Circumfusicillium cavernae gen. et sp. nov. (Bionectriaceae, Hypocreales) Isolated from a Hypogean Roman Cryptoporticus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Fungal Isolation
2.2. DNA Extraction, PCR Amplification, Sequencing and Phylogenetic Analyses
2.3. Morphological Analysis
3. Results
3.1. Phylogenetic Analyses
3.2. Morphological Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Albertano, P. Epilithic Algal communities in hypogean environments. G. Bot. Ital. 1993, 127, 386–392. [Google Scholar] [CrossRef]
- Caneva, G.; Nugari, M.P.; Salvadori, O. Plant Biology for Cultural Heritage; The Getty Conservation Institute: Los Angeles, CA, USA, 2008; ISBN 5N81-606-CB10. [Google Scholar]
- Caneva, G.; Isola, D.; Lee, H.J.; Chung, Y.J. Biological risk for hypogea: Shared data from Etruscan tombs in Italy and ancient Tombs of the Baekje Dynasty in Republic of Korea. Appl. Sci. 2020, 10, 6104. [Google Scholar] [CrossRef]
- Jurado, V.; Sanchez-Moral, S.; Saiz-Jimenez, C. Entomogenous fungi and the conservation of the cultural heritage: A review. Int. Biodeterior. Biodegrad. 2008, 62, 325–330. [Google Scholar] [CrossRef] [Green Version]
- Griffin, D.W.; Gray, M.A.; Lyles, M.B.; Northup, D.E. The Transport of nonindigenous microorganisms into caves by Human visitation: A Case Study at Carlsbad Caverns National Park. Geomicrobiol. J. 2014, 31, 175–185. [Google Scholar] [CrossRef]
- Joshi, S.R.; Chettri, U. Fungi in hypogean environment: Bioprospection perspective. In Advancing Frontiers in Mycology & Mycotechnology: Basic and Applied Aspects of Fungi; Satyanarayana, T., Deshmukh, S.K., Deshpande, M.V., Eds.; Springer: Singapore, 2019; pp. 539–561. [Google Scholar]
- Sterflinger, K. Fungi as geologic agents. Geomicrobiol. J. 2000, 17, 97–124. [Google Scholar] [CrossRef]
- Sterflinger, K. Fungi: Their role in deterioration of cultural heritage. Fungal Biol. Rev. 2010, 24, 47–55. [Google Scholar] [CrossRef]
- Sterflinger, K.; Piñar, G. Microbial deterioration of cultural heritage and works of art—Tilting at windmills? Appl. Microbiol. Biotechnol. 2013, 97, 9637–9646. [Google Scholar] [CrossRef] [Green Version]
