fairy Chemical, Imidazole-4-carboxamide, is Produced on a Novel Purine Metabolic Pathway in Rice. Sci. Rep. 2019, 9, 9899. [Google Scholar] [CrossRef]
- Zhu, J.; Sun, X.; Tang, Q.Y.; Zhang, Z.D. Seasonal Dynamics and Persistency of Endophyte Communities in Kalidium schrenkianum Shifts Under Radiation Stress. Front. Microbiol. 2021, 12, 778327. [Google Scholar] [CrossRef]
- Zuo, Y.; Li, X.; Yang, J.; Liu, J.; Zhao, L.; He, X. Fungal Endophytic Community and Diversity Associated with Desert Shrubs Driven by Plant Identity and Organ Differentiation in Extremely Arid Desert Ecosystem. J. Fungi 2021, 7, 578. [Google Scholar] [CrossRef]
- Safni, I.; Subandiyah, S.; Fegan, M. Ecology, Epidemiology and Disease Management of Ralstonia syzygii in Indonesia. Front. Microbiol. 2018, 9, 419. [Google Scholar] [CrossRef]
- Georgoulis, S.J.; Shalvarjian, K.E.; Helmann, T.C.; Hamilton, C.D.; Carlson, H.K.; Deutschbauer, A.M.; Lowe-Power, T.M. Genome-Wide Identification of Tomato Xylem Sap Fitness Factors for Three Plant-Pathogenic Ralstonia Species. mSystems 2021, 6, e0122921. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xie, J.; Fu, Y.; Cheng, J.; Qu, Z.; Zhao, Z.; Cheng, S.; Chen, T.; Li, B.; Wang, Q.; et al. A 2-kb Mycovirus Converts a Pathogenic Fungus into a Beneficial Endophyte for Brassica Protection and Yield Enhancement. Mol. Plant 2020, 13, 1420–1433. [Google Scholar] [CrossRef]
- Hiltunen, M.; Grudzinska-Sterno, M.; Wallerman, O.; Ryberg, M.; Johannesson, H. Maintenance of High Genome Integrity over Vegetative Growth in the Fairy-Ring Mushroom Marasmius oreades. Curr. Biol. 2019, 29, 2758–2765.e2756. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Su, X.; Xia, Y.; Jiang, W.; Shen, G.; Pang, Y. GbMYBR1 from Ginkgo biloba represses phenylpropanoid biosynthesis and trichome development in Arabidopsis. Planta 2020, 252, 68. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Mei, W.; Gong, M.; Zuo, W.; Bai, H.; Dai, H. Antibacterial Activity of the Flavonoids from Dalbergia odorifera on Ralstonia solanacearum. Molecules 2011, 16, 9775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Halim, V.A.; Vess, A.; Scheel, D.; Rosahl, S. The role of salicylic acid and jasmonic acid in pathogen defence. Plant Biol. 2006, 8, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.; Prats, E.; Pierre, S.; Hall, M.; Hebelstrup, K.H. Integrating nitric oxide into salicylic acid and jasmonic acid/ ethylene plant defense pathways. Front. Plant Sci. 2013, 4, 215. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Liu, H.; Jiang, D.; Wang, L.; Jiang, Y.; Tang, S.; Hou, X.; Han, X.; Liu, Z.; Zhang, M.; et al. Paecilomyces variotii extracts (ZNC) enhance plant immunity and promote plant growth. Plant Soil 2019, 441, 383–397. [Google Scholar] [CrossRef]
