Characterisation of Pythium aristosporum Oomycete—A Novel Pathogen Causing Rice Seedling Blight in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Pathogenicity of Pathogens
2.2. Identification of the Pathogens Recovered from Diseased Rice Seedlings
2.3. Biological Characteristics of P. aristosporum Isolates
2.4. Host Range Determination of P. aristosporum Isolates
2.5. Sensitivity of P. aristosporum Isolates to Fungicides
2.6. Efficacy of Metalaxyl + Propamocarb against Rice Seedling Blight Caused by P. aristosporum
2.7. Data Analysis
3. Results
3.1. Identification of Causal Organisms
3.2. Pathogenicity of P. aristosporum on Rice
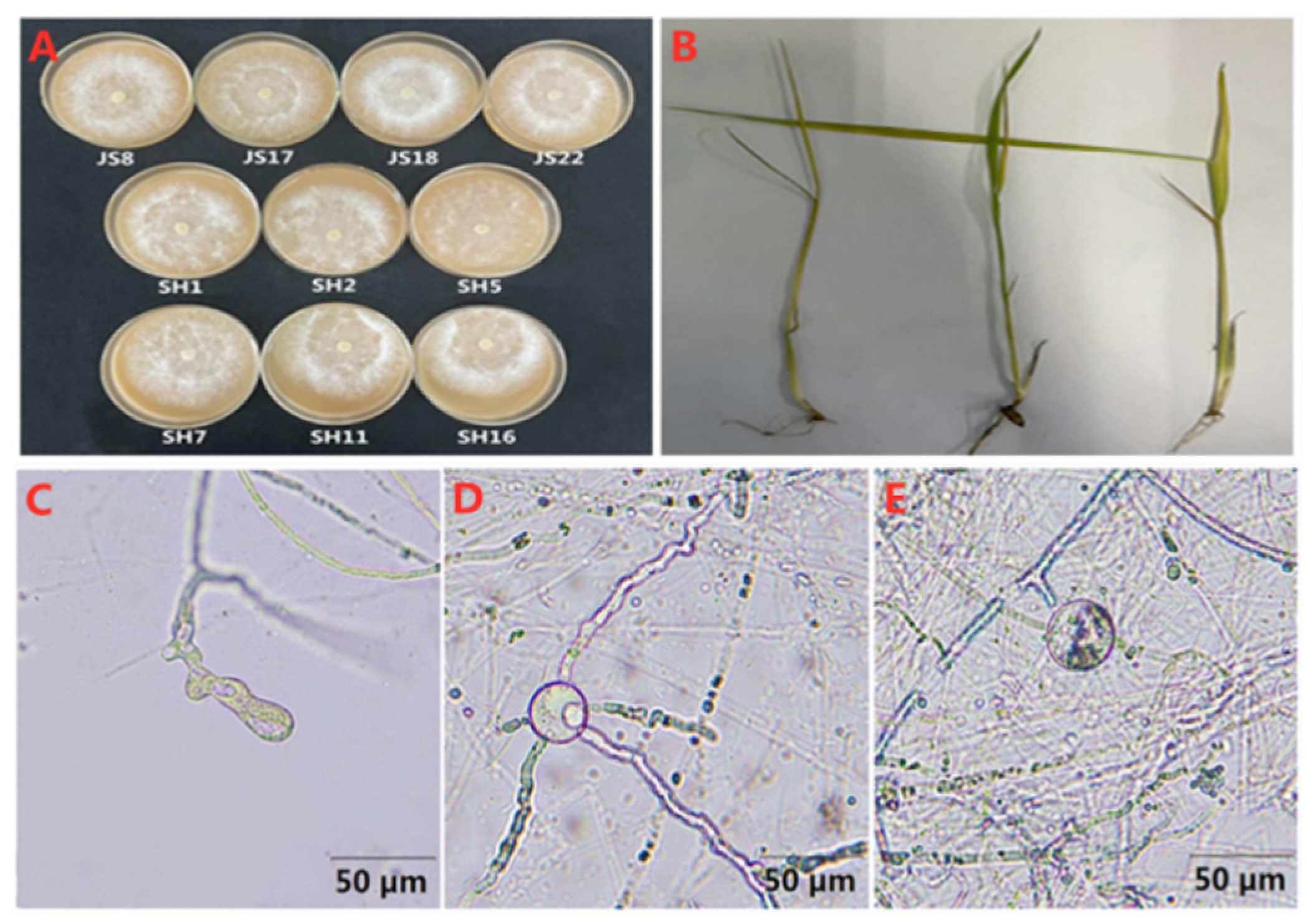
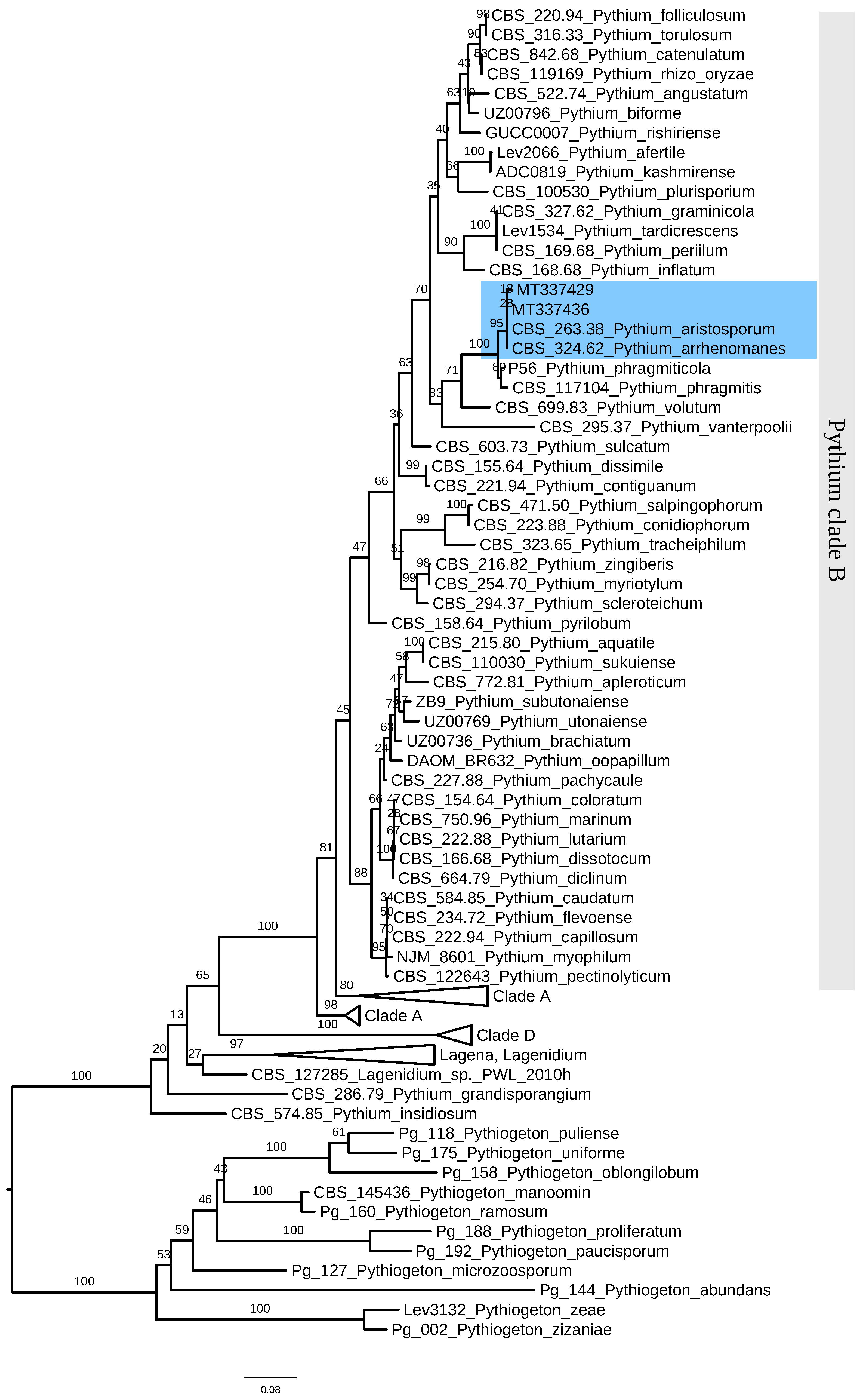
3.3. Identification of P. aristosporum
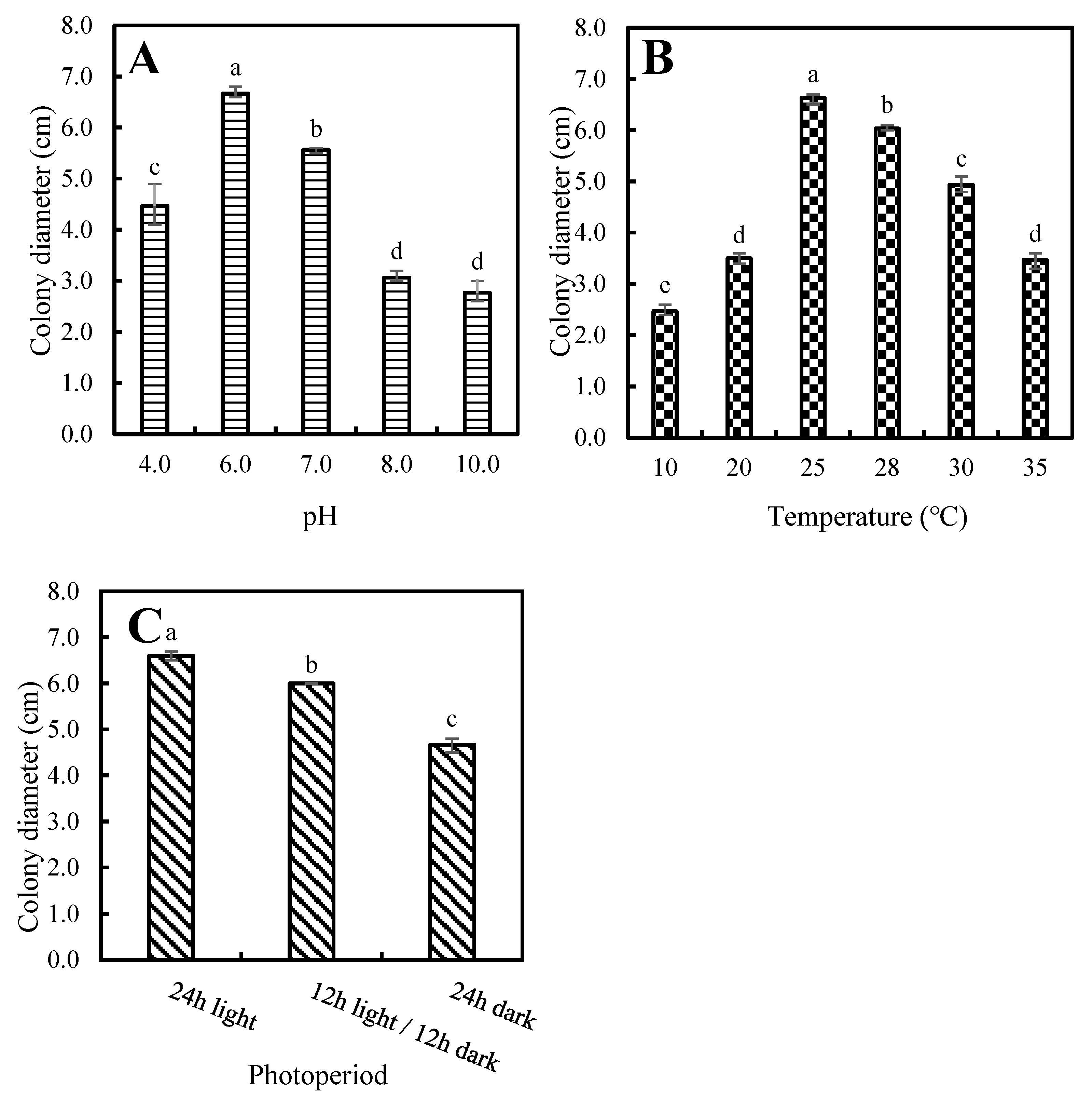
3.4. Biological Characteristics of P. aristosporum
3.5. Host Range Determination of P. aristosporum
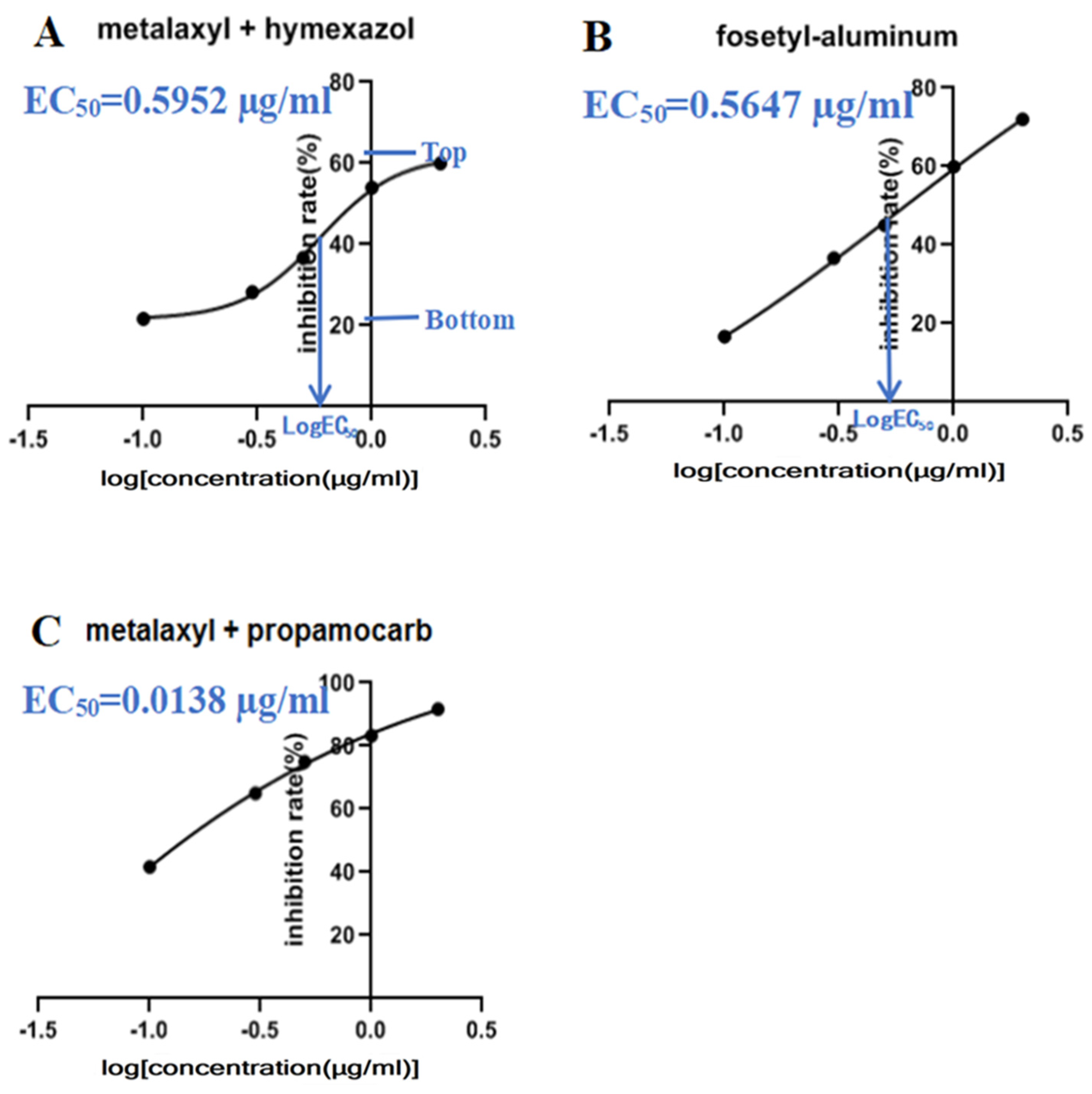
3.6. Efficacy of Chemical Fungicides
3.7. Efficacy of Metalaxyl + Propamocarb on Rice Seedling Blight Caused by P. aristosporum
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| No. | Isolates | ITS Regions | COX2 Gene |
|---|---|---|---|
| 1 | JS8 | OP050386 | OP103726 |
| 2 | JS17 | OP050387 | OP103721 |
| 3 | JS18 | OP076892 | OP103722 |
| 4 | JS22 | MT337429 | OP103727 |
| 5 | SH1 | MT337436 | OP103728 |
| 6 | SH2 | OP077086 | OP103723 |
| 7 | SH5 | OP076947 | OP103729 |
| 8 | SH7 | OP050388 | OP103720 |
| 9 | SH11 | OP076937 | OP103724 |
| 10 | SH16 | OP050389 | OP103725 |
References
- Yu, S.; Tian, L. Breeding major cereal grains through the lens of nutrition sensitivity. Mol Plant 2018, 11, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Shindell, D.; Faluvegi, G.; Kasibhatla, P.; Van, D.R. Spatial patterns of crop yield change by emitted pollutant. Earths Future 2019, 7, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Li, C.X.; Tan, K.F.; Liu, C.Z.; Xu, X.; Zhang, S.Y.; Wang, X.J.; Zhao, J.W.; Xiang, W.S. Characterization, phylogenetic analyses, and pathogenicity of Enterobacter cloacae on rice seedlings in Heilongjiang Province, China. Plant Dis. 2020, 104, 1601–1609. [Google Scholar] [CrossRef]
