A New Cryptic Lineage in Parmeliaceae (Ascomycota) with Pharmacological Properties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis
2.2. Morphological and Chemical Studies
2.3. Pharmacological Study
2.3.1. Reagents
2.3.2. Antioxidant Assays
2.3.3. Cell Culture and Cell Treatments
2.3.4. Cell Viability Assay
2.3.5. Statistical Analysis
3. Results and Discussion
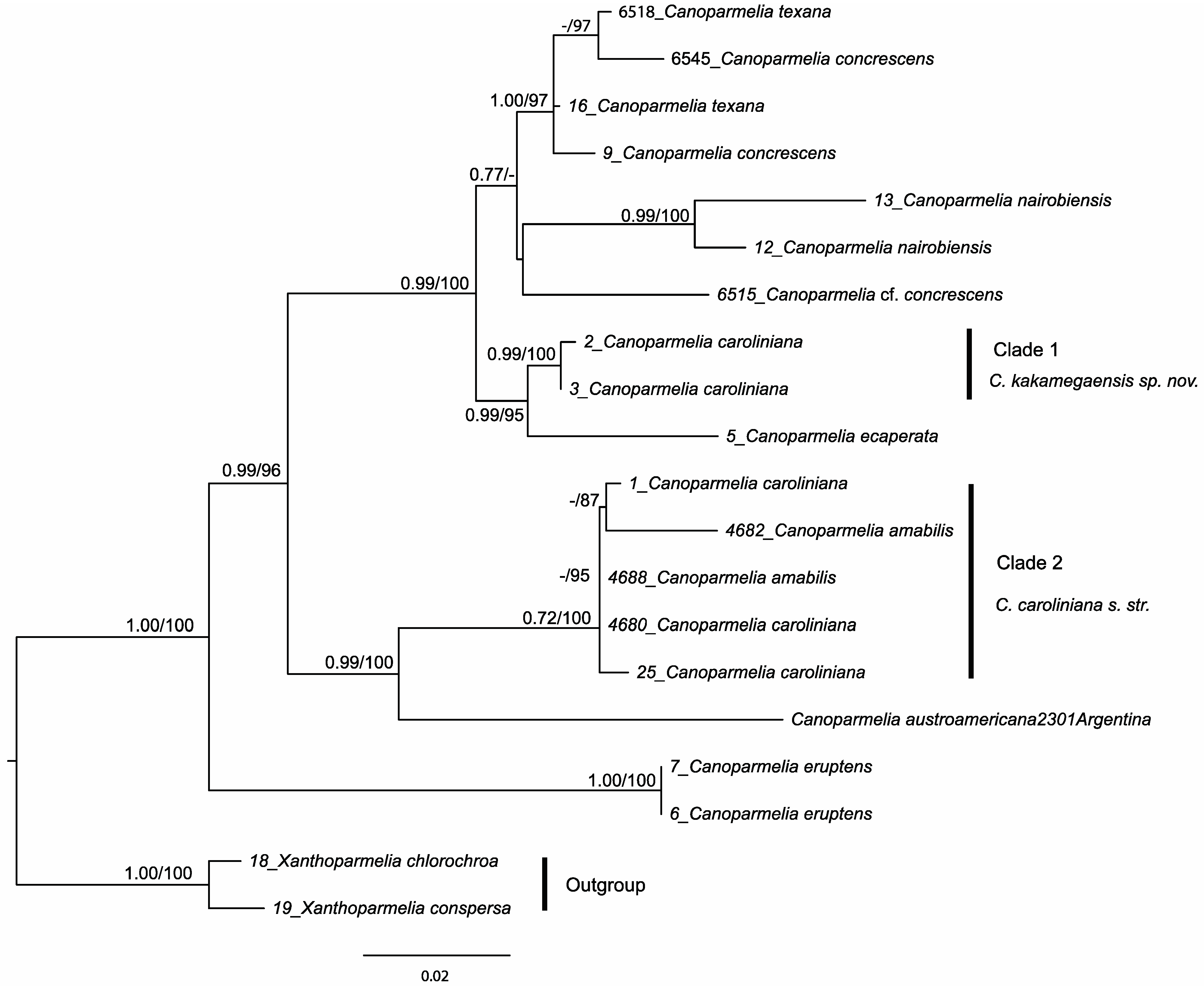
3.1. Phylogenetic Analysis
3.2. Chemical and Morphological Analysis
3.3. Taxonomy
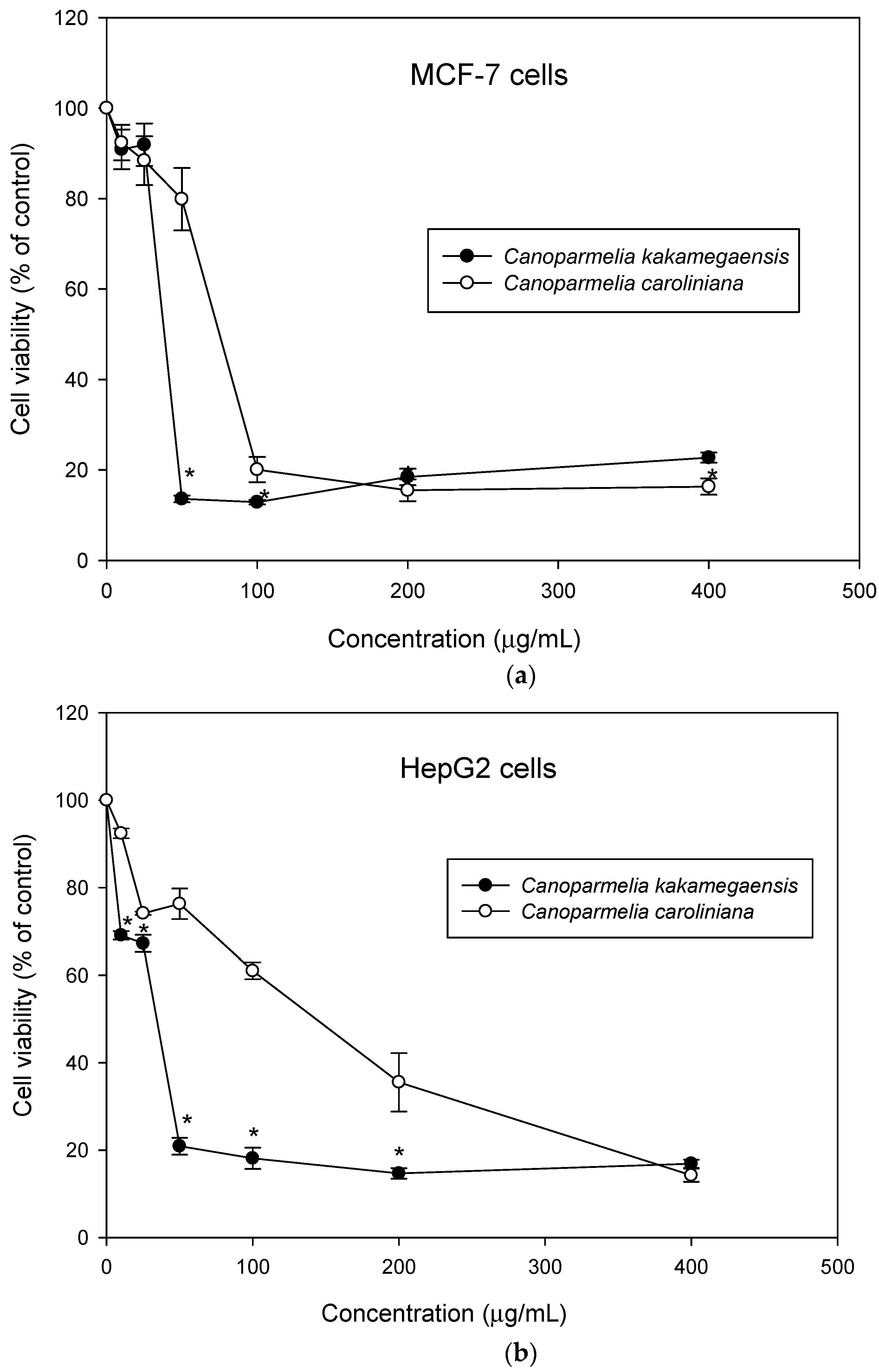
3.4. Pharmacological Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crespo, A.; Lumbsch, H.T. Cryptic species in lichen-forming fungi. IMA Fungus 2010, 1, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Crespo, A.; Pérez-Ortega, S. Cryptic species and species pairs in lichens: A discussion on the relationship between molecular phylogenies and morphological characters. An. Jardín Botánico Madr. 2009, 66, 71–81. [Google Scholar] [CrossRef]
- Crespo, A.; Ferencova, Z.; Pérez-Ortega, S.; Argüello, A.; Elix, J.A.; Divakar, P.K. Austroparmelina, a new Australasian lineage in parmelioid lichens (Parmeliaceae, Ascomycota): A multigene and morphological approach. Syst. Biodivers. 2010, 8, 209–221. [Google Scholar] [CrossRef]
- Lumbsch, H.T.; Leavitt, S.D. Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Divers. 2011, 50, 59–72. [Google Scholar] [CrossRef]
- Leavitt, S.D.; Moreau, C.S.; Lumbsch, H.T. The Dynamic Discipline of Species Delimitation: Progress toward Effectively Recognizing Species Boundaries in Natural Populations. In Recent Advances in Lichenology; Upreti, D.K., Divakar, P.K., Shukla, V., Bajpai, R., Eds.; Springer: New Delhi, India, 2015; pp. 11–44. [Google Scholar]
- Leavitt, S.D.; Esslinger, T.L.; Divakar, P.K.; Crespo, A.; Lumbsch, H.T. Hidden diversity before our eyes: Delimiting and describing cryptic lichen-forming fungal species in camouflage lichens (Parmeliaceae, Ascomycota). Fungal Biol. 2016, 120, 1374–1391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Serranillos, M.P.; Fernandez-Moriano, C.; Gonzalez-Burgos, E.; Divakar, P.K.; Crespo, A. Parmeliaceae family: Phytochemistry, pharmacological potential and phylogenetic features. Rsc Adv. 2014, 4, 59017–59047. [Google Scholar] [CrossRef]
- Crespo, A.; Kauff, F.; Divakar, P.K.; Amo, G.; Arguello, A.; Blanco, O.; Cubas, P.; del Prado, R.; Elix, J.A.; Esslinger, T.L.; et al. Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon 2010, 59, 1735–1753. [Google Scholar] [CrossRef]
