Coccidioides Species: A Review of Basic Research: 2022
Abstract
:1. Introduction
2. Coccidioides Spp.
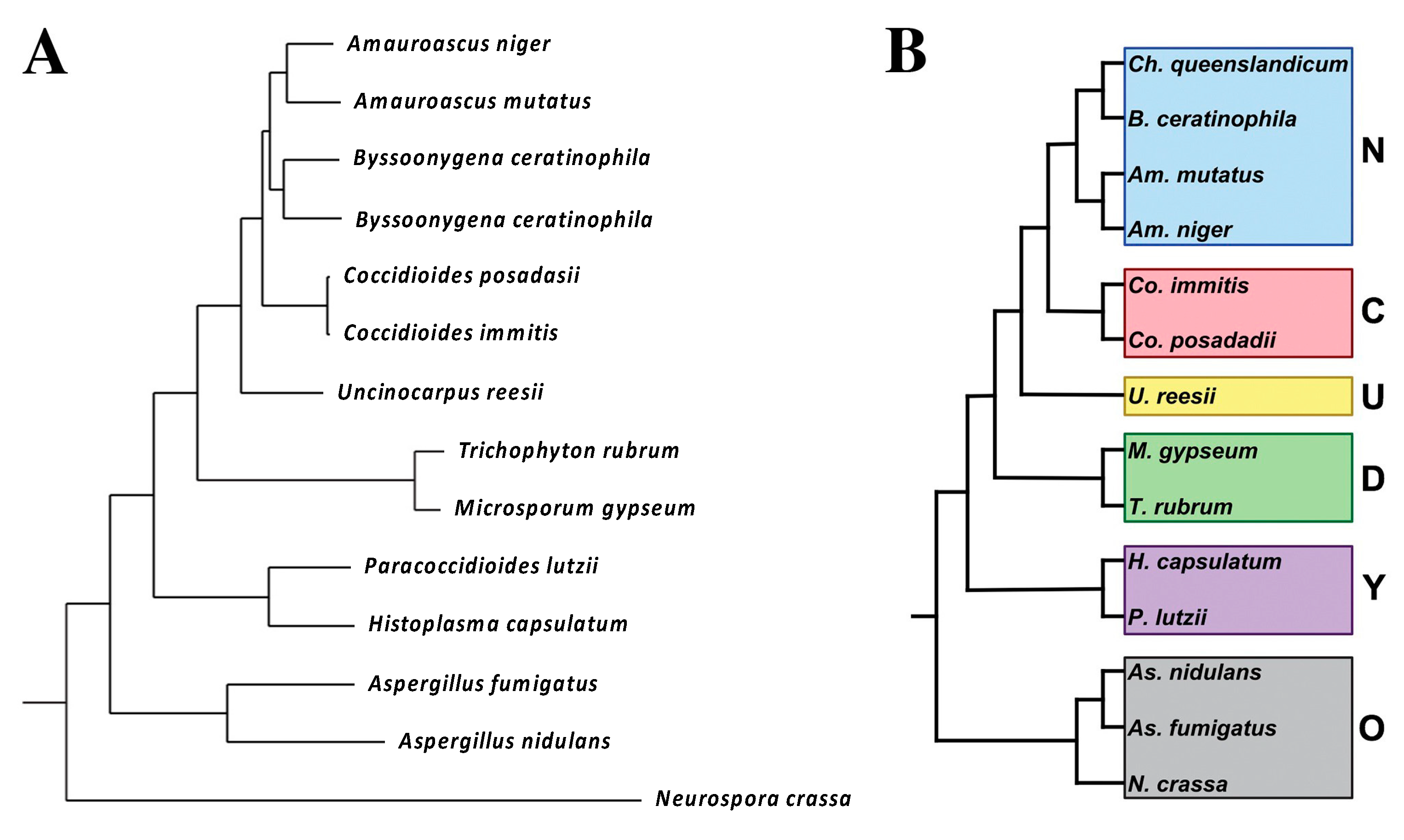
2.1. Taxonomy
C. Immitis and C. Posadasii
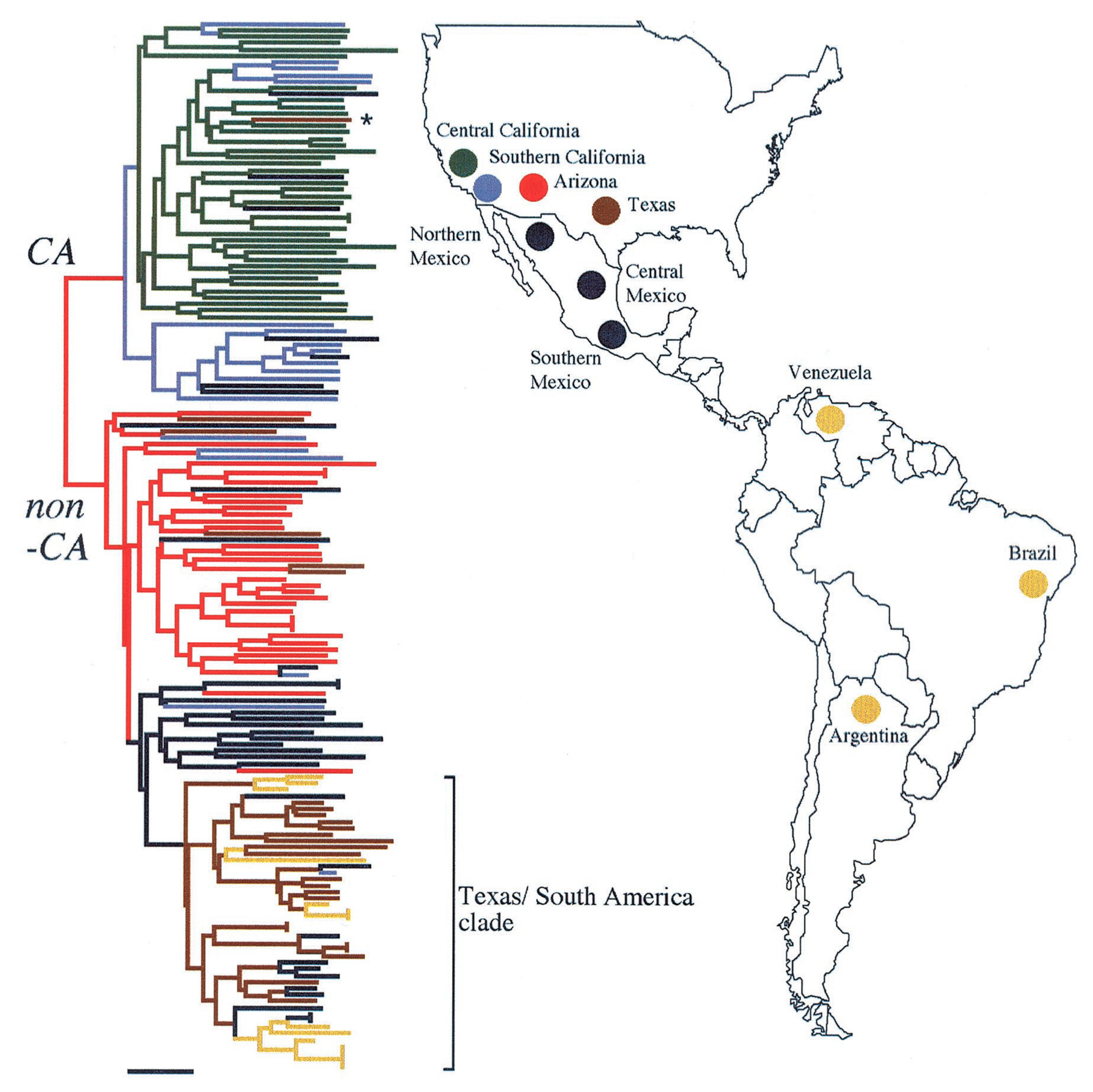
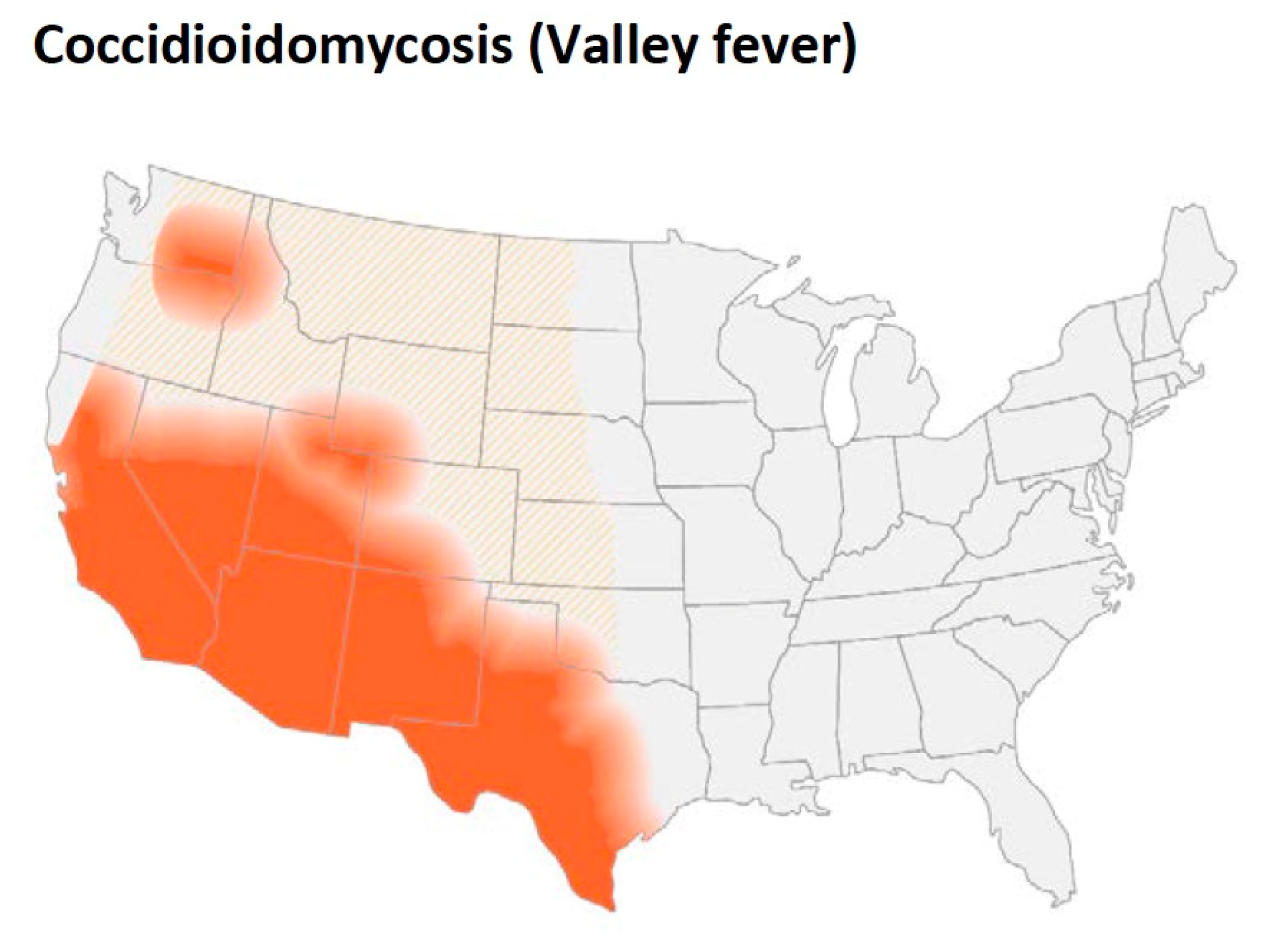
3. Geographic Distribution and Ecology
4. Culture, Detection and Morphology of Mycelia and Spherules
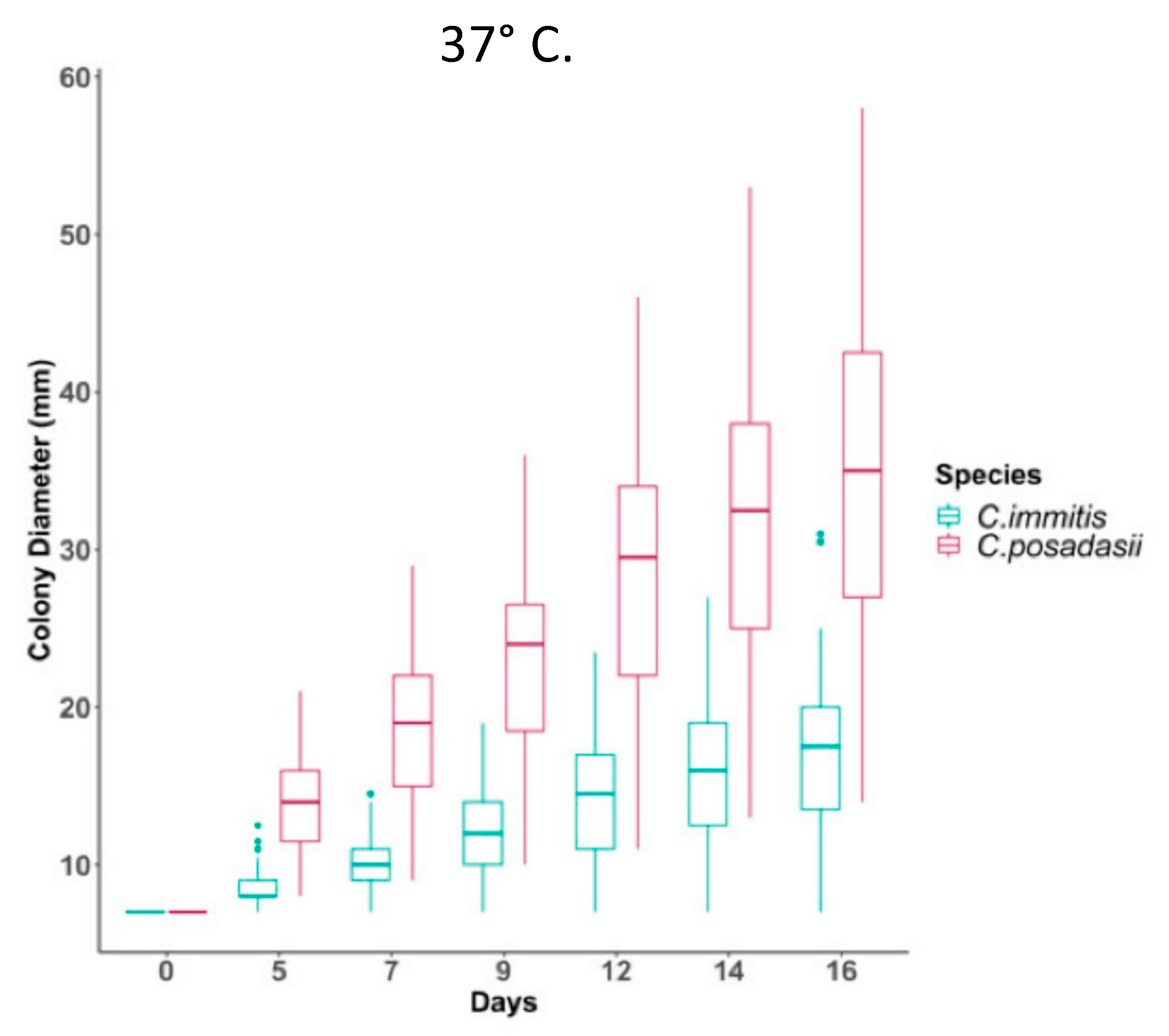
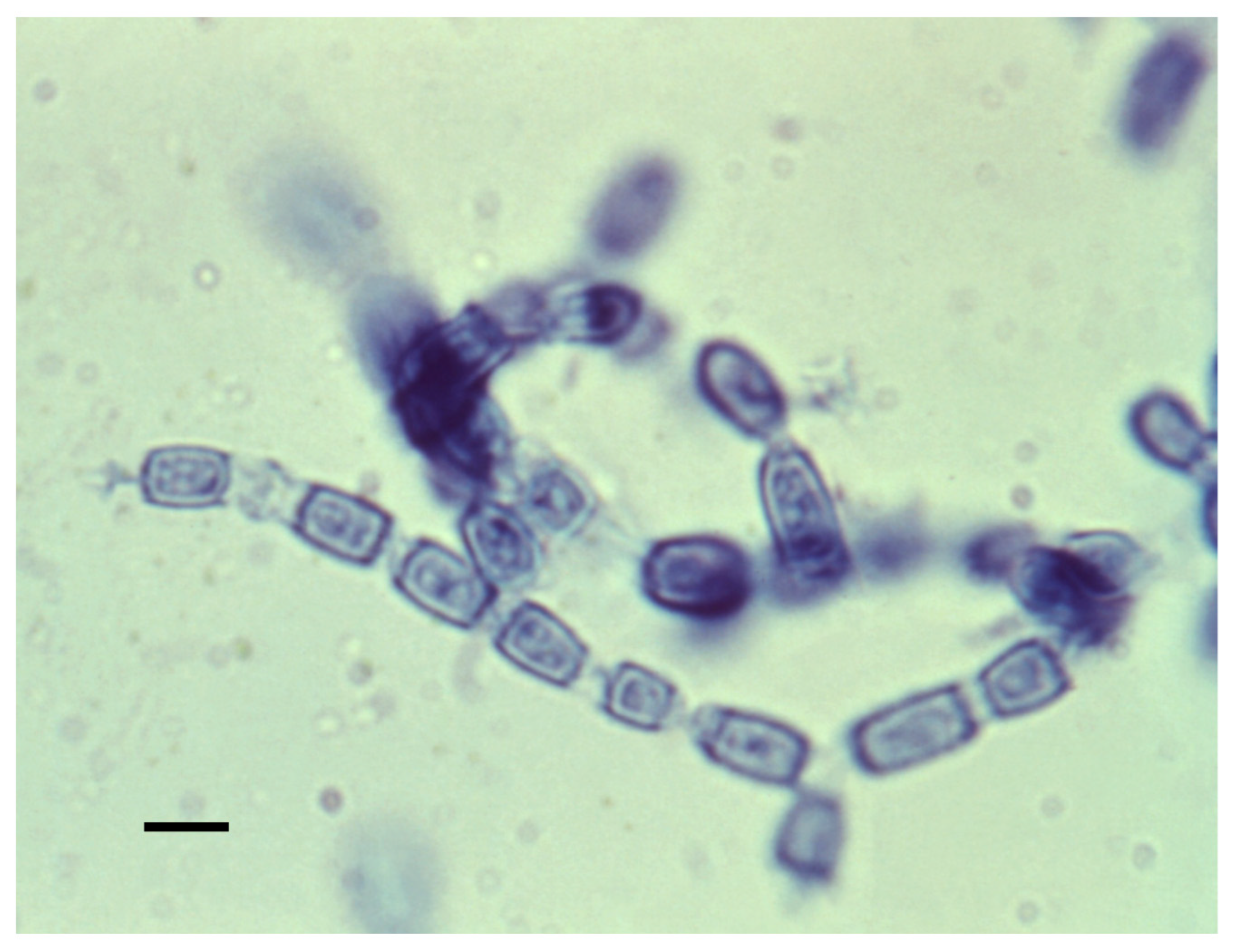
4.1. Mycelia and Arthroconidia
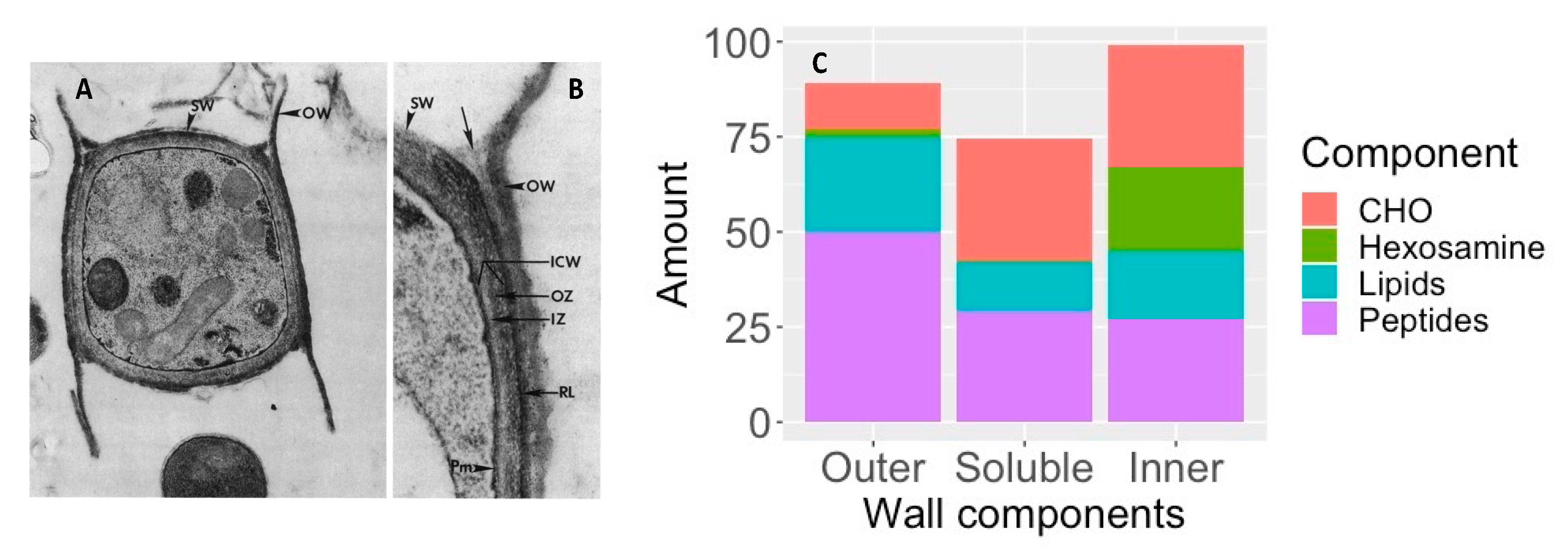
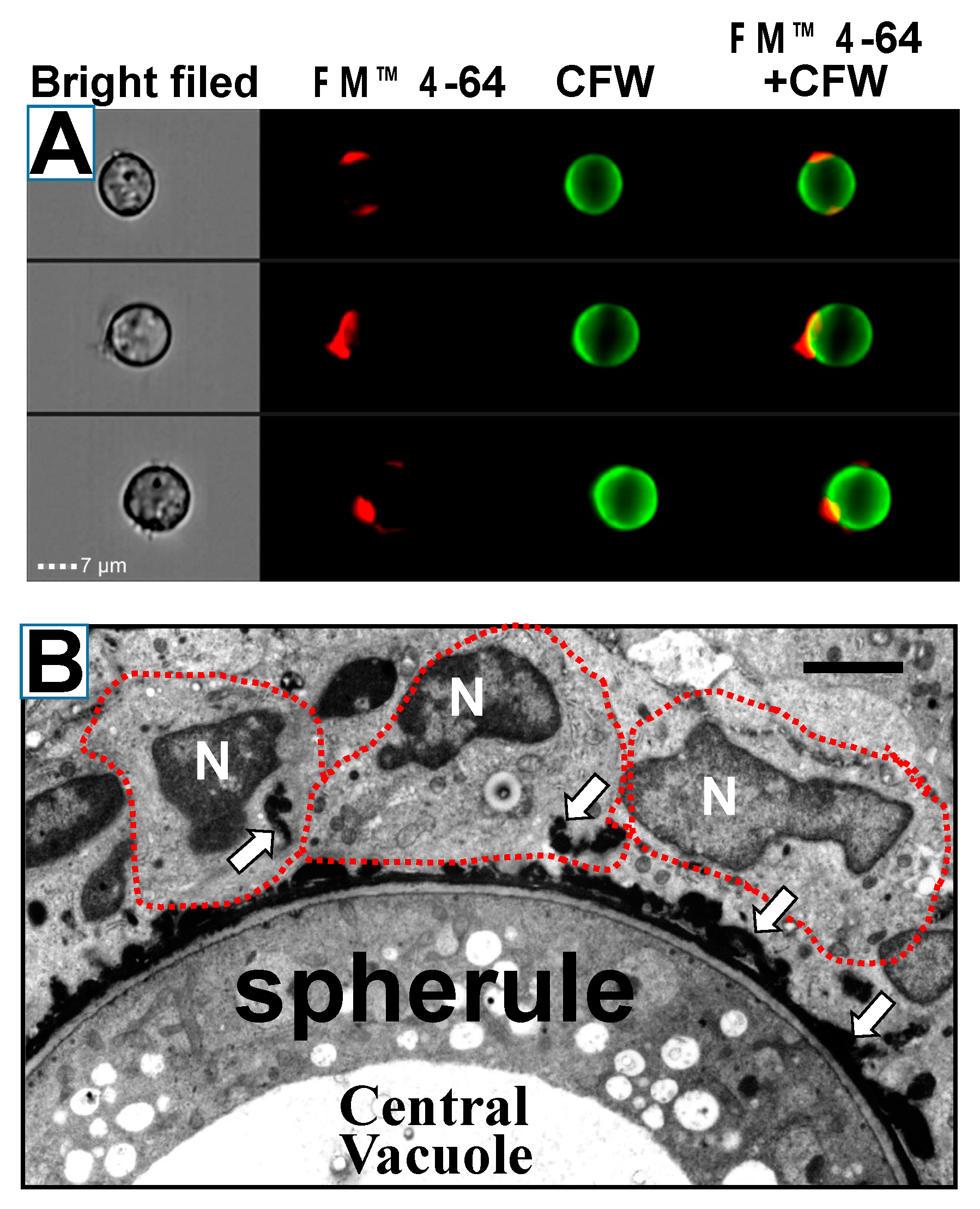
4.2. Spherules
5. Antigens
5.1. Dermal Hypersensitivity or Skin Test Antigens
5.2. Characterization and Purification of Antigens from Coccidioidin
5.3. Complement Fixation Antigen
5.4. The Precipitin Antigen
5.5. Purification of Antigens Based on the Huppert System
5.6. Antigens for In Vitro Measures of T-Cell Mediated Immunity
6. Genome Sequences
6.1. Gene Annotation by Proteomics
6.2. Transposable Elements
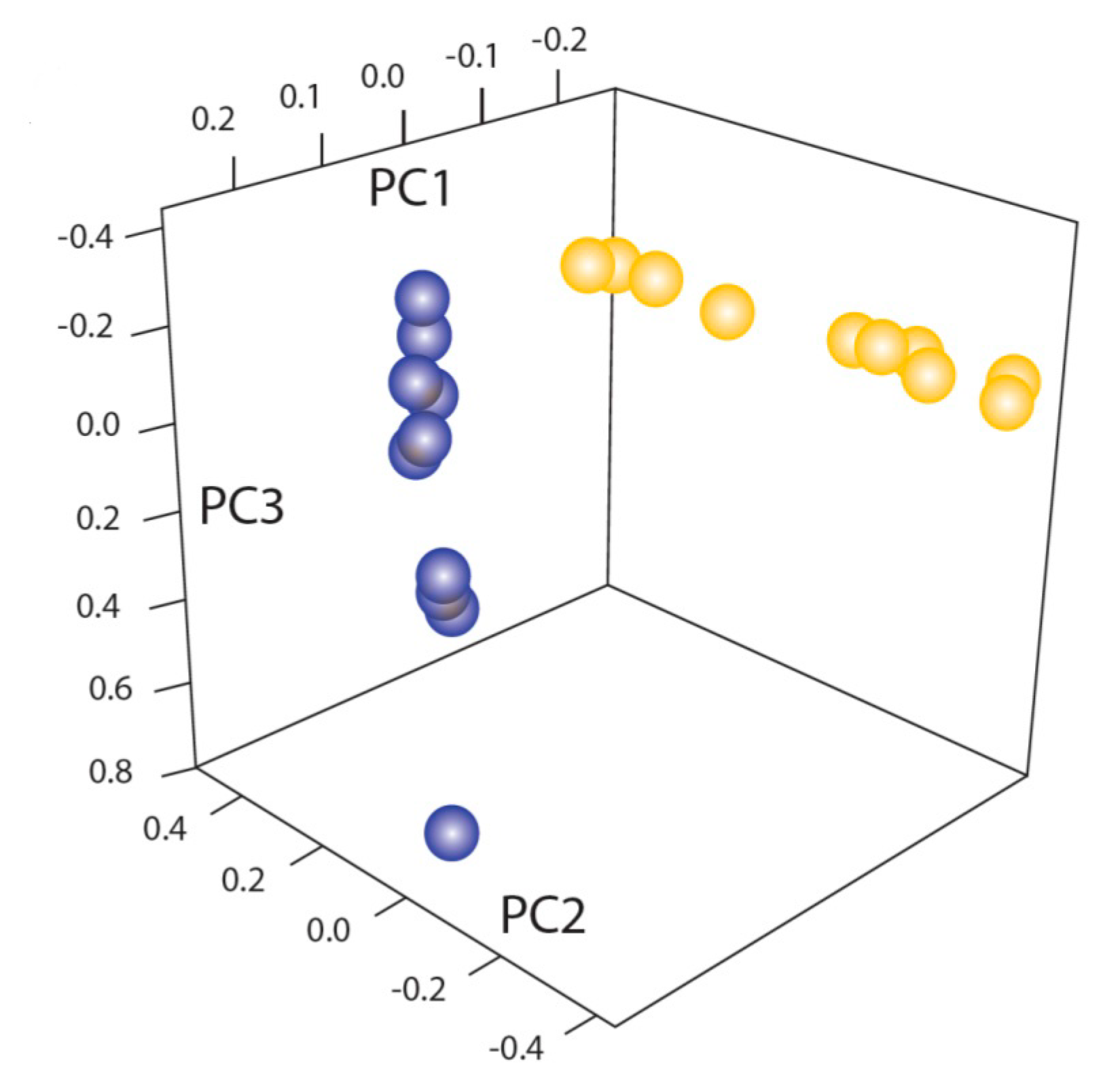
6.3. Resequencing and Phylogenomics
6.4. Recombination and Sex
7. Molecular Analysis of Spherules and Virulence-Associated Genes
7.1. Individual Genes
7.1.1. Spherule Outer Wall Glycoprotein
7.1.2. Urease and Ureidoglycolate Hydrolase
7.1.3. Metalloprotease
7.1.4. Melanin
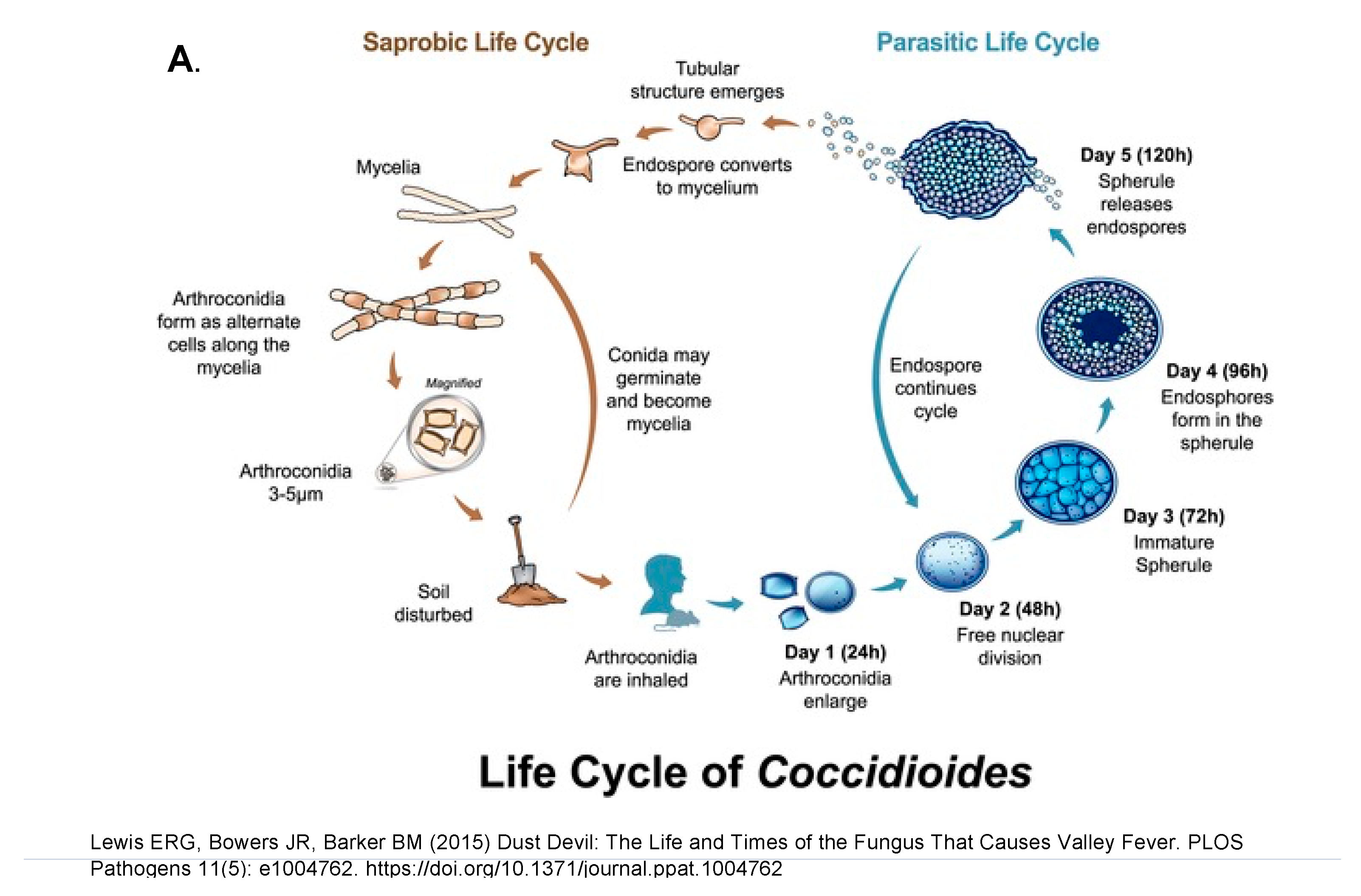
8. Comparison of the Saprobic and the Parasitic Phases
8.1. Transcriptional Studies
8.2. Comparison of Mycelia and Spherules by Proteomics
8.3. Comparison of the Metabolomics of Mycelia and Spherules
8.4. Integrating Data about Mycelia and Spherules
9. Genetic Manipulation
9.1. Engineered Avirulent Mutants
9.2. Pathogenicity of Naturally Occurring Coccidioides Spp. Strains
10. Strain Availability
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- CDC. Increase in reported coccidioidomycosis—United States, 1998–2011. Morb. Mortal. Wkly. Rep. 2013, 62, 217–221. [Google Scholar]
- Hector, R.F.; Rutherford, G.W.; Tsang, C.A.; Erhart, L.M.; McCotter, O.; Anderson, S.M.; Komatsu, K.; Tabnak, F.; Vugia, D.J.; Yang, Y.; et al. The public health impact of coccidioidomycosis in Arizona and California. Int. J. Environ. Res. Public Health 2011, 8, 1150–1173. [Google Scholar] [CrossRef]
- Valdivia, L.; Nix, D.; Wright, M.; Lindberg, E.; Fagan, T.; Lieberman, D.; Stoffer, T.P.; Ampel, N.M.; Galgiani, J.N. Coccidioidomycosis as a common cause of community-acquired pneumonia. Emerg. Inf. Dis. 2006, 12, 958–962. [Google Scholar] [CrossRef]
- Grizzle, A.J.; Wilson, L.; Nix, D.E.; Galgiani, J.N. Clinical and Economic Burden of Valley Fever in Arizona: An Incidence-Based Cost-of-Illness Analysis. Open Forum Infect. Dis. 2021, 8, ofaa623. [Google Scholar] [CrossRef] [PubMed]
- Deresinski, S.; Mirels, L.F. Coccidioidomycosis: What a long strange trip it’s been. Med. Mycol. 2019, 57, S3–S15. [Google Scholar] [CrossRef]
- Deresinski, S.C. History of coccidioidomycosis: “dust to dust”. In Coccidioidomycosis. A Text; Stevens, D.A., Ed.; Plenum Medical Book Company: New York, NY, USA, 1980; pp. 1–20. [Google Scholar]
- Ophuls, W. Futher observations on a pathogenic mold formerly described as a protozona (Coccidioides immitis, Coccidioides pyogenes). J. Exp. Med. 1905, 6, 443–485. [Google Scholar] [CrossRef] [PubMed]
- Bowman, B.H.; Taylor, J.W.; White, T.J. Molecular evolution of the fungi: Human pathogens. Mol. Biol. Evol. 1992, 9, 893–904. [Google Scholar]
- Guarro, J.; Gené, J.; Stchigel, A.M. Developments in Fungal Taxonomy. Clin. Microbiol. Rev. 1999, 12, 454–500. [Google Scholar] [CrossRef]
- De Hoog, G.S.; Bowman, B.; Graser, Y.; Haase, G.; El Fari, M.; Gerrits van den Ende, A.H.; Melzer-Krick, B.; Untereiner, W.A. Molecular phylogeny and taxonomy of medically important fungi. Med. Mycol. 1998, 36 (Suppl. 1), 52–56. [Google Scholar]
- Pan, S.; Sigler, L.; Cole, G.T. Evidence for a phylogenetic connection between Coccidioides immitis and Uncinocarpus reesii (Onygenaceae). Microbiology 1994, 140, 1481–1494. [Google Scholar] [CrossRef]
- Untereiner, W.A.; Scott, J.A.; Naveau, F.A.; Sigler, L.; Bachewich, J.; Angus, A. The Ajellomycetaceae, a new family of vertebrate-associated Onygenales. Mycologia 2004, 96, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, Z.; Gao, L.; Hao, B. A fungal phylogeny based on 82 complete genomes using the composition vector method. BMC Evol. Biol. 2009, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Whiston, E.; Taylor, J.W. Comparative Phylogenomics of Pathogenic and Nonpathogenic Species. G3 2015, 6, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Whiston, E.; Zhang Wise, H.; Sharpton, T.J.; Jui, G.; Cole, G.T.; Taylor, J.W. Comparative transcriptomics of the saprobic and parasitic growth phases in Coccidioides spp. PLoS ONE 2012, 7, e41034. [Google Scholar] [CrossRef] [PubMed]
- Carlin, A.F.; Beyhan, S.; Peña, J.F.; Stajich, J.E.; Viriyakosol, S.; Fierer, J.; Kirkland, T.N. Transcriptional Analysis of Coccidioides immitis Mycelia and Spherules by RNA Sequencing. JoF 2021, 7, 366. [Google Scholar] [CrossRef]
- Garcia Garces, H.; Hamae Yamauchi, D.; Theodoro, R.C.; Bagagli, E. PRP8 Intein in Onygenales: Distribution and Phylogenetic Aspects. Mycopathologia 2020, 185, 37–49. [Google Scholar] [CrossRef]