- Gadd, G.M. Geomycology: Biogeochemical transformations of rocks, minerals, metals and radionuclides by Fungi, Bioweathering and Bioremediation. Mycol. Res. 2007, 111, 3–49. [Google Scholar] [CrossRef]
- Gadd, G.M. Geomicrobiology of the built environment. Nat. Microbiol. 2017, 2, 16275. [Google Scholar] [CrossRef] [Green Version]
- Gadd, G.M. Fungi, rocks, and minerals. Elements 2017, 13, 171–176. [Google Scholar] [CrossRef]
- Isola, D.; Zucconi, L.; Cecchini, A.; Caneva, G. Dark-pigmented biodeteriogenic fungi in Etruscan hypogeal tombs: New data on their culture-dependent diversity, favouring conditions, and resistance to biocidal treatments. Fungal Biol. 2021, 125, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.-J.; Bao, D.-F.; Boonmee, S.; Calabon, M.S.; et al. Refined Families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Wijayawardene, N.; Hyde, K.; Dai, D.; Sánchez-García, M.; Goto, B.; Saxena, R.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.; Aptroot, A.; et al. Outline of Fungi and Fungus-like Taxa–2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Rossman, A.Y. Morphological and molecular perspectives on systematics of the Hypocreales. Mycologia 1996, 88, 1–19. [Google Scholar] [CrossRef]
- Rossman, A.Y.; Samuels, G.J.; Rogerson, C.T.; Lowen, R. Genera of Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales, Ascomycetes); Centraalbureau voor Schimmelcultures (CBS): Utrecht, The Netherlands, 1999; Volume 42, pp. 1–238. [Google Scholar]
- Zare, R.; Gams, W. More white Verticillium-like anamorphs with erect conidiophores. Mycol. Prog. 2016, 15, 993–1030. [Google Scholar] [CrossRef]
- Summerbell, R.C.; Gueidan, C.; Guarro, J.; Eskalen, A.; Crous, P.W.; Gupta, A.K.; Gené, J.; Cano-Lira, J.F.; van Iperen, A.; Starink, M.; et al. The Protean Acremonium. A. Sclerotigenum/Egyptiacum: Revision, food contaminant, and human disease. Microorganisms 2018, 6, 88. [Google Scholar] [CrossRef] [Green Version]