- Divakar, P.K.; Kauff, F.; Crespo, A.; Leavitt, S.D.; Lumbsch, H.T. Understanding phenotypical character evolution in parmelioid lichenized fungi (Parmeliaceae, Ascomycota). PLoS ONE 2013, 8, e83115. [Google Scholar] [CrossRef]
- Alastruey-Izquierdo, A.; Alcazar-Fuoli, L.; Cuenca-Estrella, M. Antifungal susceptibility profile of cryptic species of Aspergillus. Mycopathologia 2014, 178, 427–433. [Google Scholar] [CrossRef]
- Hawksworth, D.L. Lichen Chemotaxonomy. In Lichenology: Progress and Problems; Brown, D.H., Hawksworth, D.L., Bailey, R.H., Eds.; Academic Press: London, UK, 1976; pp. 139–184. [Google Scholar]
- Feige, G.B.; Lumbsch, H.T. Some types of chemical variation in lichens. Cryptogam. Bot. 1995, 5, 31–35. [Google Scholar]
- Lumbsch, H.T. The taxonomic use of metabolic data in lichen-forming fungi. In Chemical Fungal Taxonomy; Frisvad, J.C., Bridge, P.D., Arora, D.K., Eds.; M. Dekker: New York, NY, USA, 1998; pp. 345–387. [Google Scholar]
- Lumbsch, H.T. The use of metabolic data in lichenology at the species and subspecific levels. Lichenologist 1998, 30, 357–367. [Google Scholar] [CrossRef]
- Ristíc, S.; Rankovic, B.; Kosanic, M.; Stanojkovic, T.; Stamenkovic, S.; Vasiljevic, P.; Manojlovic, I.; Manojlovic, N.T. Phytochemical study and antioxidant, antimicrobial and anticancer activities of Melanelia subaurifera and Melanelia fuliginosa lichens. J. Food Sci. Technol. 2016, 53, 2804–2816. [Google Scholar] [CrossRef] [Green Version]
- Brisdelli, F.; Perilli, M.; Sellitri, D.; Piovano, M.; Garbarino, J.A.; Nicoletti, M.; Bozzi, A.; Amicosante, G.; Celenza, G. Cytotoxic activity and antioxidant capacity of purified lichen metabolites: An in vitro study. Phytother. Res. 2013, 27, 431–437. [Google Scholar] [CrossRef]
- Brandão, L.F.G.; Alcantara, G.B.; Matos, M.D.F.C.; Bogo, D.; Freitas, D.D.S.; Oyama, N.M.; Honda, N.K. Cytotoxic evaluation of phenolic compounds from lichens against melanoma cells. Chem. Pharm. Bull. 2013, 61, 176–183. [Google Scholar]
- Bézivin, C.; Tomasi, F.; Lohézic-Le Dévéhat, F.; Boustie, J. Cytotoxic activity of some lichen extracts on murine and human cancer cell lines. Phytomedicine 2003, 10, 499–503. [Google Scholar] [CrossRef]
- Chandra, P.; Sharma, R.K.; Arora, D.S. Antioxidant compounds from microbial sources: A review. Food Res. Int. 2020, 129, 108849. [Google Scholar] [CrossRef]
- Hidalgo, M.E.; Fernández, E.; Quilhot, W.; Lissi, E. Antioxidant activity of depsides and depsidones. Phytochemistry 1994, 37, 1585–1587. [Google Scholar] [CrossRef]