- Sil, A.; Andrianopoulos, A. Thermally Dimorphic Human Fungal Pathogens—Polyphyletic Pathogens with a Convergent Pathogenicity Trait. Cold Spring Harb. Perspect. Med. 2014, 5, a019794. [Google Scholar] [CrossRef]
- Koufopanou, V.; Burt, A.; Taylor, J.W. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 1997, 94, 5478–5482. [Google Scholar] [CrossRef]
- Burt, A.; Carter, D.; Koenig, G.; Whute, T.; Taylor, J.W. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc. Natl. Acad. Sci. USA 1996, 93, 700–773. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Koenig, G.L.; White, T.J.; San-Blas, G.; Negroni, R.; Alvarez, I.G.; Wanke, B.; Taylor, J.W. Biogeographic range expansion into South America by Coccidioides immitis mirrors New world patterns of human migration. Proc. Natl. Acad. Sci. USA 2001, 98, 4558–4562. [Google Scholar] [CrossRef]
- Fisher, M.C.; Koenig, G.L.; White, T.J.; Taylor, J.W. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 2002, 94, 73–84. [Google Scholar] [CrossRef]
- Mead, H.L.; Hamm, P.S.; Shaffer, I.N.; Teixeira, M.M.; Wendel, C.S.; Wiederhold, N.P.; Thompson, G.R., 3rd; Muniz-Salazar, R.; Castanon-Olivares, L.R.; Keim, P.; et al. Differential Thermotolerance Adaptation between Species of Coccidioides. J. Fungi 2020, 6, 366. [Google Scholar] [CrossRef]
- Barker, B.M.; Tabor, J.A.; Shubitz, L.F.; Perrill, R.; Orbach, M.J. Detection and phylogenetic analysis of Coccidioides posadasii in Arizona soil samples. Fungal Ecol. 2012, 5, 163–176. [Google Scholar] [CrossRef]
- Swatek, F.E. Ecology of Coccidioides immitis. Mycopathol. Mycol. Appl. 1970, 41, 3–12. [Google Scholar] [CrossRef]
- Smith, C.E. Diagnosis of pulmonary coccidioidal infections. Calif. Med. 1951, 75, 385–394. [Google Scholar] [PubMed]
- Edwards, P.Q.; Palmer, C.E. Prevalence of sensitivity to coccidioidin, with special reference to specific and nonspecific reactions to coccidioidin and to histoplasmin. Dis. Chest 1957, 31, 35–60. [Google Scholar] [CrossRef]
- Huckabone, S.E.; Gulland, F.M.; Johnson, S.M.; Colegrove, K.M.; Dodd, E.M.; Pappagianis, D.; Dunkin, R.C.; Casper, D.; Carlson, E.L.; Sykes, J.E.; et al. Coccidioidomycosis and other systemic mycoses of marine mammals stranding along the central California, USA coast: 1998–2012. J. Wildl. Dis. 2015, 51, 295–308. [Google Scholar] [CrossRef]
- Johnson, S.M.; Carlson, E.L.; Fisher, F.S.; Pappagianis, D. Demonstration of Coccidioides immitis and Coccidioides posadasii DNA in soil samples collected from Dinosaur National Monument, Utah. Med. Mycol. 2014, 52, 610–617. [Google Scholar] [CrossRef]
- Marsden-Haug, N.; Goldoft, M.; Ralston, C.; Limaye, A.P.; Chua, J.; Hill, H.; Jecha, L.; Thompson, G.R., 3rd; Chiller, T. Coccidioidomycosis acquired in Washington State. Clin. Infect. Dis. 2013, 56, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Marsden-Haug, N.; Hill, H.; Litvintseva, A.P.; Engelthaler, D.M.; Driebe, E.M.; Roe, C.C.; Ralston, C.; Hurst, S.; Goldoft, M.; Gade, L.; et al. Coccidioides immitis identified in soil outside of its known range—Washington, 2013. Morb. Mortal. Wkly. Rep. 2014, 63, 450. [Google Scholar]
- Litvintseva, A.P.; Marsden-Haug, N.; Hurst, S.; Hill, H.; Gade, L.; Driebe, E.M.; Ralston, C.; Roe, C.; Barker, B.M.; Goldoft, M.; et al. Valley fever: Finding new places for an old disease: Coccidioides immitis found in Washington State soil associated with recent human infection. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2015, 60, e1–e3. [Google Scholar] [CrossRef]
- Kollath, D.R.; Teixeira, M.M.; Funke, A.; Miller, K.J.; Barker, B.M. Investigating the Role of Animal Burrows on the Ecology and Distribution of Coccidioides spp. in Arizona Soils. Mycopathologia 2020, 185, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Emmons, C.W.; Ashburn, L.L. The isolation of Haplosporangium parvum n. sp. and Coccidioides immitis from wild rodents. Their relationship to coccidioidomycosis. Public Health Rep. 1942, 57, 1715–1727. [Google Scholar] [CrossRef]
- Kollath, D.R.; Mihalejevic, J.R.; Barker, B.M. PM10 and Other Climatic Variables Are Important Predictors of Seasonal Variability of Coccidioidomycosis in Arizona. Microbiol. Spectr. 2022, 10, e0148321. [Google Scholar] [CrossRef]
- Weaver, E.A.; Kolivras, K.N. Investigating the Relationship Between Climate and Valley Fever (Coccidioidomycosis). Ecohealth 2018, 15, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Coopersmith, E.J.; Bell, J.E.; Benedict, K.; Shriber, J.; McCotter, O.; Cosh, M.H. Relating coccidioidomycosis (valley fever) incidence to soil moisture conditions. Geohealth 2017, 1, 51–63. [Google Scholar] [CrossRef]
- Elconin, A.F.; Egeberg, R.O.; Egeberg, M.C. Significance of soil salinity on the ecology of Coccidioides immitis. J. Bacteriol. 1964, 87, 500–503. [Google Scholar] [CrossRef]
- Maddy, K.T. Observations on Coccidioides immitis found growing naturally in soil. Ariz. Med. 1965, 22, 281–288. [Google Scholar]
- Lauer, A.; Etyemezian, V.; Nikolich, G.; Kloock, C.; Arzate, A.F.; Sadiq Batcha, F.; Kaur, M.; Garcia, E.; Mander, J.; Kayes Passaglia, A. Valley Fever: Environmental Risk Factors and Exposure Pathways Deduced from Field Measurements in California. Int. J. Environ. Res. Public Health 2020, 17, 5285. [Google Scholar] [CrossRef]
- McCurdy, S.A.; Portillo-Silva, C.; Sipan, C.L.; Bang, H.; Emery, K.W. Risk for Coccidioidomycosis among Hispanic Farm Workers, California, USA, 2018. Emerg. Infect. Dis. 2020, 26, 1430–1437. [Google Scholar] [CrossRef]