- Liu, H.L. Assessment of artificial selection in maize (Zea mays L.) and Asian rice (Oryza sativa L.) using QTL data. Genet. Resour. Crop Evol. 2017, 64, 1561–1568. [Google Scholar] [CrossRef]
- Meng, Y.; Hui, Z.B.; Wu, S.; Li, S.M.; Zheng, Z.J.; Zheng, F.; Wang, M.S.; Nan, P. Main rice diseases and their control in Heilongjiang Province. Mod. Agric. 2008, 3, 6–8. [Google Scholar]
- Motschenbacher, J.M.; Brye, K.R.; Anders, M.M.; Gbur, E.E. Long-term rice rotation, tillage, and fertility effects on near-surface chemical properties in a silt-loam soil. Nutr. Cycl. Agroecosyst. 2014, 100, 77–94. [Google Scholar] [CrossRef]
- Liu, F.; Mu, W.; Zhang, W.J.; Zhang, J. Control of rice blight on dry nursery seedling by fungicide and physiological regulation on rice seedling. Chin. J. Pestic. Sci. 2004, 6, 37–42. [Google Scholar] [CrossRef]
- Liu, J.X.; Cai, Y.N.; Jiang, W.Y.; Li, Y.G.; Zhang, Q.F.; Pan, H.Y. Population structure and genetic diversity of fungi causing rice seedling blight in Northeast China based on microsatellite markers. Plant Dis. 2020, 104, 868–874. [Google Scholar] [CrossRef]
- Li, W.X.; Li, H.Y.; Cai, D.L.; Sang, H.Y.; Chen, T.Y. The induced-resistance of harpin on rice seedling blight. J. Anhui Agric. Sci. 2016, 44, 186–187. [Google Scholar]
- Yu, W.Q.; Li, P.; Wang, X.; Yan, F.C.; Huang, X.W.; Chen, P.; Liu, W.Z.; Zheng, G. Bio-control of Paenibacillus terrae NK3-4 against rice seedling blight. Mol. Plant Breed. 2021, 41, 1–11. [Google Scholar]
- Wang, W.; Ma, S.H.; Hu, P.; Ji, Y.H.; Sun, F. Genome editing of rice eIF4G loci confers partial resistance to rice black-streaked dwarf virus. Viruses 2021, 13, 2100. [Google Scholar] [CrossRef] [PubMed]
- Dossou, B.; Silue, D. Rice pathogens intercepted on seeds originating from 11 African countries and from the USA. Seed Sci. Technol. 2018, 46, 31–40. [Google Scholar] [CrossRef]
- Riera-Ruiz, C.A.; Castro-Lara, J.; Feijoo, M.I.J.; Cevallos-Cevallos, J. Burkholderia gladioli can inhibit Burkholderia glumae in rice seedlings affected with bacterial panicle blight. Phytopathology 2018, 108, 142. [Google Scholar]