- Fernández-Moriano, C.; Gómez-Serranillos, M.P.; Crespo, A. Antioxidant potential of lichen species and their secondary metabolites. A systematic review. Pharm. Biol. 2016, 54, 1–17. [Google Scholar] [CrossRef]
- Ingelfinger, R.; Henke, M.; Roser, L.; Ulshöfer, T.; Calchera, A.; Singh, G.; Parnham, M.J.; Geisslinger, G.; Fürst, R.; Schmitt, I.; et al. Unraveling the Pharmacological Potential of Lichen Extracts in the Context of Cancer and Inflammation with a Broad Screening Approach. Front. Pharmacol. 2020, 11, 1322. [Google Scholar] [CrossRef]
- Lopes, A.C.; Peixe, T.S.; Mesas, A.E.; Paoliello, M.M. Lead Exposure and Oxidative Stress: A Systematic Review. Rev. Environ. Contam. Toxicol. 2016, 236, 193–238. [Google Scholar]
- Liu, Z.; Ren, Z.; Zhang, J.; Chuang, C.C.; Kandaswamy, E.; Zhou, T.; Zuo, L. Role of ROS and Nutritional Antioxidants in Human Diseases. Front. Physiol. 2018, 9, 477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, P.A.; Oliveira, R.C.; Oliveira, A.P.; Serafini, M.R.; Araujo, A.A.; Gelain, D.P.; Moreira, J.C.; Almeida, J.R.; Quintans, J.S.; Quintains-Junior, L.J.; et al. Antioxidant activity and mechanisms of action of natural compounds isolated from lichens: A systematic review. Molecules 2014, 19, 14496–14527. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Moriano, C.; Gonzáles-Burgos, E.; Divakar, P.K.; Crespo, A.; Gómez-Serranillos, M.P. Evaluation of the antioxidant capacities and cytotoxic effects of the Parmleiaceae lichen species. Evid. Based Complementary Altern. Med. 2016, 2016, 3169751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsen, B.; Aslan, A.; Togar, B.; Turkez, H. In vitro antitumor activities of the lichen compounds olivetoric, physodic and psoromic acid in rat neuron and glioblastoma cells. Pharm. Biol. 2016, 54, 1748–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studzińska-Sroka, E.; Piotrowska, H.; Kucińska, M.; Murias, M.; Bylka, W. Cytotoxic activity of physodic acid and acetone extract from Hypogymnia physodes against breast cancer cell lines. Pharm. Biol. 2016, 54, 2480–2485. [Google Scholar] [CrossRef] [Green Version]
- Divakar, P.K.; Crespo, A.; Wedin, M.; Leavitt, S.D.; Hawksworth, D.L.; Myllys, L.; McCune, B.; Randlane, T.; Bjerke, J.W.; Ohmura, Y.; et al. Evolution of complex symbiotic relationships in a morphologically derived family of lichen-forming fungi. New Phytol. 2015, 208, 1217–1226. [Google Scholar] [CrossRef] [Green Version]
