Comparative Copper Resistance Strategies of Rhodonia placenta and Phanerochaete chrysosporium in a Copper/Azole-Treated Wood Microcosm
Abstract
1. Introduction
2. Material and Methods
2.1. Microcosm Set-Up
2.2. Respiration Tests
2.3. X-ray Fluorescence Spectroscopy and Inductively Coupled Plasma Atomic Emission Spectrometry for Copper Quantification in the Liquid Phase
2.4. Scanning Electron Microscopy (SEM) and Electron Dispersive Spectroscopy (EDS) Microanalyses for Copper Quantification in Sawdust and Fungal Hyphae
2.5. High Performance Liquid Chromatography for Soluble Oxalate Quantification
2.6. Statistical Analyses
2.7. Genome Mining to Compare Copper Related Genes in R. placenta and P. chrysosporium
3. Results
3.1. Efficient Growth of Both the Brown-Rot Rhodonia Placenta and the White-Rot Phanerochaete Chrysosporium on Tanalith E3474-Treated Wood
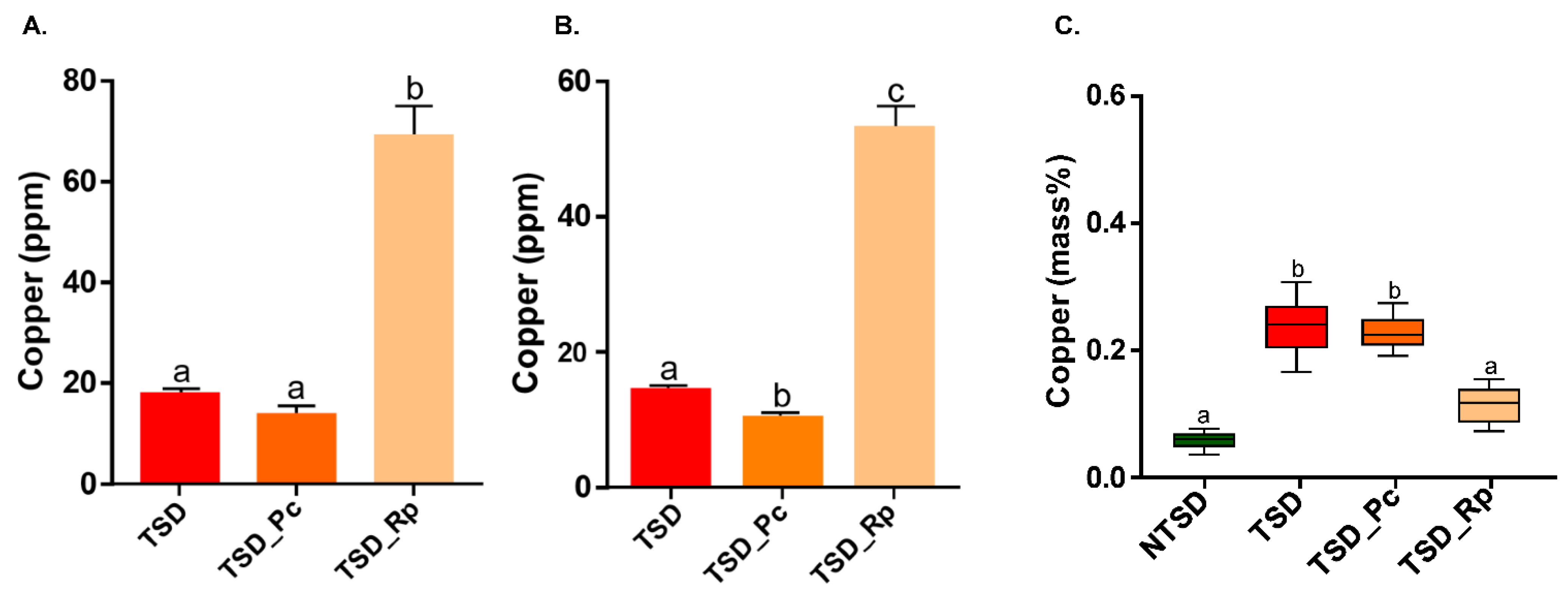
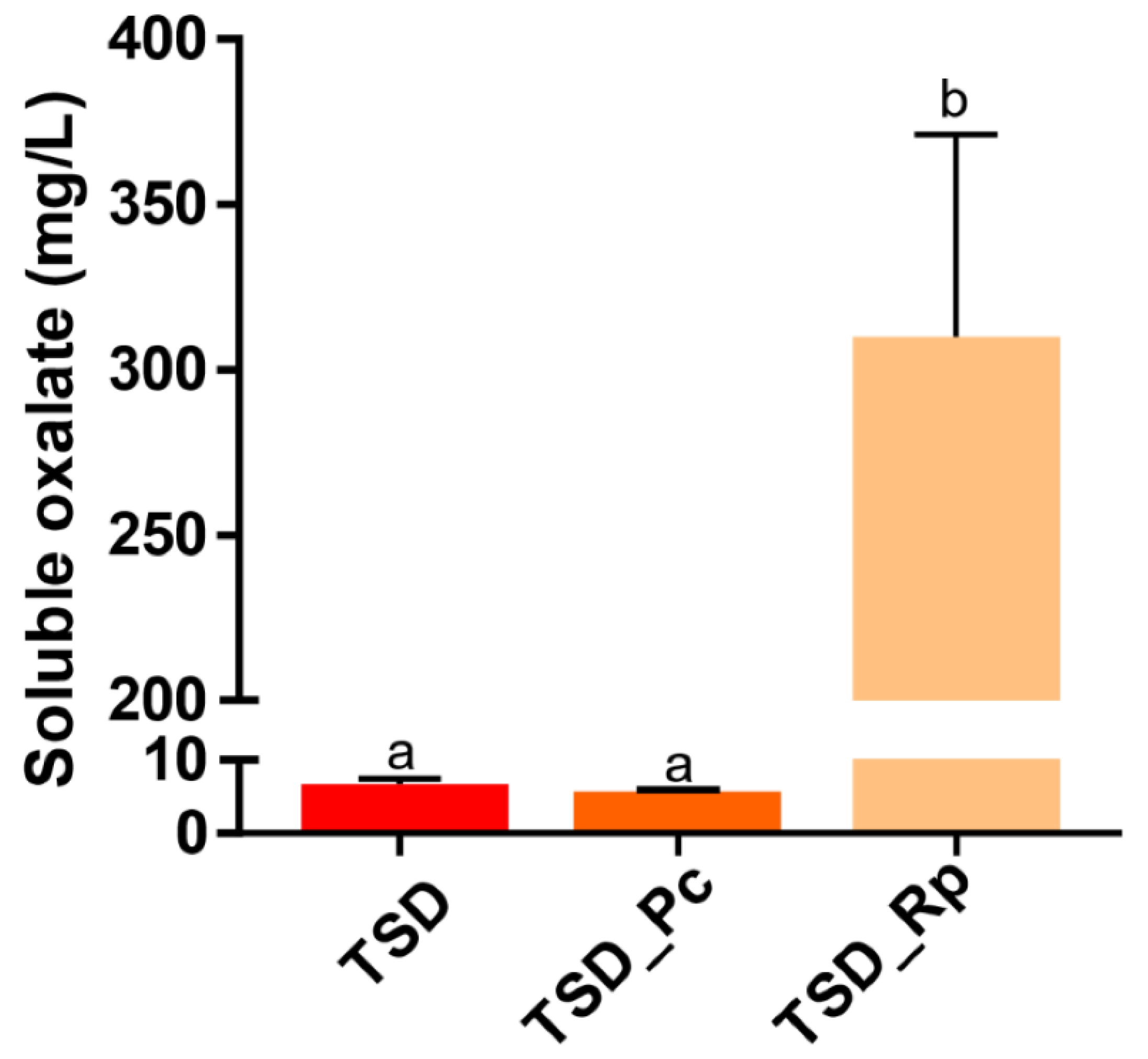
3.2. Comparative Copper Bioleaching Ability of R. placenta and P. chrysosporium from Tanalith E3474 Treated Wood
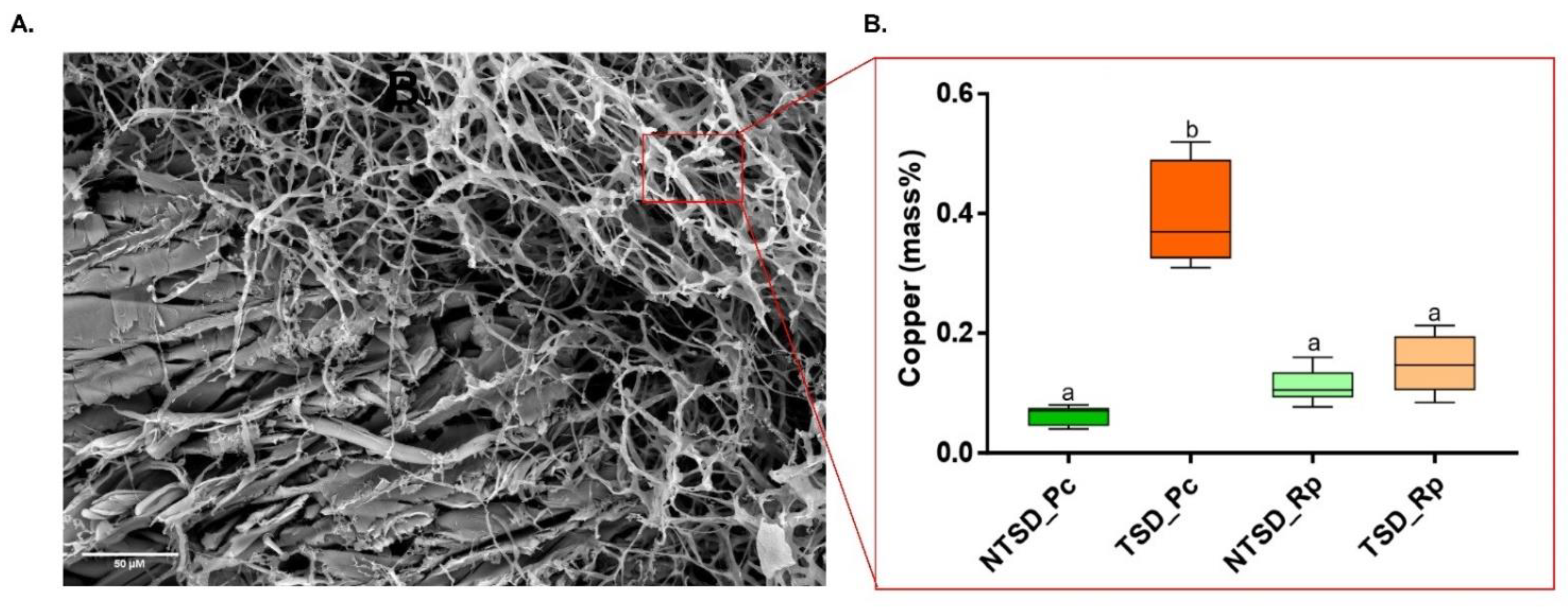
3.3. SEM-EDS Based Comparative Analysis of Copper Detoxification Strategies between R. placenta and P. chrysosporium
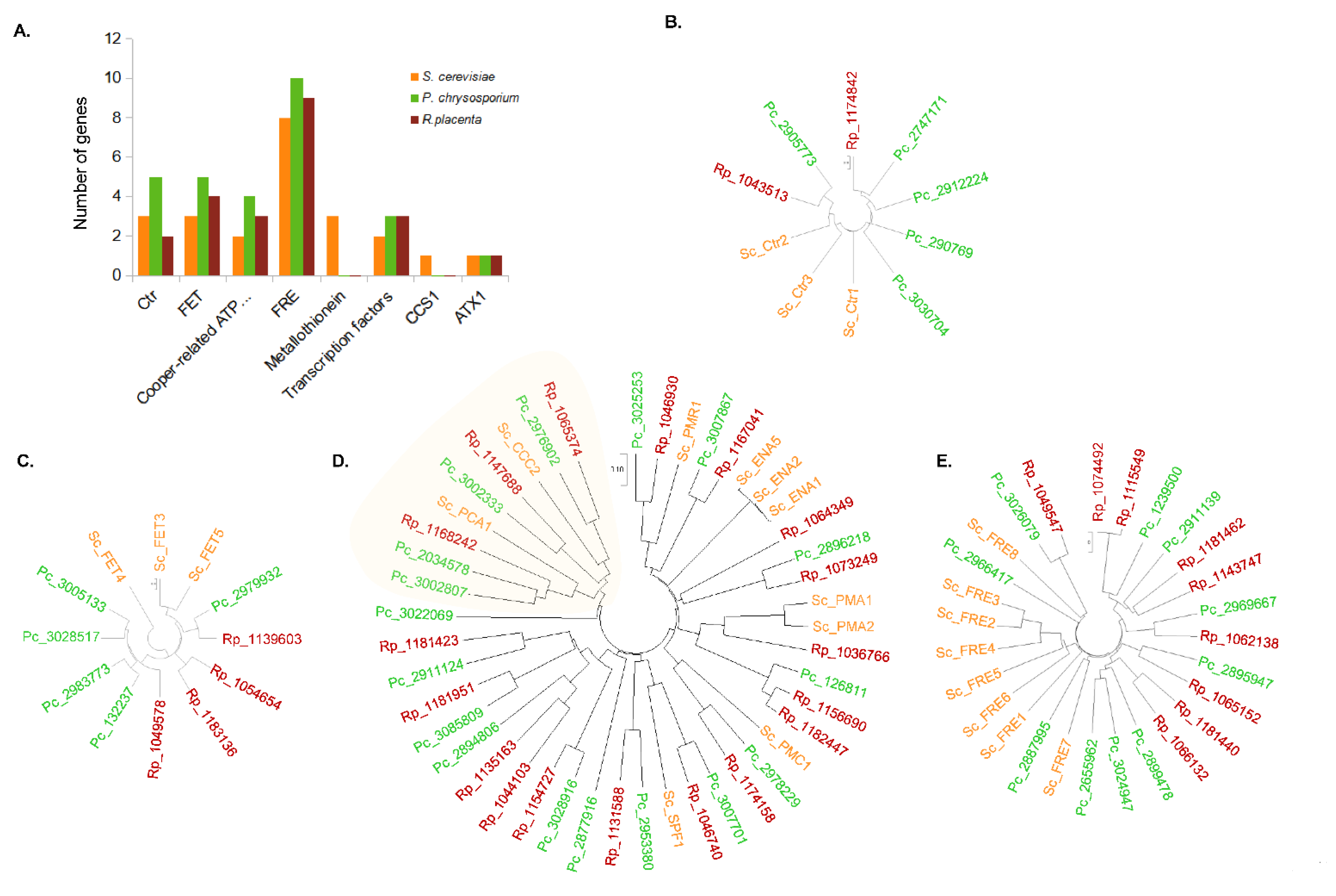
3.4. Comparative Genomics of Copper-Related Genes in R. placenta and P. chrysosporium
4. Discussion
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Verbist, M.; Nunes, L.; Jones, D.; Branco, J.M. Service Life Design of Timber Structures; Woodhead Publishing: Sawston, UK, 2019; Volume 11, pp. 311–336. [Google Scholar] [CrossRef]