- Ma, B.; Wang, J.H.; Liu, C.Z.; Hu, J.F.; Tan, K.F.; Zhao, F.Y.; Yuan, M.; Zhang, J.H.; Gai, Z.J. Preventive effects of fluoro-substituted benzothiadiazole derivatives and chitosan oligosaccharide against the rice seedling blight induced by Fusarium oxysporum. Plants 2019, 8, 538. [Google Scholar] [CrossRef]
- Van, B.E.; Hofte, M. Pythium species from rice roots differ in virulence, host colonization and nutritional profile. BMC Plant Biol. 2013, 13, 203. [Google Scholar]
- Ling, Y.; Xia, J.; Koji, K.; Zhang, X.; Li, Z. First report of damping-off caused by Pythium arrhenomanes on rice in China. Plant Dis. 2018, 102, 2382–2383. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Chon, T.S.; Kim, J.; Seo, Y.S.; Heo, M. Comparative and bioinformatics analyses of pathogenic bacterial secretomes identified by mass spectrometry in Burkholderia species. J. Microbiol. 2017, 55, 568–582. [Google Scholar] [CrossRef]
- Park, J.; Lee, H.H.; Jung, H.; Seo, Y.S. Transcriptome analysis to understand the effects of the toxoflavin and tropolone produced by phytopathogenic Burkholderia on Escherichia coli. J. Microbiol. 2019, 57, 781–794. [Google Scholar] [CrossRef]
- Spurlock, T.N.; Rothrock, C.S.; Monfort, W.S.; Griffin, T.W. The distribution and colonization of soybean by Rhizoctonia solani AG11 in fields rotated with rice. Soil Biol. Biochem. 2016, 94, 29–36. [Google Scholar] [CrossRef]
- Gaire, S.P.; Zhou, X.G.; Jo, Y.K.; Shi, J. First Report of Rhizoctonia solani AG-4 Causing Seedling Disease in Rice. Plant Dis. 2020, 104, 1546. [Google Scholar] [CrossRef]
- Gaire, S.P.; Zhou, X.G.; Jo, Y.K. Sterile White Basidiomycete Fungus Marasmius graminum: A New Pathogen Causing Seedling Blight in Rice. Plant Dis. 2020, 105, 702. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Du, M.L.; Liu, P.; Tang, Y.Q.; Li, H.; Yuan, Q.H.; Ruan, Y.Z.; Meng, L.; Zhang, J.C.; Lin, M.; et al. Alternation of soil bacterial and fungal communities by tomato-rice rotation in Hainan Island in Southeast of China. Arch. Microbiol. 2021, 203, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Yan, T.M.; Xue, F.; Yang, L.Z.; Lu, P. Reduction of nitrogen fertilizer application under different crop rotation systems in paddy fields of Taihu Area. Chin. J. Eco Agric. 2011, 19, 24–31. [Google Scholar] [CrossRef]