- Peng, C.; Zhang, A.; Wang, Q.; Song, Y.; Zhang, M.; Ding, X.; Li, Y.; Geng, Q.; Zhu, C. Ultrahigh-activity immune inducer from Endophytic Fungi induces tobacco resistance to virus by SA pathway and RNA silencing. BMC Plant Biol. 2020, 20, 169. [Google Scholar] [CrossRef] [Green Version]
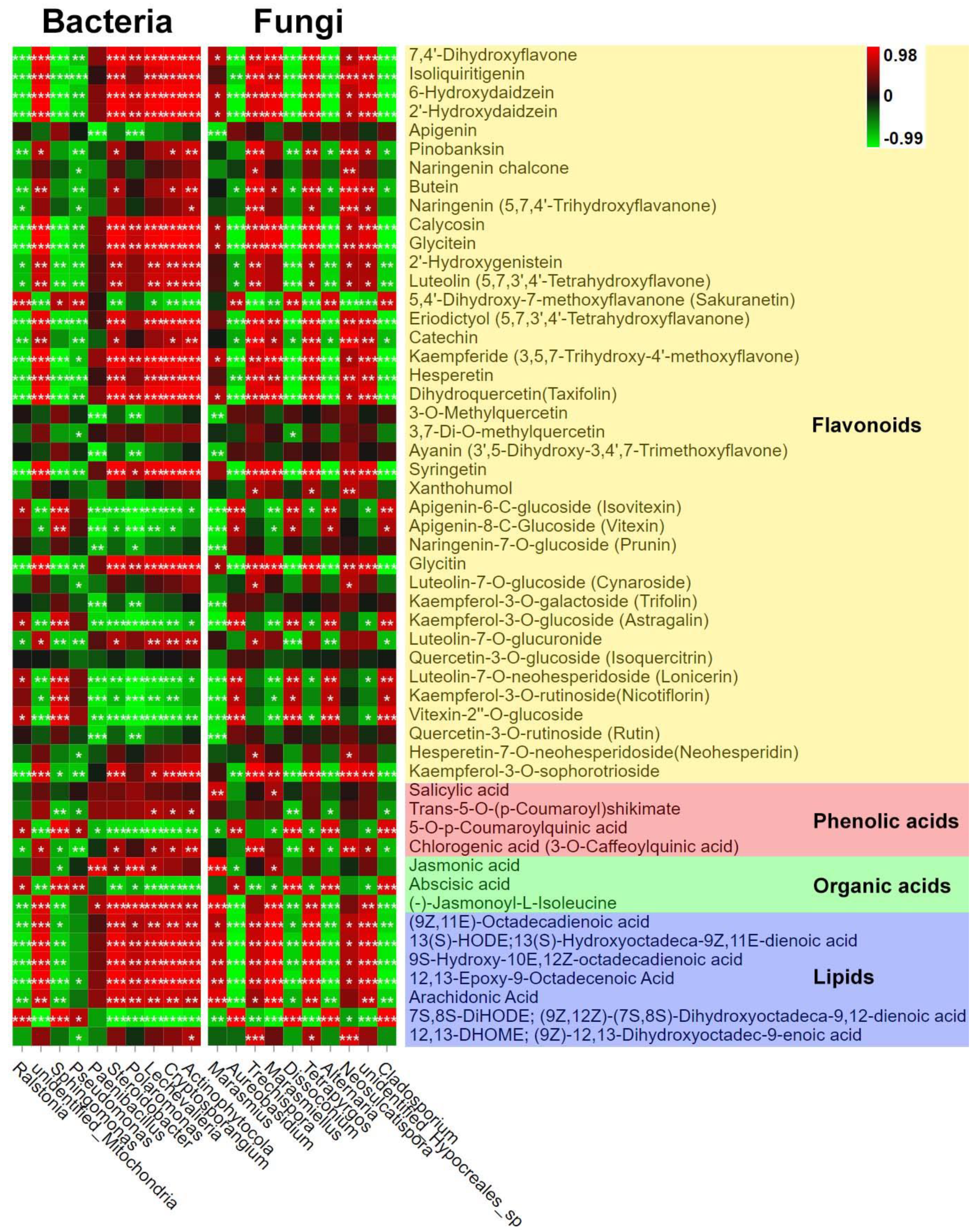
| Sample ID | Bacteria | Fungi | ||||
|---|---|---|---|---|---|---|
| Clean Tags | Average Length (nt) | OTU Number | Clean Tags | Average Length (nt) | OTU Number | |
| Leaf.CK1 | 104,742 | 374 | 362 | 84,184 | 233 | 485 |
| Leaf.CK2 | 80,975 | 374 | 277 | 81,380 | 234 | 515 |
| Leaf.CK3 | 98,878 | 374 | 283 | 84,101 | 239 | 500 |
| Leaf.FR1 | 95,560 | 374 | 365 | 74,920 | 237 | 1038 |
| Leaf.FR2 | 96,290 | 375 | 418 | 77,076 | 235 | 951 |
| Leaf.FR3 | 91,267 | 375 | 459 | 82,657 | 237 | 868 |
| Root.CK1 | 90,050 | 382 | 964 | 85,607 | 267 | 402 |
| Root.CK2 | 92,448 | 384 | 1058 | 76,933 | 261 | 407 |
| Root.CK3 | 95,095 | 392 | 988 | 79,489 | 264 | 504 |
| Root.FR1 | 77,247 | 381 | 906 | 75,696 | 279 | 706 |
| Root.FR2 | 85,075 | 379 | 803 | 68,124 | 267 | 895 |
| Root.FR3 | 97,973 | 378 | 803 | 87,868 | 272 | 714 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, M.; Lu, J.; Yang, W.; Lu, M.; Wang, J.; Li, S.; Chen, Y.; Hu, L.; Wang, L. Metabarcoding and Metabolome Analyses Reveal Mechanisms of Leymus chinensis Growth Promotion by Fairy Ring of Leucocalocybe mongolica. J. Fungi 2022, 8, 944. https://doi.org/10.3390/jof8090944
Duan M, Lu J, Yang W, Lu M, Wang J, Li S, Chen Y, Hu L, Wang L. Metabarcoding and Metabolome Analyses Reveal Mechanisms of Leymus chinensis Growth Promotion by Fairy Ring of Leucocalocybe mongolica. Journal of Fungi. 2022; 8(9):944. https://doi.org/10.3390/jof8090944
Chicago/Turabian StyleDuan, Mingzheng, Jia Lu, Wenjing Yang, Meiling Lu, Jibin Wang, Suli Li, Yiting Chen, Lihua Hu, and Lingqiang Wang. 2022. "Metabarcoding and Metabolome Analyses Reveal Mechanisms of Leymus chinensis Growth Promotion by Fairy Ring of Leucocalocybe mongolica" Journal of Fungi 8, no. 9: 944. https://doi.org/10.3390/jof8090944
APA StyleDuan, M., Lu, J., Yang, W., Lu, M., Wang, J., Li, S., Chen, Y., Hu, L., & Wang, L. (2022). Metabarcoding and Metabolome Analyses Reveal Mechanisms of Leymus chinensis Growth Promotion by Fairy Ring of Leucocalocybe mongolica. Journal of Fungi, 8(9), 944. https://doi.org/10.3390/jof8090944