- Ramage, M.H.; Henry, B.; Marta, B.-W.; George, F.; Thomas, R.; Darshil, U.S.; Guanglu, W.; Li, Y.; Patrick, F.; Danielle, D.-T.; et al. The wood from the trees: The use of timber in construction. Renew. Sustain. Energy Rev. 2017, 68, 333–359. [Google Scholar] [CrossRef]
- Rowell, R.M. (Ed.) Handbook of Wood Chemistry and Wood Composites, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar] [CrossRef]
- Marais, B.N.; Brischke, C.; Militz, H. Wood durability in terrestrial and aquatic environments–A review of biotic and abiotic influence factors. Wood Mater. Sci. Eng. 2020, 17, 82–105. [Google Scholar] [CrossRef]
- Civardi, C.; van den Bulcke, J.; Schubert, M.; Michel, E.; Butron, M.I.; Boone, M.N.; Dierick, M.; Van Acker, J.; Wick, P.; Schwarze, F.W.M.R. Penetration and Effectiveness of Micronized Copper in Refractory Wood Species. PLoS ONE 2016, 11, e0163124. [Google Scholar] [CrossRef][Green Version]
- Freeman, B.M.H.; Mcintyre, C.R. Copper-based wood preservatives. For. Prod. J. 2008, 58, 6–27. [Google Scholar]
- Kartal, S.N.; Terzi, E.; Yilmaz, H.; Goodell, B. Bioremediation and decay of wood treated with ACQ, micronized ACQ, nano-CuO and CCA wood preservatives. Int. Biodeterior. Biodegrad. 2015, 99, 95–101. [Google Scholar] [CrossRef]
- Helsen, L.; Hardy, A.; Van Bael, M.K.; Mullens, J. Tanalith E 3494 impregnated wood: Characterisation and thermal behaviour. J. Anal. Appl. Pyrolysis 2007, 78, 133–139. [Google Scholar] [CrossRef]
- Woo, C.; Daniels, B.; Stirling, R.; Morris, P. Tebuconazole and propiconazole tolerance and possible degradation by Basidiomycetes: A wood-based bioassay. Int. Biodeterior. Biodegrad. 2010, 64, 403–408. [Google Scholar] [CrossRef]