- Jin, W.H.; Yang, J.S.; Hou, X.J.; Yao, R.J.; Yu, S.P.; Wang, X.P.; Xie, W.P. Effects of rotation systems on soil organic carbon and aggregates in light salinized farmland in north Jiangsu province. Soils 2013, 48, 1195–1201. [Google Scholar]
- Wu, Y.X.Y.; Jiang, Z.H.; Yang, J.P.; Lin, J.D.; Liu, Y.Z. Nitrogen content and enzyme activity in rhizosphere and non-rhizosphere soils of paddy field under maize-rice rotation and rice continuous mono-cropping. J. Plant Nutr. Fertil. 2019, 25, 535–543. [Google Scholar]
- Grijalba, P.E.; Palmucci, H.E.; Guillin, E. Identification and characterization of Pythium graminicola, causal agent of kikuyu yellows in Argentina. Trop. Plant Pathol. 2017, 42, 284–290. [Google Scholar] [CrossRef]
- Wang, S.; Sun, L.; Li, W.Q.; Liu, J.X.; Li, Y.G.; Wei, D. First report of seedling blight caused by Fusarium redolens on rice in Northeast China. Plant Dis. 2019, 103, 1418. [Google Scholar] [CrossRef]
- Feng, D.X.; Zheng, A.P.; Wang, S.Q.; Xiang, X.Z.; Li, P. Genetic diversity and pathogenicity variation of different Rhizoctonia solani isolates in Sichuan province. Acta Phytopathol. Sin. 2005, 35, 520–525. [Google Scholar]
- Wei, J.C. Handbook of Fungus Identification; Shanghai Scientific: Shanghai, China, 1979. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Lookabaugh, E.; Shew, B.; Cowger, C. Three Pythium species isolated from severely stunted wheat during an outbreak in North Carolina. Plant Health Prog. 2017, 18, 169–173. [Google Scholar] [CrossRef]
- Deborah, S.S.H.; Steven, A.N.; Michael, E.S.H. A COX2 molecular phylogeny of the Peronosporomycetes. Mycologia 2000, 92, 674–684. [Google Scholar]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Li, L.H.; Kageyama, K.; Kinoshita, N.; Yu, W.J.; Fukui, H. Development of bioassay for screening of resistant roses against root rot disease caused by Pythium helicoides Drechsler. J. Jpn. Soc. Hortic. Sci. 2007, 76, 79–84. [Google Scholar] [CrossRef][Green Version]
- Jiang, N.; Hu, F.Y.; Ye, Y.F. Pathogen identification of a new disease in Siraitia grosvenorii and screening of effective fungicides. Plant Prot. 2015, 41, 173–177. [Google Scholar]