- Kraichak, E.; Divakar, P.K.; Crespo, A.; Leavitt, S.D.; Nelsen, M.P.; Lücking, R.; Lumbsch, H.T. A tale of two hyper-diversities: Diversification dynamics of the two largest families of lichenized fungi. Sci. Rep. 2015, 5, e10028. [Google Scholar] [CrossRef] [Green Version]
- Elix, J.A. Progress in the generic delimitation of Parmelia sensu lato lichens (Ascomycotina: Parmeliaceae) and a synoptic key to the Parmeliaceae. Bryologist 1993, 96, 359–383. [Google Scholar] [CrossRef]
- Elix, J.A.; Johnston, J.; Verdon, D. Canoparmelia, Paraparmelia and Relicinopsis, three new genera in the Parmeliaceae (lichenized Ascomycotina). Mycotaxon 1986, 27, 271–282. [Google Scholar]
- Lendemer, J.C.; Hodkinson, B.P. Recognition of the Parmelia crozalsiana group as the genus Crespoa. N. Am. Fungi 2012, 7, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Kirika, P.; Divakar, P.K.; Crespo, A.; Leavitt, S.D.; Mugambi, G.K.; Gatheri, G.W.; Lumbsch, H.T. Polyphyly of the genus Canoparmelia—Another example of incongruence between phenotype-based classification and molecular phylogeny within lichenized Ascomycota (Parmeliaceae). Phytotaxa 2016, 289, 36–48. [Google Scholar] [CrossRef] [Green Version]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Larena, I.; Salazar, O.; González, V.; Julián, M.C.; Rubio, V. Design of a primer for ribosomal DNA internal transcribed spacer with enhanced specificity for ascomycetes. J. Biotechnol. 1999, 75, 187–194. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Zoller, S.; Scheidegger, C.; Sperisen, C. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 1999, 31, 511–516. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [Green Version]
- Wiens, J.J. Combining data sets with different phylogenetic histories. Syst. Biol. 1998, 47, 568–581. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Arup, U.; Ekman, S.; Lindblom, L.; Mattsson, J.-E. High performance thin layer chromatography (HPTLC), an improved technique for screening lichen substances. Lichenologist 1993, 25, 61–71. [Google Scholar] [CrossRef]
- Gadea, A.; Le Lamer, A.C.; Le Gall, S.; Jonard, C.; Ferron, S.; Catheline, D.; Ertz, D.; Le Pogam, P.; Boustie, J.; Lohezic-Le Devehat, F.; et al. Intrathalline metabolite profiles in the lichen Argopsis friesiana shape gastropod grazing patterns. J. Chem. Ecol. 2018, 44, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef] [PubMed]