- Hitchcock, C.A.; Pye, G.W.; Troke, P.F.; Johnson, E.M.; Warnock, D.W. Fluconazole resistance in Candida glabrata. Antimicrob. Agents Chemother. 1993, 37, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.R.; Isikhuemhen, O.S.; Anike, F.N. Fungal–metal interactions: A review of toxicity and homeostasis. J. Fungi 2021, 7, 225. [Google Scholar] [CrossRef]
- Macomber, L.; Imlay, J.A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 8344–8349. [Google Scholar] [CrossRef]
- Bridžiuvienė, D.; Levinskaitė, L. Fungal tolerance towards copper-based wood preservatives. Biologija 2007, 53, 54–61. [Google Scholar] [CrossRef]
- Clausen, C.A.; Green, F. Oxalic acid overproduction by copper-tolerant brown-rot basidiomycetes on southern yellow pine treated with copper-based preservatives. Int. Biodeterior. Biodegrad. 2003, 51, 139–144. [Google Scholar] [CrossRef]
- Xing, D.; Magdouli, S.; Zhang, J.; Koubaa, A. Microbial remediation for the removal of inorganic contaminants from treated wood: Recent trends and challenges. Chemosphere 2020, 258, 127429. [Google Scholar] [CrossRef]
- Deshmukh, R.; Khardenavis, A.A.; Purohit, H.J. Diverse Metabolic Capacities of Fungi for Bioremediation. Indian J. Microbiol. 2016, 56, 247–264. [Google Scholar] [CrossRef]
- Smith, A.D.; Logeman, B.L.; Thiele, D.J. Copper Acquisition and Utilization in Fungi. Annu. Rev. Microbiol. 2017, 71, 597–623. [Google Scholar] [CrossRef]