- Soares, F.; Trovão, J.; Portugal, A. Phototrophic and fungal communities inhabiting the roman cryptoporticus of the National Museum Machado de Castro (UNESCO Site, Coimbra, Portugal). World J. Microbiol. Biotechnol. 2022, 38, 157. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A.; Soares, F.; Paiva, D.S.; Mesquita, N.; Coelho, C.; Pinheiro, A.C.; Catarino, L.; Gil, F.; Tiago, I. Fungal diversity and distribution across distinct biodeterioration phenomena in limestone walls of the Old Cathedral of Coimbra, UNESCO World Heritage Site. Int. Biodeterior. Biodegrad. 2019, 142, 91–102. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Crous, P.W.; Schoch, C.L.; Hyde, K.D.; Wood, A.R.; Gueidan, C.; de Hoog, G.S.; Groenewald, J.Z. Phylogenetic lineages in the Capnodiales. Stud. Mycol. 2009, 64, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voglmayr, H.; Jaklitsch, W.M. Stilbocrea walteri sp. nov., an Unusual Species of Bionectriaceae. Mycol. Prog. 2019, 18, 91–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolboli, Z.; Tavakolian, B.; Mostowfizadeh-Ghalamfarsa, R.; Jafari, M.; Cacciola, S.O. Stilbocrea banihashemiana sp. nov. a new fungal pathogen causing stem cankers and twig dieback of fruit trees. J. Fungi 2022, 8, 694. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M. UGENE team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Nylander, J.A.A. MrModeltest v2 Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. RaxmlGUI 2.0: A Graphical Interface and Toolkit for Phylogenetic Analyses Using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J. FigTree: Tree Figure Drawing Tool, Version 1.2.2. 2008. Available online: https://tree.bio.ed.ac.uk/software/figtree/ (accessed on 10 July 2022).
- Rambaut, A.; Drummond, A.J. Tracer v 1.4. 2007. Available online: https://beast.bio.ed.ac.uk/Tracer (accessed on 10 July 2022).
- Díaz Herráiz, M. Caracterización de Comunidades Microbianas en Tumbas Etruscas y Romanas. Ph.D. Thesis, Universidad de Sevilla, Sevilla, Spain, 2015; p. 226. [Google Scholar]
- Mascaro, M.E.; Pellegrino, G.; Palermo, A.M. Analysis of biodeteriogens on architectural heritage. An approach of applied botany on a gothic building in southern Italy. Sustainability 2021, 14, 34. [Google Scholar] [CrossRef]
- Domínguez-Moñino, I. Evaluación y Control de Comunidades Microbianas en Cuevas Turísticas. Ph.D. Thesis, CSIC-Instituto de Recursos Naturales y Agrobiología de Sevilla (IRNAS), Sevilla, Spain, 2015; p. 368. [Google Scholar]