- Ferron, S.; Berry, O.; Olivier-Jimenez, D.; Rouaud, I.; Boustie, J.; Lohézic-Le Dévéhat, F.; Poncet, R. Chemical diversity of five coastal Roccella species from mainland France, the Scattered Islands, and São Tomé and Príncipe. Plant Fungal Syst. 2020, 65, 247–260. [Google Scholar] [CrossRef]
- Amarowicz, R.; Pegg, R.B.; Rahimi-Moghaddam, P.; Barl, B.; Weil, J.A. Free-radical scavenging capacity and antioxidant activity of selected plant species from the Canadian prairies. Food Chem. 2004, 84, 551–562. [Google Scholar] [CrossRef]
- Davalos, A.; Gomez-Cordoves, C.; Bartolome, B. Extending applicability of the oxygen radical absorbance capacity (ORAC-fluorescein) assay. J. Agric. Food Chem. 2004, 52, 48–54. [Google Scholar] [CrossRef]
- Avan, A.N.; Cekic, S.D.; Uzunboy, S.; Apak, R. Spectrophotometric Determination of Phenolic Antioxidants in the Presence of Thiols and Proteins. Int. J. Mol. Sci. 2016, 17, 1325. [Google Scholar] [CrossRef]
- Folin, O.; Ciocalteu, V. On tyrosine and tryptophane determinations in proteins. J. Biol. Chem. 1927, 73, 627–650. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival—Application to prolifertaion and cyto-toxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Lendemer, J.C.; Ruiz, A.M. Molecular data confirm morphological variability the widespread foliose lichen Canoparmelia carolinana (Parmeliaceae). Castania 2015, 80, 29–36. [Google Scholar] [CrossRef]
- Kirika, P.; Lumbsch, H.; Garrido Huéscar, E.; Quedensley, T.; Divakar, P.K. Canoparmelia texana (Parmeliaceae, Ascomycota) consists of two independent lineages. Lichenologist 2022, 54, 183–194. [Google Scholar] [CrossRef]
- Culberson, C.F.; Culberson, W.L. Chemosyndromic variation in lichens. Syst. Bot. 1976, 1, 325–339. [Google Scholar] [CrossRef]
- Aoussar, N.; Laasri, F.E.; Bourhia, M.; Manoljovic, N.; Mhand, R.A.; Rhallabi, N.; Ullah, R.; Shahat, A.A.; Noman, O.M.; Nasr, F.A.; et al. Phytochemical Analysis, Cytotoxic, Antioxidant, and Antibacterial Activities of Lichens. Evid. Based Complementary Altern. Med. 2020, 2020, 8104538. [Google Scholar] [CrossRef]
- Sies, H. Oxidative Stress: Concept and Some Practical Aspects. Antioxidants 2020, 9, 852. [Google Scholar] [CrossRef]
- Liang, N.J.; Kitts, D.D. Antioxidant Property of Coffee Components: Assessment of Methods that Define Mechanisms of Action. Molecules 2014, 19, 19180–19208. [Google Scholar] [CrossRef] [Green Version]
- Comsa, S.; Cîmpean, A.; Raica, M. The Story of MCF-7 Breast Cancer Cell Line: 40 years of Experience in Research. Anticancer Res. 2015, 35, 3147. [Google Scholar]