- Yasokawa, D.; Murata, S.; Kitagawa, E.; Iwahashi, Y.; Nakagawa, R.; Hashido, T.; Iwahashi, H. Mechanisms of copper toxicity in Saccharomyces cerevisiae determined by microarray analysis. Environ. Toxicol. Int. J. 2008, 23, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Antsotegi-Uskola, M.; Markina-Iñarrairaegui, A.; Ugalde, U. Copper Homeostasis in Aspergillus nidulans Involves Coordinated Transporter Function, Expression and Cellular Dynamics. Front. Microbiol. 2020, 11, 555306. [Google Scholar] [CrossRef]
- Suzuki, M.; Gitlin, G.D. Intracellular localization of the Menkes and Wilson’s disease proteins and their role in intracellular copper transport. Pediatr. Int. 1999, 41, 436–442. [Google Scholar] [CrossRef]
- Pohleven, F.; Humar, M.; Amartey, S.; Benedik, J. Tolerance of Wood Decay Fungi to Commercial Copper based Wood Preservatives; IRG/WP 02-30291; International Research Group on Wood Preservation: Stockholm, Sweden, 2002. [Google Scholar]
- Say, R.; Denizli, A.; Yakup Arica, M. Biosorption of cadmium(II), lead(II) and copper(II) with the filamentous fungus Phanerochaete chrysosporium. Bioresour. Technol. 2001, 76, 67–70. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Dutton, M.V.; Evans, C.S. Oxalate production by fungi: Its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 1996, 42, 881–895. [Google Scholar] [CrossRef]
- Gadd, G.M.; Bahri-Esfahani, J.; Li, Q.; Rhee, Y.J.; Wei, Z.; Fomina, M.; Liang, X. Oxalate production by fungi: Significance in geomycology, biodeterioration and bioremediation. Fungal Biol. Rev. 2014, 28, 36–55. [Google Scholar] [CrossRef]