- Chow, N.A.; Kangiser, D.; Gade, L.; McCotter, O.Z.; Hurst, S.; Salamone, A.; Wohrle, R.; Clifford, W.; Kim, S.; Salah, Z.; et al. Factors Influencing Distribution of Coccidioides immitis in Soil, Washington State, 2016. Msphere 2021, 6, e0059821. [Google Scholar] [CrossRef] [PubMed]
- Litvintseva, A.P.; Chow, N.A.; Salah, Z. The Association between Coccidioides immitis and Rodent Habitats in Washington State Remains Unresolved. Msphere 2022. Ahead of Print. [Google Scholar] [CrossRef] [PubMed]
- Lauer, A.; Baal, J.D.; Mendes, S.D.; Casimiro, K.N.; Passaglia, A.K.; Valenzuela, A.H.; Guibert, G. Valley Fever on the Rise-Searching for Microbial Antagonists to the Fungal Pathogen Coccidioides immitis. Microorganisms 2019, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Saubolle, M.A.; Wojack, B.R.; Wertheimer, A.M.; Fuayagem, A.Z.; Young, S.; Koeneman, B.A. Multicenter Clinical Validation of a Cartridge-Based Real-Time PCR System for Detection of Coccidioides spp. in Lower Respiratory Specimens. J. Clin. Microbiol. 2018, 56, e01277-17. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.; Dizon, D.; Libke, R.; Peterson, M.; Slater, D.; Dhillon, A. Development of a real-time PCR Assay for identification of Coccidioides immitis by use of the BD Max system. J. Clin. Microbiol. 2015, 53, 926–929. [Google Scholar] [CrossRef]
- Binnicker, M.J.; Buckwalter, S.P.; Eisberner, J.J.; Stewart, R.A.; McCullough, A.E.; Wohlfiel, S.L.; Wengenack, N.L. Detection of Coccidioides species in clinical specimens by real-time PCR. J. Clin. Microbiol. 2007, 45, 173–178. [Google Scholar] [CrossRef]
- Gastelum-Cano, J.M.; Dautt-Castro, M.; Garcia-Galaz, A.; Felix-Murray, K.; Rascon-Careaga, A.; Cano-Rangel, M.A.; Islas-Osuna, M.A. The clinical laboratory evolution in coccidioidomycosis detection: Future perspectives. J. Mycol. Med. 2021, 31, 101159. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Victor, T.R.; Marathe, A.; Sidamonidze, K.; Crucillo, K.L.; Chaturvedi, V. Real-time PCR assay for detection and differentiation of Coccidioides immitis and Coccidioides posadasii from culture and clinical specimens. PLoS Negl. Trop. Dis. 2021, 15, e0009765. [Google Scholar] [CrossRef]
- Bowers, J.R.; Parise, K.L.; Kelley, E.J.; Lemmer, D.; Schupp, J.M.; Driebe, E.M.; Engelthaler, D.M.; Keim, P.; Barker, B.M. Direct detection of Coccidioides from Arizona soils using CocciENV, a highly sensitive and specific real-time PCR assay. Med. Mycol. 2019, 57, 246–255. [Google Scholar] [CrossRef]
- Chow, N.A.; Griffin, D.W.; Barker, B.M.; Loparev, V.N.; Litvintseva, A.P. Molecular detection of airborne Coccidioides in Tucson, Arizona. Med. Mycol. 2016, 54, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Lauer, A.; Talamantes, J.; Castañón Olivares, L.R.; Medina, L.J.; Baal, J.D.H.; Casimiro, K.; Shroff, N.; Emery, K.W. Combining forces—The use of Landsat TM satellite imagery, soil parameter information, and multiplex PCR to detect Coccidioides immitis growth sites in Kern County, California. PLoS ONE 2014, 9, e111921. [Google Scholar] [CrossRef]
- Ocampo-Chavira, P.; Eaton-Gonzalez, R.; Riquelme, M. Of Mice and Fungi: Coccidioides spp. Distribution Models. J. Fungi 2020, 6, 320. [Google Scholar] [CrossRef]
- Dobos, R.R.; Benedict, K.; Jackson, B.R.; McCotter, O.Z. Using soil survey data to model potential Coccidioides soil habitat and inform Valley fever epidemiology. PLoS ONE 2021, 16, e0247263. [Google Scholar] [CrossRef]
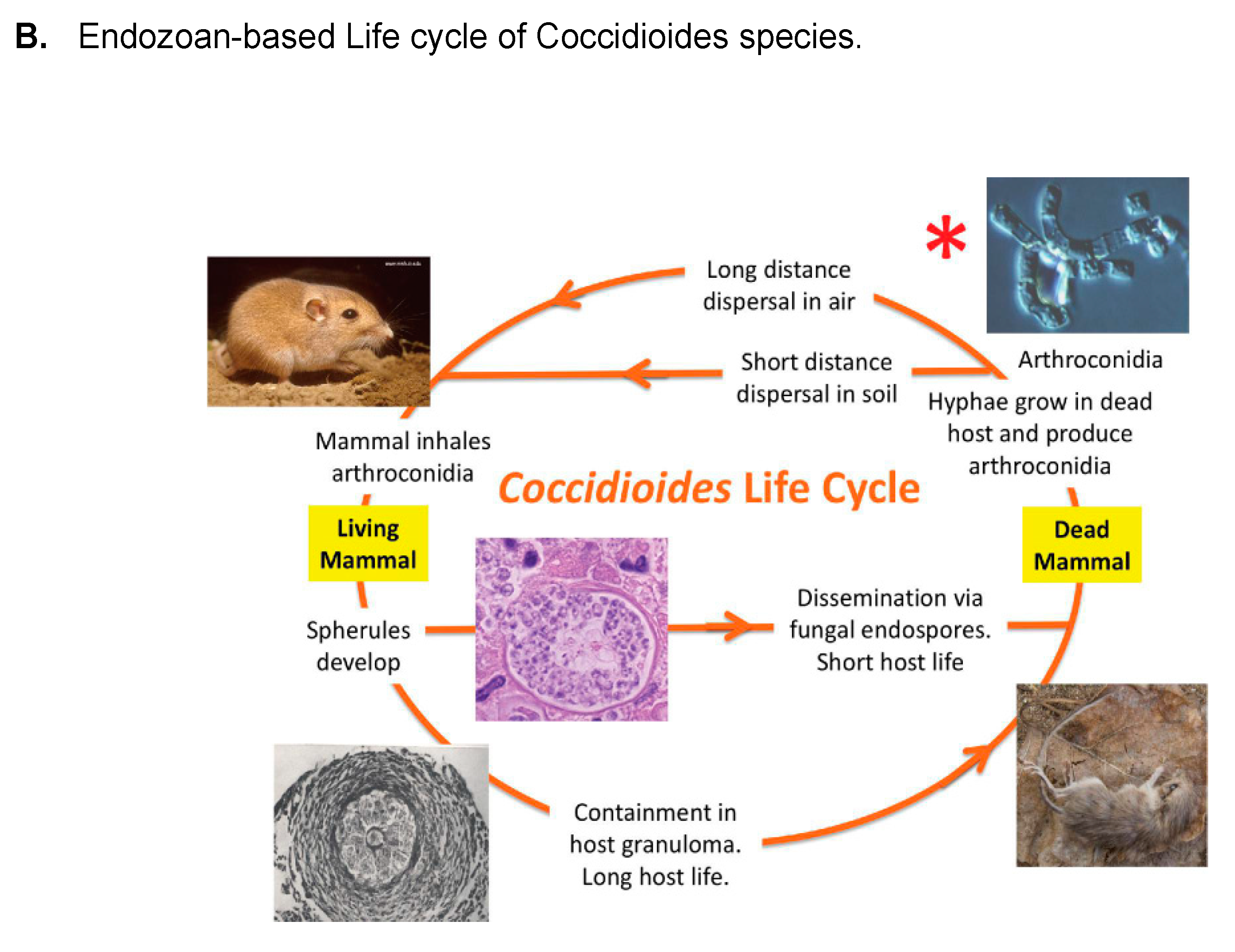
- Taylor, J.W.; Barker, B.M. The endozoan, small-mammal reservoir hypothesis and the life cycle of Coccidioides species. Med. Mycol. 2019, 57, S16–S20. [Google Scholar] [CrossRef]
- Del Rocio Reyes-Montes, M.; Perez-Huitron, M.A.; Ocana-Monroy, J.L.; Frias-De-Leon, M.G.; Martinez-Herrera, E.; Arenas, R.; Duarte-Escalante, E. The habitat of Coccidioides spp. and the role of animals as reservoirs and disseminators in nature. BMC Infect. Dis. 2016, 16, 550. [Google Scholar] [CrossRef]
- de Macêdo, R.C.; Rosado, A.S.; da Mota, F.F.; Cavalcante, M.A.; Eulálio, K.D.; Filho, A.D.; Martins, L.M.; Lazéra, M.S.; Wanke, B. Molecular identification of Coccidioides spp. in soil samples from Brazil. BMC Microbiol. 2011, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Eulálio, K.D.; de Macedo, R.L.; Cavalcanti, M.A.; Martins, L.M.; Lazéra, M.S.; Wanke, B. Coccidioides immitis isolated from armadillos (Dasypus novemcinctus) in the state of Piauí, northeast Brazil. Mycopathologia 2001, 149, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Sharpton, T.J.; Stajich, J.E.; Rounsley, S.D.; Gardner, M.J.; Wortman, J.R.; Jordar, V.S.; Maiti, R.; Kodira, C.D.; Neafsey, D.E.; Zeng, Q.; et al. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 2009, 19, 1722–1731. [Google Scholar] [CrossRef]
- Chow, E.D.; Liu, O.W.; O’Brien, S.; Madhani, H.D. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: Nitric oxide stress and body temperature. Curr. Genet. 2007, 52, 137–148. [Google Scholar] [CrossRef]
- Huppert, M.; Sun, S.H. Overview of Mycology, and the Mycology of Coccidioides immitis. In Coccidioidomycosis: A Text; Stevens, D.A., Ed.; Springer: Boston, MA, USA, 1980; pp. 21–46. [Google Scholar]