- Ji, X.X.; Li, J.J.; Meng, Z.; Zhang, S.A.; Dong, B.; Qiao, K. Synergistic effect of combined application of a new fungicide fluopimomide with a biocontrol agent Bacillus methylotrophicus TA-1 for management of gray mold in tomato. Plant Dis. 2019, 103, 1991–1997. [Google Scholar] [CrossRef]
- Zhang, Y.K.; Zhai, H.W.; Liu, Z.F. Efficacy of 1 × 106 zoospores/g Pythium oligodendrum wettable powder against rice seedling blight. North Rice 2013, 43, 68–70. [Google Scholar]
- van der Plaats-Niterink, A.J. Monograph of the genus pythium. Stud. Mycol. 1981, 21, 1–242. [Google Scholar]
- Imolehin, E.D. Rice seed borne fungi and their effect on seed germination. Plant Dis. 1983, 67, 1334–1336. [Google Scholar] [CrossRef]
- Baek, I.; Kim, M.S.; Cho, B.K.; Mo, C.; Barnaby, J.Y.; McClung, A.M.; Oh, M. Selection of optimal hyperspectral wavebands for detection of discolored, diseased rice seeds. Appl. Sci. 2019, 9, 1027. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, X.H.; Han, Y.T.; Zhang, L.L.; Zhang, J.H. Studies on identification of pathogenicity of rice seedling blight in Heilongjiang province. J. Northeast Agric. Sci. 2015, 40, 75–78. [Google Scholar]
- Mostowfizadeh-Ghalamfarsa, R.; Banihashemi, Z. Identification of soil Pythium species in Fars province of Iran. Iran. J. Sci. Technol. Trans. A 2005, 29, 79–87. [Google Scholar]
- Alaei, H.; Rostami, F. Identification of cucumber damping-off based on morphological and molecular characterizations in Rafsanjan. Iran. J. Plant Pathol. 2013, 48, 177–182. [Google Scholar]
- Naima, B.; Najwa, B.; Souli, M. Pythium species associated with die-back apple trees and citrus gummosis in Tunisia. In Pythium Diagnosis, Diseases, and Management, 1st ed.; CRC Press: Boca Raton, FL, USA, 2020; Chapter 6. [Google Scholar]
- Klemsdal, S.S.; Herrero, M.L.; Wanner, L.A.; Lund, G.; Hermansen, A. PCR-based identification of Pythium spp. causing cavity spot in carrots and sensitive detection in soil samples. Plant Pathol. 2008, 57, 877–886. [Google Scholar] [CrossRef]