- Shrestha, G.; Clair, L.L.S. Lichens: A promising source of antibiotic and anticancer drugs. Phytochem. Rev. 2013, 12, 229–244. [Google Scholar] [CrossRef]

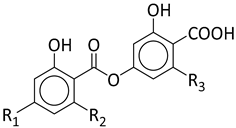 | 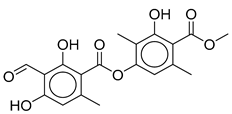 | ||||||
|---|---|---|---|---|---|---|---|
| Stenosporic Acid | Perlatolic Acid | Glomelliferic Acid | Divaricatic Acid | Nordivaricatic Acid | Subdivaricatic ACID | Atranorin | |
| R1 | OCH3 | OCH3 | OCH3 | OCH3 | OH | OCH3 | |
| R2 | C3H7 | C4H9 | (CH2-CO-C3H7) | C3H7 | C3H7 | CH3 | |
| R3 | C4H9 | C4H9 | C4H9 | C3H7 | C3H7 | C3H7 | |
| C. amabilis (n = 2) | 45.8 | 34.5 | 16.8 | - | - | - | 1.9 |
| C. caroliniana (n = 8) | 46.2 | 31.8 | 15.9 | - | - | - | 5.6 |
| C. kakamegaensis * | 60.2 | 23.6 | 15.8 | - | - | - | 0.4 |
| C. concrescens (n = 2) | 4.9 | - | - | 85.3 | 4.5 | 5.3 | - |
| Lichen Species | Yields (% w/w) | Total Phenolic Contents (µg GA/mg) | DPPH EC50 (µg/mL) | ORAC Value (µmol TE/mg Dry Extract) | FRAP (µmol of Fe2+ eq/g Sample) |
|---|---|---|---|---|---|
| Canoparmelia kakamegaensis | 18.5 ± 2.0 | 53.7 ± 2.8 * | 400.2 ± 34.9 * | 3.2 ± 0.1 * | 9.6 ± 0.2 |
| Canoparmelia caroliniana | 4.9 ± 0.7 | 44.7 ± 2.2 | 1004.3 ± 112.8 | 1.3 ± 0.04 | 10.8 ± 1.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrido-Huéscar, E.; González-Burgos, E.; Kirika, P.M.; Boustie, J.; Ferron, S.; Gómez-Serranillos, M.P.; Lumbsch, H.T.; Divakar, P.K. A New Cryptic Lineage in Parmeliaceae (Ascomycota) with Pharmacological Properties. J. Fungi 2022, 8, 826. https://doi.org/10.3390/jof8080826
Garrido-Huéscar E, González-Burgos E, Kirika PM, Boustie J, Ferron S, Gómez-Serranillos MP, Lumbsch HT, Divakar PK. A New Cryptic Lineage in Parmeliaceae (Ascomycota) with Pharmacological Properties. Journal of Fungi. 2022; 8(8):826. https://doi.org/10.3390/jof8080826
Chicago/Turabian StyleGarrido-Huéscar, Elisa, Elena González-Burgos, Paul M. Kirika, Joël Boustie, Solenn Ferron, M. Pilar Gómez-Serranillos, Helge Thorsten Lumbsch, and Pradeep K. Divakar. 2022. "A New Cryptic Lineage in Parmeliaceae (Ascomycota) with Pharmacological Properties" Journal of Fungi 8, no. 8: 826. https://doi.org/10.3390/jof8080826
APA StyleGarrido-Huéscar, E., González-Burgos, E., Kirika, P. M., Boustie, J., Ferron, S., Gómez-Serranillos, M. P., Lumbsch, H. T., & Divakar, P. K. (2022). A New Cryptic Lineage in Parmeliaceae (Ascomycota) with Pharmacological Properties. Journal of Fungi, 8(8), 826. https://doi.org/10.3390/jof8080826