- Mead, H.L.; Van Dyke, M.C.C.; Barker, B.M. Proper Care and Feeding of Coccidioides: A Laboratorian’s Guide to Cultivating the Dimorphic Stages of C. immitis and C. posadasii. Curr. Protoc. Microbiol. 2020, 58, e113. [Google Scholar] [CrossRef] [PubMed]
- Cole, G.T. Models of cell differentiation in Conidial fungi. Microbiol. Rev. 1986, 50, 95–132. [Google Scholar] [CrossRef]
- Cole, G.T.; Sun, S.H.; Huppert, M. Isolation and ultrastructural examination of conidial wall components of Coccidioides and Aspergillus. Scan. Electron Microsc. 1982, 1677–1685. [Google Scholar]
- Lewis, E.R.; Bowers, J.R.; Barker, B.M. Dust devil: The life and times of the fungus that causes valley Fever. PLoS Pathog. 2015, 11, e1004762. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.J.; Holbrook, E.; Liao, Y.R.; Zarnowski, R.; Andes, D.R.; Wheat, L.J.; Malo, J.; Hung, C.Y. Characterization of an Uncinocarpus reesii-expressed recombinant tube precipitin antigen of Coccidioides posadasii for serodiagnosis. PLoS ONE 2019, 14, e0221228. [Google Scholar] [CrossRef] [PubMed]
- Drutz, D.J.; Huppert, M. Coccidioidomycosis: Factors affecting the host-parasite interaction. J. Infect. Dis. 1983, 147, 372–390. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Drutz, D.J.; Huppert, M.; Sun, S.H.; DeMarsh, P.L. The critical role of CO2 in the morphogenesis of Coccidioides immitis in cell-free subcutaneous chambers. J. Infect. Dis. 1984, 150, 127–134. [Google Scholar] [CrossRef]
- Converse, J.L.; Besemer, A.R. Nutrition of the Parasitic Phase of Coccidioides immitis in a Chemically Defined Liquid Medium. J. Bacteriol. 1959, 78, 231–239. [Google Scholar] [CrossRef]
- Converse, J.L. Effect of surface active agents on endosporulation of Coccidioides immitis in a chemically defined medium. J. Bacteriol. 1957, 74, 106–107. [Google Scholar] [CrossRef]
- Viriyakosol, S.; Singhania, A.; Fierer, J.; Goldberg, J.; Kirkland, T.N.; Woelk, C.H. Gene expression in human fungal pathogen Coccidioides immitis changes as arthroconidia differentiate into spherules and mature. BMC Microbiol. 2013, 13, 121. [Google Scholar] [CrossRef]
- Mead, H.L.; Teixeira, M.M.; Galgiani, J.N.; Barker, B.M. Characterizing in vitro spherule morphogenesis of multiple strains of both species of Coccidioides. Med. Mycol. 2019, 57, 478–488. [Google Scholar] [CrossRef]
- Brooks, L.D.; Northey, W.T. Studies on Coccidioides immitis. II. Physiological studies on in vitro spherulation. J. Bacteriol. 1963, 85, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Drutz, D.J.; Huppert, M.; Sun, S.H.; McGuire, W.L. Human sex hormones stimulate the growth and maturation of Coccidioides immitis. Infect. Immun. 1981, 32, 897–907. [Google Scholar] [CrossRef]
- Powell, B.L.; Drutz, D.J. Identification of a high-affinity binder for estradiol and a low-affinity binder for testosterone in Coccidioides immitis. Infect. Immun. 1984, 45, 784–786. [Google Scholar] [CrossRef] [PubMed]
- Petkus, A.F.; Baum, L.L.; Ellis, R.B.; Stern, M.; Danley, D.L. Pure spherules of Coccidioides immitis in continuous culture. J. Clin. Microbiol. 1985, 22, 165–167. [Google Scholar] [CrossRef]
- Mitchell, N.M.; Grys, T.E.; Lake, D.F. Carbo-loading in Coccidioides spp.: A quantitative analysis of CAZyme abundance and resulting glycan populations. Glycobiology 2020, 30, 186–197. [Google Scholar] [CrossRef]
- Mitchell, N.M.; Dasari, S.; Grys, T.E.; Lake, D.F. Laser Capture Microdissection-Assisted Protein Biomarker Discovery from Coccidioides-Infected Lung Tissue. J. Fungi 2020, 6, 365. [Google Scholar] [CrossRef]
- Smith, C.E. Epidemiology of Acute Coccidioidomycosis with Erythema Nodosum (“San Joaquin” or “Valley Fever”). Am. J. Public Health Nations Health 1940, 30, 600–611. [Google Scholar] [CrossRef]
- Smith, C.E.; Whiting, E.G.; Baker, E.E.; Rosenberger, H.G.; Beard, R.R.; Saito, M.T. The use of coccidioidin. Am. Rev. Tuberc. 1948, 57, 330–351. [Google Scholar]
- Anderson, K.L.; Wheat, R.W.; Conant, N.F. Fractionation and composition studies of skin test-active components of sensitins from Coccidioides immitis. Appl. Microbiol. 1971, 22, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Chaparas, S.D.; Levine, H.B.; Papagianis, D.; Scalarone, G.; Baer, H. Comparison of skin reactivity of spherule and mycelial coccidioidins produced by different strains of Coccidioides immitis. Proc. Soc. Exp. Biol. Med. 1974, 145, 806–810. [Google Scholar] [CrossRef]
- Williams, P.L.; Sable, D.L.; Sorgen, S.P.; Pappagianis, D.; Levine, H.B.; Brodine, S.K.; Brown, B.W.; Grumet, F.C.; Stevens, D.A. Immunologic responsiveness and safety associated with the Coccidioides immitis spherule vaccine in volunteers of white, black, and Filipino ancestry. Am. J. Epidemiol. 1984, 119, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.C.; Savage, D.C.; Kong, L.N. Delayed dermal hypersensitivity in mice to spherule and mycelial extracts of Coccidioides immitis. J. Bacteriol. 1966, 91, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.A.; Brummer, E.; Lecara, G. In vitro lymphocyte responses of coccidioidin skin test-positive and -negative persons to coccidioidin, spherulin, and a coccidioides cell wall antigen. Infect. Immun. 1977, 15, 751–755. [Google Scholar] [CrossRef]
- Galgiani, J.N.; Dugger, K.O.; Ampel, N.M.; Sun, S.H.; Law, J.H. Extraction of serologic and delayed hypersensitivity antigens from spherules of Coccidioides immitis. Diagn. Microbiol. Infect. Dis. 1988, 11, 65–80. [Google Scholar] [CrossRef]