- Leplat, J.; François, A.; Touron, S.; Frouin, M.; Portais, J.-C.; Bousta, F. Aerobiological behavior of paleolithic rock art sites in dordogne (france): A comparative study in protected sites ranging from rock shelters to caves, with and without public access. Aerobiologia 2020, 36, 355–374. [Google Scholar] [CrossRef]
- Ashrafi, S.; Helaly, S.; Schroers, H.-J.; Stadler, M.; Richert-Poeggeler, K.R.; Dababat, A.A.; Maier, W. Ijuhya vitellina sp. nov., a novel source for chaetoglobosin a, is a destructive parasite of the cereal cyst nematode Heterodera filipjevi. PLoS ONE 2017, 12, e0180032. [Google Scholar] [CrossRef] [PubMed]
- Pyzik, A.; Ciuchcinski, K.; Dziurzynski, M.; Dziewit, L. The Bad and the Good—Microorganisms in cultural heritage environments—An update on biodeterioration and biotreatment approaches. Materials 2021, 14, 177. [Google Scholar] [CrossRef] [PubMed]
- Selbmann, L.; Stoppiello, G.A.; Onofri, S.; Stajich, J.E.; Coleine, C. Culture-dependent and amplicon sequencing approaches reveal diversity and distribution of black fungi in antarctic cryptoendolithic communities. J. Fungi 2021, 7, 213. [Google Scholar] [CrossRef] [PubMed]
- Savković, Ž.; Unković, N.; Stupar, M.; Franković, M.; Jovanović, M.; Erić, S.; Šarić, K.; Stanković, S.; Dimkić, I.; Vukojević, J.; et al. Diversity and biodeteriorative potential of fungal dwellers on ancient stone stela. Int. Biodeterior. Biodegrad. 2016, 115, 212–223. [Google Scholar] [CrossRef]
- Jansson, H.; Friman, E. Infection-related surface proteins on conidia of the nematophagous fungus Drechmeria coniospora. Mycol. Res. 1999, 103, 249–256. [Google Scholar] [CrossRef]

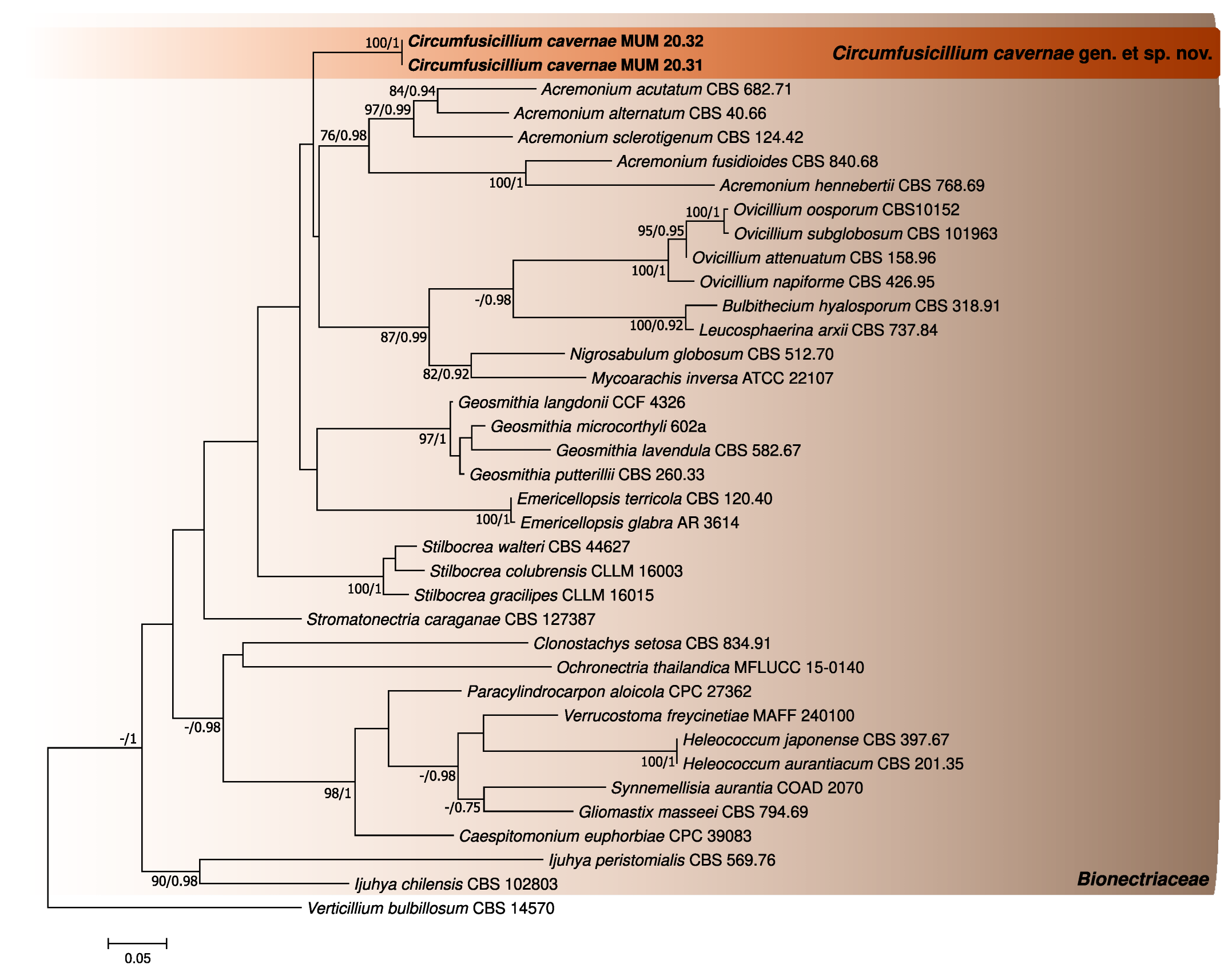
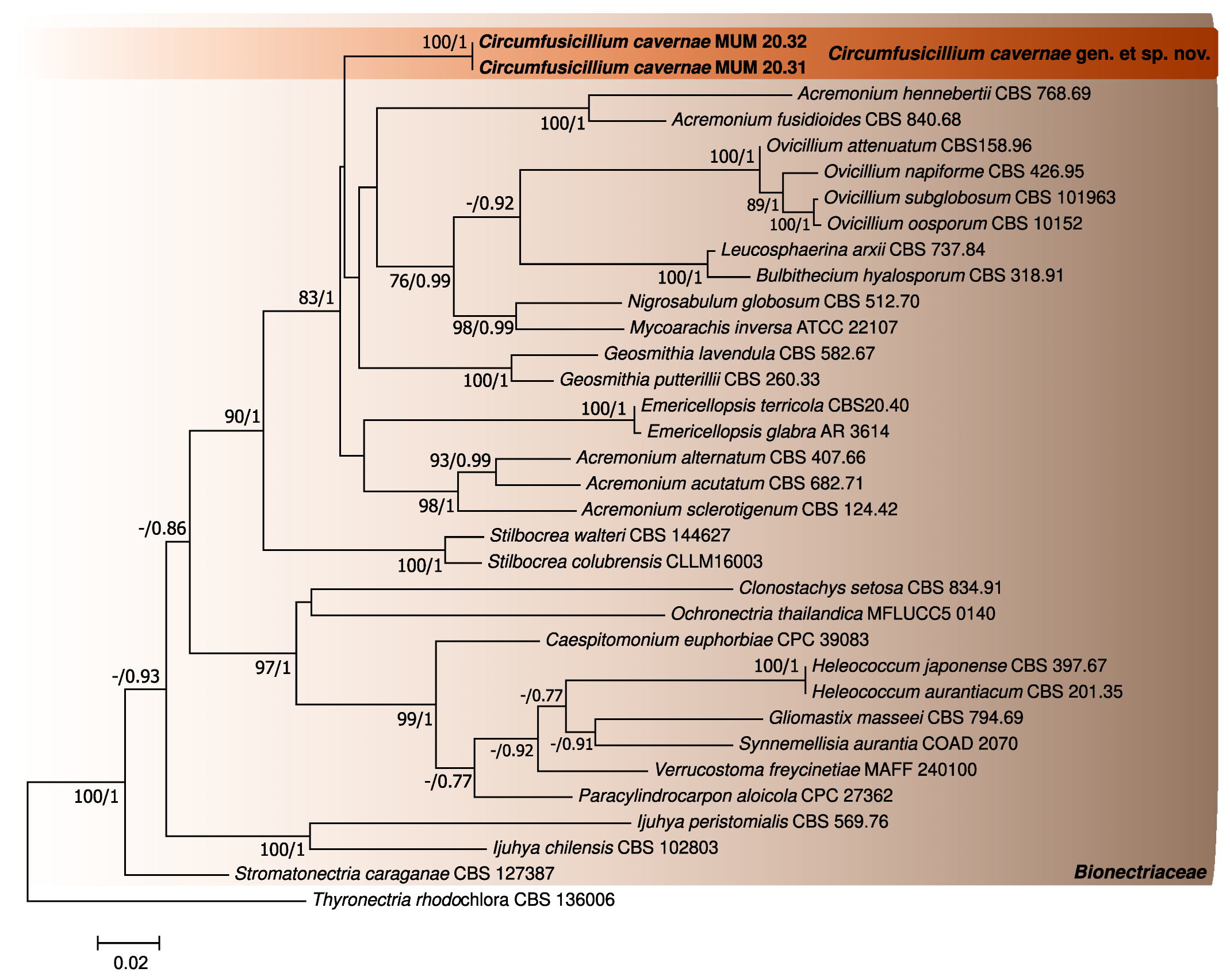


| Species | Isolate Reference | LSU Accession Number | ITS Accession Number |
|---|---|---|---|
| Acremonium acutatum | CBS 682.71 | NG056976 | MH860300 |
| Acremonium alternatum | CBS 407.66 | NG056977 | MH424672 |
| Acremonium fusidioides | CBS 840.68 | NG056984 | FN706542 |
| Acremonium hennebertii | CBS 768.69 | NG056987 | MH859420 |
| Acremonium sclerotigenum | CBS 124.42 | NG057139 | MH856101 |
| Acremonium zeylanicum | CBS 746.73 | HQ232154 | - |
| Bryocentria brongniartii | M190 | EU940125 | - |
| Bryocentria metzgeriae | M140 | EU940106 | - |