- Rees, E.M.; Lee, J.; Thiele, D.J. Mobilization of intracellular copper stores by the Ctr2 vacuolar copper transporter. J. Biol. Chem. 2004, 279, 54221–54229. [Google Scholar] [CrossRef]
- Kwok, E.Y.; Severance, S.; Kosman, D.J. Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane. Biochemistry 2006, 45, 6317–6327. [Google Scholar] [CrossRef]
- Hassett, R.; Dix, D.R.; Eide, D.J.; Kosman, D.J. The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 2000, 351, 477–484. [Google Scholar] [CrossRef]
- Adle, D.J.; Sinani, D.; Kim, H.; Lee, J. A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 947–955. [Google Scholar] [CrossRef]
- Georgatsou, E.; Mavrogiannis, L.A.; Fragiadakis, G.S.; Alexandraki, D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 1997, 272, 13786–13792. [Google Scholar] [CrossRef]
- Tascioglu, C.; Tsunoda, K. Biological performance of copper azole-treated wood and wood-based composites. Holzforschung 2010, 64, 399–406. [Google Scholar] [CrossRef]
- Bak, J.S. Lignocellulose depolymerization occurs via an environmentally adapted metabolic cascades in the wood-rotting basidiomycete Phanerochaete chrysosporium. Microbiologyopen 2015, 4, 151–166. [Google Scholar] [CrossRef]
- Sierra-Alvarez, R. Removal of copper, chromium and arsenic from preservative-treated wood by chemical extraction-fungal bioleaching. Waste Manag. 2009, 29, 1885–1891. [Google Scholar] [CrossRef] [PubMed]
- Schilling, J.S.; Jellison, J. Metal accumulation without enhanced oxalate secretion in wood degraded by brown rot fungi. Appl. Environ. Microbiol. 2006, 72, 5662–5665. [Google Scholar] [CrossRef] [PubMed]