- Johnson, R.; Kernerman, S.M.; Sawtelle, B.G.; Rastogi, S.C.; Nielsen, H.S.; Ampel, N.M. A reformulated spherule-derive coccidioidin (Spherusol) to detect delayed-type hypersensitivity in coccidiodomycosis. Mycopathologia 2012, 174, 353–358. [Google Scholar] [CrossRef]
- Wack, E.E.; Ampel, N.M.; Sunenshine, R.H.; Galgiani, J.N. The Return of Delayed-Type Hypersensitivity Skin Testing for Coccidioidomycosis. Clin. Infect. Dis. 2015, 61, 787–791. [Google Scholar] [CrossRef]
- Huppert, M.; Spratt, N.S.; Vukovich, K.R.; Sun, S.H.; Rice, E.H. Antigenic analysis of coccidioidin and spherulin determined by two-dimensional immunoelectrophoresis. Infect. Immun. 1978, 20, 541–551. [Google Scholar] [CrossRef]
- Cole, G.T.; Kirkland, T.N.; Sun, S.H. An immunoreactive, water-soluble conidial wall fraction of Coccidioides immitis. Infect. Immun. 1987, 55, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, N.M.; Sherrard, A.L.; Dasari, S.; Magee, D.M.; Grys, T.E.; Lake, D.F. Proteogenomic Re-Annotation of Coccidioides posadasii Strain Silveira. Proteomics 2018, 18, 1700173. [Google Scholar] [CrossRef]
- Cox, R.A.; Britt, L.A. Antigenic heterogeneity of an alkali-soluble, water-soluble cell wall extract of Coccidioides immitis. Infect. Immun. 1985, 50, 365–369. [Google Scholar] [CrossRef]
- Dugger, K.O.; Galgiani, J.N.; Ampel, N.M.; Sun, S.H.; Magee, D.M.; Harrison, J.L.; Law, J.H. An immunoreactive apoglycoprotein purified from Coccidioides immitis. Infect. Immun. 1991, 59, 2245–2251. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Cole, G.T. Isolation and characterization of an extracellular proteinase of Coccidioides immitis. Infect. Immun. 1987, 55, 1970–1978. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yang, C.; Magee, D.M.; Cox, R.A. Molecular cloning and characterization of Coccidioides immitis antigen 2 cDNA. Infect. Immun. 1996, 64, 2695–2699. [Google Scholar] [CrossRef] [PubMed]
- Dugger, K.O.; Villareal, K.M.; Nguyen, A.; Zimmermann, C.R.; Law, J.H.; Galgiani, J.N. Cloning and sequence analysis of the cDNA for a protein from Coccidioides immitis with immunogenic potential. Biochem. Biophys. Res. Commun. 1996, 218, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Cole, G.T. Molecular and Biochemical Characterization of a Coccidioides immitis-Specific Antigen. Infect. Immun. 1995, 63, 3994–4002. [Google Scholar] [CrossRef]
- Pappagianis, D.; Zimmer, B.L. Serology of coccidioidomycosis. Clin. Microbiol. Rev. 1990, 3, 247–268. [Google Scholar] [CrossRef]
- Johnson, S.M.; Pappagianis, D. The coccidioidal complement fixation and immunodiffusion-complement fixation antigen is a chitinase. Infect. Immun. 1992, 60, 2588–2592. [Google Scholar] [CrossRef]
- Johnson, S.M.; Zimmermann, C.R.; Pappagianis, D. Amino-terminal sequence analysis of the Coccidioides immitis chitinase/immunodiffusion-complement fixation protein. Infect. Immun. 1993, 61, 3090–3092. [Google Scholar] [CrossRef]
- Reichard, U.; Hung, C.Y.; Thomas, P.W.; Cole, G.T. Disruption of the gene which encodes a serodiagnostic antigen and chitinase of the human fungal pathogen Coccidioides immitis. Infect. Immun. 2000, 68, 5830–5838. [Google Scholar] [CrossRef]
- Xue, J.; Chen, X.; Selby, D.; Hung, C.Y.; Yu, J.J.; Cole, G.T. A genetically engineered live attenuated vaccine of Coccidioides posadasii protects BALB/c mice against coccidioidomycosis. Infect. Immun. 2009, 77, 3196–3208. [Google Scholar] [CrossRef]
- Yu, J.J.; Kirkland, T.N.; Hall, L.K.; Wopschall, J.; Smith, R.C.; Hung, C.Y.; Chen, X.; Tarcha, E.; Thomas, P.W.; Cole, G.T. Characterization of a serodiagnostic complement fixation antigen of Coccidioides posadasii expressed in the nonpathogenic Fungus Uncinocarpus reesii. J. Clin. Microbiol. 2005, 43, 5462–5469. [Google Scholar] [CrossRef] [PubMed]
- Kruse, D.; Cole, G.T. Isolation of tube precipitin antibody-reactive fractions of Coccidioides immitis. Infect. Immun. 1990, 58, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Cole, G.T.; Kruse, D.; Zhu, S.W.; Seshan, K.R.; Wheat, R.W. Composition, serologic reactivity, and immunolocalization of a 120-kilodalton tube precipitin antigen of Coccidioides immitis. Infect. Immun. 1990, 58, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Kruse, D.; Cole, G.T. A seroreactive 120-kilodalton b-(1,3)-glucanase of Coccidioides immitis which may participate in spherule morphogenesis. Infect. Immun. 1992, 60, 4350–4363. [Google Scholar] [CrossRef]
- Hung, C.Y.; Yu, J.J.; Lehmann, P.F.; Cole, G.T. Cloning and expression of the gene which encodes a tube precipitin antigen and wall-associated beta-glucosidase of Coccidioides immitis. Infect. Immun. 2001, 69, 2211–2222. [Google Scholar] [CrossRef]
- Kirkland, T.N.; Finley, F.; Orsborn, K.I.; Galgiani, J.N. Evaluation of the proline-rich antigen of Coccidioides immitis as a vaccine candidate in mice. Infect. Immun. 1998, 66, 3519–3522. [Google Scholar] [CrossRef]
- Abuodeh, R.O.; Shubitz, L.F.; Siegel, E.; Snyder, S.; Peng, T.; Orsborn, K.I.; Brummer, E.; Stevens, D.A.; Galgiani, J.N. Resistance to Coccidioides immitis in mice after immunization with recombinant protein or a DNA vaccine of a proline-rich antigen. Infect. Immun. 1999, 67, 2935–2940. [Google Scholar] [CrossRef]