- Robideau, G.P.; de Cock, A.W.A.M.; Coffey, M.D.; Voglmayr, H.; Brouwer, H.; Bala, K.; Chitty, D.W.; Désaulniers, N.; Eggertson, Q.A.; Gachon, C.M.M.; et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 2011, 11, 1002–1011. [Google Scholar] [CrossRef]
- Rojas, J.A.; Jacobs, J.L.; Napieralski Jardine, D.J.; Malvick, D.K.; Markell, S.G.; Nelson, B.D.; Robertson, A.E.; Rupe, J.C.; Smith, D.L.; Sweets, L.E.; et al. Oomycete species associated with soybean seedlings in North America-Part I: Identification and pathogenicity characterisation. Phytopathology 2016, 107, 280–292. [Google Scholar] [CrossRef]
- Li, S.F. Analysis on major diseases during rice seedlings in Northeast China. Agric. Dev. Equip. 2015, 11, 124. [Google Scholar]
- Cai, S.J.; Xu, F.Z.; Zhang, X.Y.; Wang, K.C.; Wei, Q. Temporal and spatial variation of rice suitable growing region in Heilongjiang Province under climate warming. China Rural. Water Hydropower 2020, 448, 138–142. [Google Scholar]
- Wu, Y.; Sun, B.; Xu, L.X. Study on sulphur status in Heilongjiang paddy soil and effect of sulphur. Heilongjiang Agric. Sci. 2002, 5–7. [Google Scholar]
- Huang, S.W.; Su, X.Q. Biological studies on Pythium guiyangense, a fungal pathogen of mosquito larvae. Mygosystema 2007, 26, 380–388. [Google Scholar]
- Guo, M.; Zhou, Y.N.; Xiao, Q.; Li, B.D.; Liang, C. The pathogen causing Pythium root rot of Chinese cabbage. Mygosystema 2019, 38, 761–767. [Google Scholar]
- Li, J.X.; Zhang, Y.W. Biological characteristics and isolation of Pythium ultimum causing rot of Chinese cabbage. Australas. Plant Pathol. 2020, 49, 201–207. [Google Scholar] [CrossRef]
- Hendrix, F.F.; Campbell, W.A. Pythium as plant pathogens. Annu. Rev. Phytopathol. 1973, 11, 77–98. [Google Scholar] [CrossRef]
- Santosh, K.B. The pathogenicity of different Pythium spp. on aerobic rice seedlings. J. Bangladesh Soc. Agric. Sci. Technol. 2012, 9, 83–87. [Google Scholar]
- Kageyama, K. Molecular taxonomy and its application to ecological studies of Pythium species. J. Gen. Plant Pathol. 2014, 80, 314–326. [Google Scholar] [CrossRef]
- Vanterpool, T.C. Some species of Pythium parasitic on wheat in Canada and England. Ann. Appl. Biol. 1938, 25, 528–543. [Google Scholar] [CrossRef]