| Bulbithecium hyalosporum | CBS 318.91 | AF096187 | NR_137155 |
| Bullanockia australis | CPC 28976 | KY173506 | - |
| Caespitomonium euphorbiae | CPC 39083 | OK663737 | OK664698 |
| Circumfusicillium cavernae | MUM 20.31 | MT012542 | MT012542 |
| Circumfusicillium cavernae | MUM 20.32 | MT012543 | MT012543 |
| Clonostachys epichloe | CBS 118752 | DQ363259 | - |
| Clonostachys grammicospora | GJS 85-218 | AF193238 | - |
| Clonostachys ochroleuca | CCFC 226708 | AY283558 | - |
| Clonostachys pityrodes | GJS 95 | AY489728 | - |
| Clonostachys setosa | CBS 834.91 | AF210670 | AF210670 |
| Emericellopsis glabra | AR 3614 | GQ505993 | HM484860 |
| Emericellopsis maritima | AFTOLID 999 | FJ176861 | - |
| Emericellopsis terricola | CBS 120.40 | U57082 | MH856058 |
| Geonectria subalpina | CBS 143540 | MH155487 | - |
| Geosmithia langdonii | CCF 4326 | - | KF808298 |
| Geosmithia lavendula | CBS 582.67 | KT155289 | MH85905 |
| Geosmithia microcorthyli | 602a | - | MT955334 |
| Geosmithia putterillii | CBS 260.33 | KT155185 | MH855435 |
| Gliomastix masseei | CBS 794.69 | HQ232060 | MH859431 |
| Gliomastix roseogrisea | CBS 279.79 | HQ232122 | - |
| Heleococcum aurantiacum | CBS 201.35 | JX158441 | MH855645 |
| Heleococcum japonense | CBS 397.67 | JX158442 | JX158420 |
| Heleococcum japonicum | ATCC 18157 | U17429 | - |
| Hydropisphaera peziza | GJS 92101 | AY489730 | - |
| Hydropisphaera suffulta | CLLMAR 13023 | KU237207 | - |
| Ijuhya chilensis | CBS 102803 | KY607553 | KY607538 |
| Ijuhya fournieri | CLLG10113 | KP899118 | - |
| Ijuhya peristomialis | CBS 569.76 | KY607559 | KY607544 |
| Kallichroma glabrum | JK5123 | AF193233 | - |
| Kallichroma tethys | JK5181 | AF193234 | - |
| Lasionectriella rubioi | CBS 140157 | KU593581 | - |
| Leucosphaerina arxii | CBS 737.84 | NG057892 | NR_145040 |
| Mycoarachis inversa | ATCC 22107 | NG059437 | HM484861 |
| Nectriopsis violacea | MUCL 40056 | AF193242 | - |
| Nigrosabulum globosum | ATCC 22102 | AF096195 | NR_160124 |
| Ochronectria calami | ATCC46692 | AF193243 | - |
| Ochronectria thailandica | MFLUCC 15-0140 | KU564069 | KU564071 |
| Ovicillium attenuatum | CBS 158.96 | KU382232 | KU382186 |