- Cole, G.T.; Zhu, S.W.; Pan, S.C.; Yuan, L.; Kruse, D.; Sun, S.H. Isolation of antigens with proteolytic activity from Coccidioides immitis. Infect. Immun. 1989, 57, 1524–1534. [Google Scholar] [CrossRef]
- Cole, G.T.; Kirkland, T.N.; Franco, M.; Zhu, S.; Yuan, L.; Sun, S.H.; Hearn, V.M. Immunoreactivity of a surface wall fraction produced by spherules of Coccidioides immitis. Infect. Immun. 1988, 56, 2695–2701. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Cole, G.T.; Sun, S.H. Possible role of a proteinase in endosporulation of Coccidioides immitis. Infect. Immun. 1988, 56, 1551–1559. [Google Scholar] [CrossRef]
- Cole, G.T.; Zhu, S.W.; Hsu, L.L.; Kruse, D.; Seshan, K.R.; Wang, F. Isolation and expression of a gene which encodes a wall-associated proteinase of Coccidioides immitis. Infect. Immun. 1992, 60, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Tarcha, E.J.; Basrur, V.; Hung, C.Y.; Gardner, M.J.; Cole, G.T. Multivalent recombinant protein vaccine against coccidioidomycosis. Infect. Immun. 2006, 74, 5802–5813. [Google Scholar] [CrossRef] [PubMed]
- Deresinski, S.C.; Levine, H.B.; Stevens, D.A. Soluble antigens of mycelia and spherules in the in vitro detection of immunity to Coccidioides immitis. Infect. Immun. 1974, 10, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Wyckoff, E.E.; Pishko, E.J.; Kirkland, T.N.; Cole, G.T. Cloning and expression of a gene encoding a T-cell reactive protein from Coccidioides immitis: Homology to 4-hydroxyphenylpyruvate dioxygenase and the mammalian F antigen. Gene 1995, 161, 107–111. [Google Scholar] [CrossRef]
- Pan, S.; Cole, G.T. Electrophoretic karyotypes of clinical isolates of Coccidioides immitis. Infect. Immun. 1992, 60, 4872–4880. [Google Scholar] [CrossRef]
- De Melo Teixeira, M.; Lang, B.F.; Matute, D.R.; Stajich, J.E.; Barker, B. The mitochondrial genomes of the human pathogens Coccidioides immitis and C. posadasii. G3 2021, 11, jkab132. [Google Scholar] [CrossRef]
- De Melo Teixeira, M.; Stajich, J.E.; Sahl, J.W.; Thompson, G.R.; Brem, R.B.; Dubin, C.A.; Blackmon, A.V.; Mead, H.L.; Keim, P.; Barker, B.M. A chromosomal-level reference genome of the widely utilized Coccidioides posadasii laboratory strain "Silveira". G3 2022, 12, jkac031. [Google Scholar] [CrossRef]
- Neafsey, D.E.; Barker, B.M.; Sharpton, T.J.; Stajich, J.E.; Park, D.J.; Whiston, E.; Hung, C.Y.; McMahan, C.; White, J.; Sykes, S.; et al. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 2010, 20, 938–946. [Google Scholar] [CrossRef]
- Teixeira, M.M.; Barker, B.M.; Stajich, J.E. Improved Reference Genome Sequence of Coccidioides immitis Strain WA_211, Isolated in Washington State. Microbiol. Resour. Announc. 2019, 8, e00149-19. [Google Scholar] [CrossRef]
- Shang, Y.; Xiao, G.; Zheng, P.; Cen, K.; Zhan, S.; Wang, C. Divergent and Convergent Evolution of Fungal Pathogenicity. Genome Biol. Evol. 2016, 8, 1374–1387. [Google Scholar] [CrossRef]
- Owens, R.A.; Hammel, S.; Sheridan, K.J.; Jones, G.W.; Doyle, S. A proteomic approach to investigating gene cluster expression and secondary metabolite functionality in Aspergillus fumigatus. PLoS ONE 2014, 9, e106942. [Google Scholar] [CrossRef]
- Kirkland, T.N.; Muszewska, A.; Stajich, J.E. Analysis of Transposable Elements in Coccidioides Species. J. Fungi 2018, 4, 13. [Google Scholar] [CrossRef]
- Engelthaler, D.M.; Roe, C.C.; Hepp, C.M.; Teixeira, M.; Driebe, E.M.; Schupp, J.M.; Gade, L.; Waddell, V.; Komatsu, K.; Arathoon, E.; et al. Local Population Structure and Patterns of Western Hemisphere Dispersal for Coccidioides spp., the Fungal Cause of Valley Fever. MBio 2016, 7, e00550-16. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.M.; Alvarado, P.; Roe, C.C.; Thompson, G.R.; Patané, J.S.L.; Sahl, J.W.; Keim, P.; Galgiani, J.N.; Litvintseva, A.P.; Matute, D.R.; et al. Population Structure and Genetic Diversity among Isolates of Coccidioides posadasii in Venezuela and Surrounding Regions. MBio 2019, 10, e01976-19. [Google Scholar] [CrossRef] [PubMed]
- Mandel, M.A.; Galgiani, J.N.; Kroken, S.; Orbach, M.J. Coccidioides posadasii contains single chitin synthase genes corresponding to classes I to VII. Fungal Genet Biol. 2006, 43, 775–788. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Hall, C.R.; Kowbel, D.; Welch, J.; Taylor, J.W.; Brem, R.B.; Glass, N.L. Genome wide association identifies novel loci involved in fungal communication. PLoS Genet. 2013, 9, e1003669. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Olvera, L.; Hung, C.Y.; Yu, J.J.; Cole, G.T. Sequence, expression and functional analysis of the Coccidioides immitis ODC (ornithine decarboxylase) gene. Gene 2000, 242, 437–448. [Google Scholar] [CrossRef]
- Frey, C.L.; Drutz, D.J. Influence of fungal surface components on the interaction of Coccidioides immitis with polymorphonuclear neutrophils. J. Infect. Dis. 1986, 153, 933–943. [Google Scholar] [CrossRef]
- Maxwell, C.S.; Mattox, K.; Turissini, D.A.; Teixeira, M.M.; Barker, B.M.; Matute, D.R. Gene exchange between two divergent species of the fungal human pathogen, Coccidioides. Evolution 2018, 73, 42–58. [Google Scholar] [CrossRef]
- Fraser, J.A.; Heitman, J. Evolution of fungal sex chromosomes. Mol. Microbiol. 2004, 51, 299–306. [Google Scholar] [CrossRef]