- Lipps, P.E.; Bruehl, G.W. Snow rot of winter wheat in Washington. Phytopathology 1978, 68, 1120–1127. [Google Scholar] [CrossRef]
- Kobriger, K.; Hagedorn, D.J. Additional Pythium species associated with the bean root rot complex in Wisconsin’ s central sands. Plant Dis. 1984, 68, 595–596. [Google Scholar]
- Iwaizako, C. Study on the ecology and control of konnyaku (Amorphophallus konjac C. Koch) root rot. Spec. Bull. Ibaraki Agric. Exp. Stn. 1987, 5, 1–114, (In Japanese with English summary). [Google Scholar]
- Titone, P.; Mocioni, M.; Garibaldi, A.; Gullino, M.L. Fungicide failure to control Pythium blight on turf grass in Italy. J. Plant Dis. Prot. 2009, 116, 55–59. [Google Scholar] [CrossRef]
- Jin, L.Y.; Li, M.S.; Wang, Z.W.; Shi, J.; Guo, N.; Liu, S.S.; Zhang, H.J. Resistance identification and genetic diversity analysis of American maize inbred lines to four pathogenic stalk rot diseases. J. Plant Genet. Resour. 2019, 20, 1428–1437. [Google Scholar]
- Mazzola, M.; Cook, R.J. Effects of fungal root pathogens on the population dynamics of biocontrol strains of Fluorescent pseudomonads in the wheat rhizosphere. Appl. Environ. Microbiol. 1991, 57, 2171–2178. [Google Scholar] [CrossRef] [PubMed]
- Mocioni, M. Prodotti fitosanitari registrati per l’ impiego su tappeto erboso in Italia: Situazione e prospettive. Inf. Fitopatol. 2001, 51, 38–41. [Google Scholar]

| Pathogens | No of Isolates | Frequency (%) |
| Fusarium oxysporum | 23 | 51.1 |
| Pythium aristosporum | 10 | 22.2 |
| F. redolens | 6 | 13.3 |
| F. solani | 3 | 6.7 |
| Rhizoctonia solani | 3 | 6.7 |
| No. | Isolate | Disease Index | Pathogenicity a |
|---|---|---|---|
| 1 | JS8 | 80.00 | H |
| 2 | JS17 | 58.33 | M |
| 3 | JS18 | 80.00 | H |
| 4 | JS22 | 86.67 | H |
| 5 | SH1 | 85.00 | H |
| 6 | SH2 | 56.67 | M |
| 7 | SH5 | 51.67 | M |
| 8 | SH7 | 58.33 | M |
| 9 | SH11 | 58.33 | M |
| 10 | SH16 | 58.33 | M |
| Isolates | Wheat | Maize | Sorghum | Alfalfa | Oats | White Clover |
|---|---|---|---|---|---|---|
| JS8 a | 56.67 (M) | 41.67 (W) | 21.67 (W) | 95.00 (H) | 88.33 (H) | 78.33(H) |
| JS17 a | 51.67 (M) | 36.67 (W) | 16.67 (W) | 95.00 (H) | 91.67 (H) | 71.67 (H) |
| JS18 a | 55.00 (M) | 45.00 (W) | 21.67 (W) | 95.00 (H) | 88.33 (H) | 75.00 (H) |