| Ovicillium napiforme | CBS 426.95 | KU382233 | KU382192 |
| Ovicillium oosporum | CBS 110152 | KU382234 | KU382194 |
| Ovicillium subglobosum | CBS 101963 | KU382235 | NR_154335 |
| Paracylindrocarpon aloicola | CPC 27362 | KX228328 | KX228277 |
| Protocreopsis korfii | CLLM 14077 | KT852955 | - |
| Protocreopsis pertusa | CTR 72184 | GQ506002 | - |
| Roumegueriella rufula | GJS 91164 | EF469082 | - |
| Selinia pulchra | AR 2750 | AF193246 | - |
| Stephanonectria keithii | GJS 92133 | AY489727 | - |
| Stilbocrea colubrensis | CLLM 16003 | MN497409 | NR_173884 |
| Stilbocrea gracilipes | CLLM 16015 | - | MN497407 |
| Stilbocrea macrostoma | GJS 02125 | GQ506004 | - |
| Stilbocrea macrostoma | GJS 7326 | AY489725 | - |
| Stilbocrea macrostoma | WU 32032 | MH562718 | - |
| Stilbocrea walteri | CBS 144627 | MH562717 | NR_160063 |
| Stromatonectria caraganae | CBS 127387 | HQ112287 | HQ112287 |
| Synnemellisia aurantia | COAD 2070 | KX866396 | NR_154444 |
| Verrucostoma freycinetiae | MAFF 240100 | GQ506013 | NR_137761 |
| Verrucostoma martiniciensis | PAM 2015 | KP192672 | - |
| Xanthonectria pseudopeziza | CLL16005 | KU946964 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trovão, J.; Soares, F.; Paiva, D.S.; Tiago, I.; Portugal, A. Circumfusicillium cavernae gen. et sp. nov. (Bionectriaceae, Hypocreales) Isolated from a Hypogean Roman Cryptoporticus. J. Fungi 2022, 8, 837. https://doi.org/10.3390/jof8080837
Trovão J, Soares F, Paiva DS, Tiago I, Portugal A. Circumfusicillium cavernae gen. et sp. nov. (Bionectriaceae, Hypocreales) Isolated from a Hypogean Roman Cryptoporticus. Journal of Fungi. 2022; 8(8):837. https://doi.org/10.3390/jof8080837
Chicago/Turabian StyleTrovão, João, Fabiana Soares, Diana Sofia Paiva, Igor Tiago, and António Portugal. 2022. "Circumfusicillium cavernae gen. et sp. nov. (Bionectriaceae, Hypocreales) Isolated from a Hypogean Roman Cryptoporticus" Journal of Fungi 8, no. 8: 837. https://doi.org/10.3390/jof8080837
APA StyleTrovão, J., Soares, F., Paiva, D. S., Tiago, I., & Portugal, A. (2022). Circumfusicillium cavernae gen. et sp. nov. (Bionectriaceae, Hypocreales) Isolated from a Hypogean Roman Cryptoporticus. Journal of Fungi, 8(8), 837. https://doi.org/10.3390/jof8080837