- Fraser, J.A.; Stajich, J.E.; Tarcha, E.J.; Cole, G.T.; Inglis, D.O.; Sil, A.; Heitman, J. Evolution of the mating type locus: Insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 2007, 6, 622–629. [Google Scholar] [CrossRef]
- Mandel, M.A.; Barker, B.M.; Kroken, S.; Rounsley, S.D.; Orbach, M.J. Genomic and Population Analyses of the Mating Type Loci in Coccidioides Species Reveal Evidence for Sexual Reproduction and Gene Acquisition. Eukaryotic Cell 2007, 6, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Peláez-Jaramillo, C.A.; Jiménez-Alzate, M.D.P.; Araque-Marin, P.; Hung, C.-Y.; Castro-Lopez, N.; Cole, G.T. Lipid Secretion by Parasitic Cells of Coccidioides Contributes to Disseminated Disease. Front. Cell. Infect. Microbiol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Wise, H.Z.; Hung, C.-Y.; Whiston, E.; Taylor, J.W.; Cole, G.T. Extracellular ammonia at sites of pulmonary infection with Coccidioides posadasii contributes to severity of the respiratory disease. Microb. Pathog. 2013, 59–60, 19–28. [Google Scholar] [CrossRef]
- Cole, G.T.; Seshan, K.R.; Franco, M.; Bukownik, E.; Sun, S.H.; Hearn, V.M. Isolation and morphology of an immunoreactive outer wall fraction produced by spherules of Coccidioides immitis. Infect. Immun. 1988, 56, 2686–2694. [Google Scholar] [CrossRef]
- Cole, G.T.; Hung, C.-Y. The parasitic cell wall of Coccidioides immitis. Med. Mycol. 2001, 39, 31–40. [Google Scholar] [CrossRef]
- Hung, C.Y.; Yu, J.J.; Seshan, K.R.; Reichard, U.; Cole, G.T. A Parasitic Phase-Specific Adhesin of Coccidioides immitis Contributes to the Virulence of This Respiratory Fungal Pathogen. Infect. Immun. 2002, 70, 3443–3456. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.J.; Smithson, S.L.; Thomas, P.W.; Kirkland, T.N.; Cole, G.T. Isolation and characterization of the urease gene (URE) from the pathogenic fungus Coccidioides immitis. Gene 1997, 198, 387–391. [Google Scholar] [CrossRef]
- Mirbod, F.; Schaller, R.A.; Cole, G.T. Purification and characterization of urease isolated from the pathogenic fungus Coccidioides immitis. Med. Mycol. 2002, 40, 35–44. [Google Scholar] [CrossRef]
- Mirbod-Donovan, F.; Schaller, R.; Hung, C.-Y.; Xue, J.; Reichard, U.; Cole, G.T. Urease produced by Coccidioides posadasii contributes to the virulence of this respiratory pathogen. Infect. Immun. 2006, 74, 504–515. [Google Scholar] [CrossRef]
- Hung, C.Y.; Seshan, K.R.; Yu, J.J.; Schaller, R.; Xue, J.; Basrur, V.; Gardner, M.J.; Cole, G.T. A metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect Immun 2005, 73, 6689–6703. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Yu, J.J.; Hung, C.Y.; Casadevall, A.; Cole, G.T. Coccidioides posadasii produces melanin in vitro and during infection. Fungal Genet. Biol. 2007, 44, 517–520. [Google Scholar] [CrossRef] [PubMed]
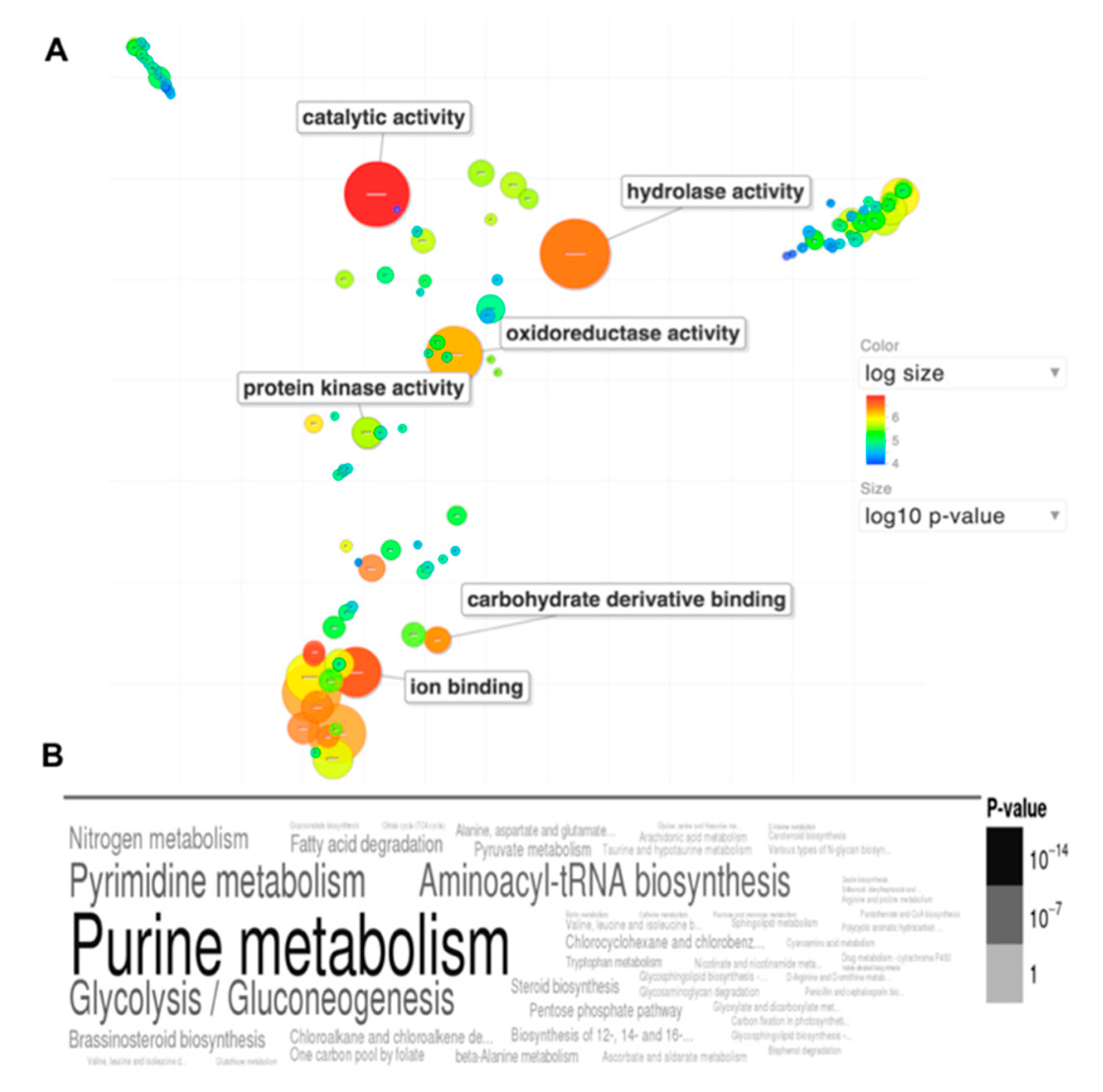
- Delgado, N.; Hung, C.Y.; Tarcha, E.; Gardner, M.J.; Cole, G.T. Profiling gene expression in Coccidioides posadasii. Med. Mycol. 2004, 42, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.B.; Costanzo, M.C.; Shah, P.; Skrzypek, M.S.; Sherlock, G. Gene Ontology and the annotation of pathogen genomes: The case of Candida albicans. Trends Microbiol. 2009, 17, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.E.; Marques Edos, R.; Malavazi, I.; Torres, I.; Restrepo, A.; Nunes, L.R.; de Oliveira, R.C.; Goldman, M.H.; Goldman, G.H. Transcriptome analysis and molecular studies on sulfur metabolism in the human pathogenic fungus Paracoccidioides brasiliensis. Mol. Genet. Genom. 2006, 276, 450–463. [Google Scholar] [CrossRef] [PubMed]
- Boyce, K.J.; McLauchlan, A.; Schreider, L.; Andrianopoulos, A. Intracellular growth is dependent on tyrosine catabolism in the dimorphic fungal pathogen Penicillium marneffei. PLoS Pathog. 2015, 11, e1004790. [Google Scholar] [CrossRef] [PubMed]
- Inglis, D.O.; Voorhies, M.; Hocking Murray, D.R.; Sil, A. Comparative transcriptomics of infectious spores from the fungal pathogen Histoplasma capsulatum reveals a core set of transcripts that specify infectious and pathogenic states. Eukaryot. Cell 2013, 12, 828–852. [Google Scholar] [CrossRef] [PubMed]
- Duttke, S.H.; Beyhan, S.; Singh, R.; Neal, S.; Viriyakosol, S.; Fierer, J.; Kirkland, T.N.; Stajich, J.E.; Benner, C.; Carlin, A.F. Decoding Transcription Regulatory Mechanisms Associated with Coccidioides immitis Phase Transition Using Total RNA. Msystems 2022, 7, e0140421. [Google Scholar] [CrossRef] [PubMed]
- Tollot, M.; Assmann, D.; Becker, C.; Altmuller, J.; Dutheil, J.Y.; Wegner, C.E.; Kahmann, R. The WOPR Protein Ros1 Is a Master Regulator of Sporogenesis and Late Effector Gene Expression in the Maize Pathogen Ustilago maydis. PLoS Pathog. 2016, 12, e1005697. [Google Scholar] [CrossRef]
- Nguyen, V.Q.; Sil, A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc. Natl. Acad. Sci. USA 2008, 105, 4880–4885. [Google Scholar] [CrossRef]
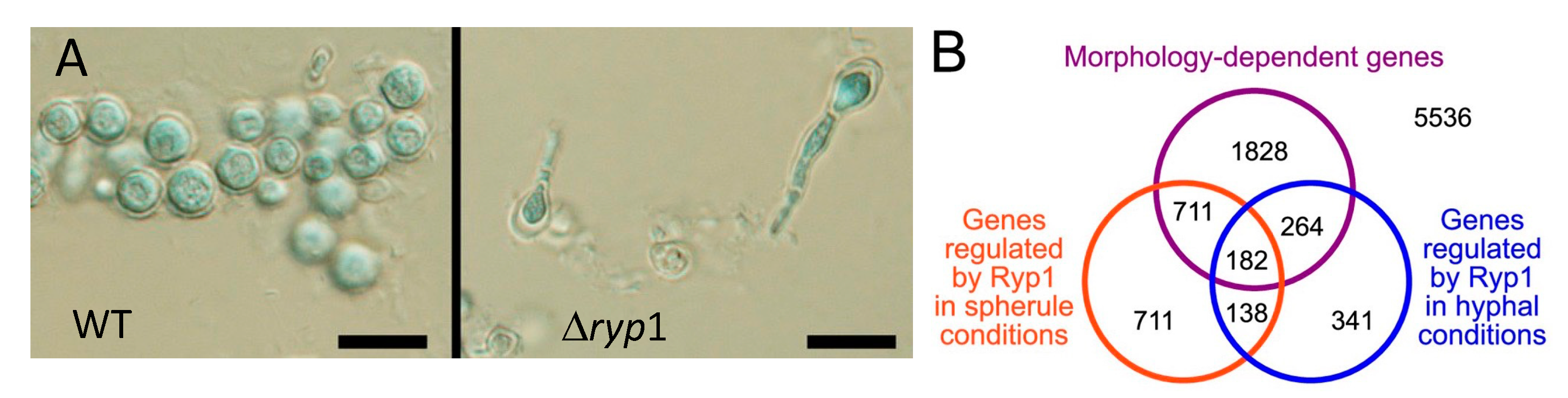
- Mandel, M.A.; Beyhan, S.; Voorhies, M.; Shubitz, L.F.; Galgiani, J.N.; Orbach, M.J.; Sil, A. The WOPR family protein Ryp1 is a key regulator of gene expression, development, and virulence in the thermally dimorphic fungal pathogen Coccidioides posadasii. PLoS Pathog. 2022, 18, e1009832. [Google Scholar] [CrossRef] [PubMed]
- Posas, F.; Takekawa, M.; Saito, H. Signal transduction by MAP kinase cascades in budding yeast. Curr. Opin. Microbiol. 1998, 1, 175–182. [Google Scholar] [CrossRef]