| JS22 a | 58.33 (M) | 48.33 (W) | 25.00 (W) | 98.33 (H) | 90.00 (H) | 75.00 (H) |
| SH1 a | 58.33 (M) | 46.67 (W) | 23.33 (W) | 96.67 (H) | 91.67 (H) | 71.67 (H) |
| SH2 a | 51.67 (M) | 36.67 (W) | 16.67 (W) | 95.00 (H) | 83.33 (H) | 66.67 (H) |
| SH5 a | 51.67 (M) | 36.67 (W) | 16.67 (W) | 91.67 (H) | 85.00 (H) | 66.67 (H) |
| SH7 a | 51.67 (M) | 36.67 (W) | 16.67 (W) | 95.00 (H) | 86.67 (H) | 66.67 (H) |
| SH11 a | 51.67 (M) | 36.67 (W) | 16.67 (W) | 95.00 (H) | 88.33 (H) | 73.33 (H) |
| SH16 a | 51.67 (M) | 36.67 (W) | 16.67 (W) | 95.00 (H) | 86.67 (H) | 66.67 (H) |
| Fungicide | Effective Dose (μg/mL) | Control Efficacy (%) a | Plant Height (cm) a | Root Length (cm) a | Fresh Weight (g) a |
|---|---|---|---|---|---|
| Metalaxyl + propamocarb (25% WP) | 313 | 84.1 ± 0.05 a | 12.7 ± 0.05 a | 4.6 ± 0.03 a | 0.235 ± 0.001 a |
| 250 | 80.4 ± 0.29 b | 12.1 ± 0.06 b | 4.5 ± 0.04 a | 0.230 ± 0.000 a | |
| 208 | 75.9 ± 0.05 c | 10.5 ± 0.03 c | 4.2 ± 0.03 b | 0.221 ± 0.001 b | |
| Control b | - | - | 8.9 ± 0.04 d | 3.7 ± 0.07 c | 0.176 ± 0.004 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Zhang, R.; Xu, C.; Liu, C.; Zheng, Y.; Zhang, X.; Liu, S.; Li, Y. Characterisation of Pythium aristosporum Oomycete—A Novel Pathogen Causing Rice Seedling Blight in China. J. Fungi 2022, 8, 890. https://doi.org/10.3390/jof8090890
Liu J, Zhang R, Xu C, Liu C, Zheng Y, Zhang X, Liu S, Li Y. Characterisation of Pythium aristosporum Oomycete—A Novel Pathogen Causing Rice Seedling Blight in China. Journal of Fungi. 2022; 8(9):890. https://doi.org/10.3390/jof8090890
Chicago/Turabian StyleLiu, Jinxin, Ruisi Zhang, Chuzhen Xu, Chunlai Liu, Yanyan Zheng, Xue Zhang, Shasha Liu, and Yonggang Li. 2022. "Characterisation of Pythium aristosporum Oomycete—A Novel Pathogen Causing Rice Seedling Blight in China" Journal of Fungi 8, no. 9: 890. https://doi.org/10.3390/jof8090890
APA StyleLiu, J., Zhang, R., Xu, C., Liu, C., Zheng, Y., Zhang, X., Liu, S., & Li, Y. (2022). Characterisation of Pythium aristosporum Oomycete—A Novel Pathogen Causing Rice Seedling Blight in China. Journal of Fungi, 8(9), 890. https://doi.org/10.3390/jof8090890