- Chen, C.; Pande, K.; French, S.D.; Tuch, B.B.; Noble, S.M. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 2011, 10, 118–135. [Google Scholar] [CrossRef] [PubMed]
- Nemecek, J.C.; Wuthrich, M.; Klein, B.S. Global control of dimorphism and virulence in fungi. Science 2006, 312, 583–588. [Google Scholar] [CrossRef]
- Mead, H.L.; Roe, C.C.; Higgins Keppler, E.A.; Van Dyke, M.C.C.; Laux, K.L.; Funke, A.L.; Miller, K.J.; Bean, H.D.; Sahl, J.W.; Barker, B.M. Defining Critical Genes During Spherule Remodeling and Endospore Development in the Fungal Pathogen, Coccidioides posadasii. Front. Genet. 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Grys, T.E.; Kaushal, S.; Chowdhury, Y.; Dasari, S.; Mitchell, N.M.; Magee, D.M.; Blair, J.E.; Colby, T.V.; Lake, D.F. Total and Lectin-Binding Proteome of Spherulin from Coccidioides posadasii. J. Proteome Res. 2016, 15, 3463–3472. [Google Scholar] [CrossRef]
- Higgins Keppler, E.A.; Mead, H.L.; Barker, B.M.; Bean, H.D. Life Cycle Dominates the Volatilome Character of Dimorphic Fungus Coccidioides spp. Msphere 2021, 6, e00040-21. [Google Scholar] [CrossRef]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microb. Cell Fact. 2017, 16, 168. [Google Scholar] [CrossRef]
- Michielse, C.B.; Hooykaas, P.J.; van den Hondel, C.A.; Ram, A.F. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005, 48, 1–17. [Google Scholar] [CrossRef]
- Hung, C.-Y.; Wise, H.Z.; Cole, G.T. Gene Disruption in Coccidioides Using Hygromycin or Phleomycin Resistance Markers. In Host-Fungus Interactions: Methods and Protocols; Brand, A.C., MacCallum, D.M., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 131–147. [Google Scholar]
- Abuodeh, R.O.; Orbach, M.J.; Mandel, M.A.; Das, A.; Galgiani, J.N. Genetic transformation of Coccidioides immitis facilitated by Agrobacterium tumefaciens. J. Infect. Dis. 2000, 181, 2106–2110. [Google Scholar] [CrossRef]
- Narra, H.P.; Shubitz, L.F.; Mandel, M.A.; Trinh, H.T.; Griffin, K.; Buntzman, A.S.; Frelinger, J.A.; Galgiani, J.N.; Orbach, M.J. A Coccidioides posadasii CPS1 Deletion Mutant Is Avirulent and Protects Mice from Lethal Infection. Infect. Immun. 2016, 84, 3007–3016. [Google Scholar] [CrossRef]
- Walch, H.A.; Kalvoda, A. Immunization of mice with induced mutants of Coccidioides immitis. I. Characterization of mutants and preliminary studies of their use as viable vaccines. Sabouraudia 1971, 9, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, T.N.; Fierer, J. Inbred mouse strains differ in resistance to lethal Coccidioides immitis infection. Infect. Immun. 1983, 40, 912–916. [Google Scholar] [CrossRef] [PubMed]
- Fierer, J.; Waters, C.; Walls, L. Both CD4+ and CD8+ T cells can medicate vaccine-induced protection against Coccidioides immitis infection in mice. J. Infect. Dis. 2006, 193, 1323–1331. [Google Scholar] [CrossRef]
- Kellner, E.M.; Orsborn, K.I.; Siegel, E.M.; Mandel, M.A.; Orbach, M.J.; Galgiani, J.N. Coccidioides posadasii contains a single 1,3-beta-glucan synthase gene that appears to be essential for growth. Eukaryot. Cell 2005, 4, 111–120. [Google Scholar] [CrossRef]
- McGinnis, M.R.; Smith, M.B.; Hinson, E. Use of the Coccidioides posadasii Deltachs5 strain for quality control in the ACCUPROBE culture identification test for Coccidioides immitis. J. Clin. Microbiol. 2006, 44, 4250–4251. [Google Scholar] [CrossRef]
- Smith, C.E.; Pappagianis, D.; Levine, H.B.; Saito, M. Human coccidioidomycosis. Bacteriol. Rev. 1961, 25, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Friedman, L.; Smith, C.E.; Gordon, L.E. The assay of virulence of Coccidioides in white mice. J. Infect. Dis. 1955, 97, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.A.; Magee, D.M. Coccidioidomycosis: Host response and vaccine development. Clin. Microbiol. Rev. 2004, 17, 804–839. [Google Scholar] [CrossRef]
- Shubitz, L.F.; Powell, D.A.; Butkiewicz, C.D.; Lewis, M.L.; Trinh, H.T.; Frelinger, J.A.; Orbach, M.J.; Galgiani, J.N. A Chronic Murine Disease Model of Coccidioidomycosis Using Coccidioides posadasii, strain 1038. J. Infect. Dis. 2020, 223, 166–173. [Google Scholar] [CrossRef]
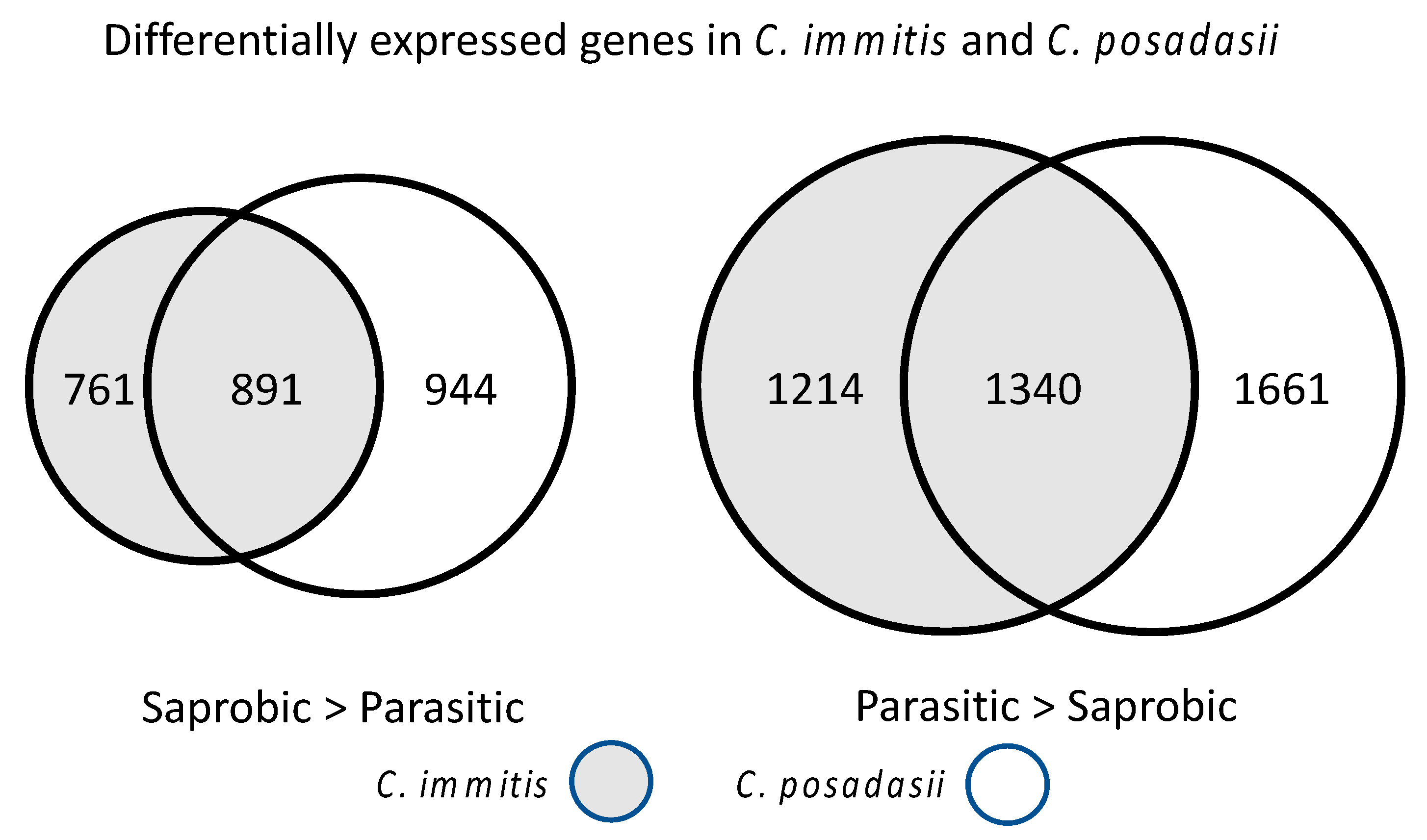

| Experiment | Deletion | % Mortality | CFU/Lung (log10) |
|---|---|---|---|
| 1 | None | 100 | NA |
| SOWgp | 40 | NA | |
| 2 | None | 100 | 7.2 |
| urease | 30 | 3.6 | |
| ugh | 25 | 4.4 | |
| urease/ugh | 10 | 2.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kirkland, T.N.; Stevens, D.A.; Hung, C.-Y.; Beyhan, S.; Taylor, J.W.; Shubitz, L.F.; Duttke, S.H.; Heidari, A.; Johnson, R.H.; Deresinski, S.C.; et al. Coccidioides Species: A Review of Basic Research: 2022. J. Fungi 2022, 8, 859. https://doi.org/10.3390/jof8080859
Kirkland TN, Stevens DA, Hung C-Y, Beyhan S, Taylor JW, Shubitz LF, Duttke SH, Heidari A, Johnson RH, Deresinski SC, et al. Coccidioides Species: A Review of Basic Research: 2022. Journal of Fungi. 2022; 8(8):859. https://doi.org/10.3390/jof8080859
Chicago/Turabian StyleKirkland, Theo N., David A. Stevens, Chiung-Yu Hung, Sinem Beyhan, John W. Taylor, Lisa F. Shubitz, Sascha H. Duttke, Arash Heidari, Royce H. Johnson, Stanley C. Deresinski, and et al. 2022. "Coccidioides Species: A Review of Basic Research: 2022" Journal of Fungi 8, no. 8: 859. https://doi.org/10.3390/jof8080859