- Alluri, H.K.; Ronda, S.R.; Settalluri, V.S.; Jayakumar Singh, B.; Suryanarayana, V.; Venkateshwar, P. Biosorption: An eco-friendly alternative for heavy metal removal. Afr. J. Biotechnol. 2007, 6, 2924–2931. [Google Scholar] [CrossRef]
- Ide, M.; Ichinose, H.; Wariishi, H. Molecular identiWcation and functional characterization of cytochrome P450 monooxygenases from the brown-rot basidiomycete Postia placenta. Arch. Microbiol. 2012, 194, 243–253. [Google Scholar] [CrossRef]
- Sing, C.; Yu, J. Copper adsorption and removal from water by living mycelium of white-rot fungus Phanerochaete chrysosporium. Water Res. 1998, 32, 2746–2752. [Google Scholar] [CrossRef]
- Alfredsen, G.; Fossdal, C.G.; Nagy, N.E.; Jellison, J.; Goodell, B. Furfurylated wood: Impact on Postia placenta gene expression and oxalate crystal formation. Holzforschung 2016, 70, 947–962. [Google Scholar] [CrossRef]
- Cragg, S.M.; Beckham, G.T.; Bruce, N.C.; Bugg, T.D.H.; Distel, D.L.; Dupree, P.; Etxabe, A.G.; Goodell, B.S.; Jellison, J.; McGeehan, J.E.; et al. Lignocellulose degradation mechanisms across the Tree of Life. Curr. Opin. Chem. Biol. 2015, 29, 108–119. [Google Scholar] [CrossRef]
- Schilling, J.S. Oxalate Production and Cation Translocation during Wood Biodegradation by Fungi. Electronic PhD Theses and Dissertations, Forestry, University of Maine, 2006; 336. Available online: https://digitalcommons.library.umaine.edu/etd/336 (accessed on 9 February 2022).
- Clausen, C.A.; Green, F.; Woodward, B.M.; Evans, J.W.; DeGroot, R.C. Correlation between oxalic acid production and copper tolerance in Wolfiporia cocos. Int. Biodeterior. Biodegrad. 2000, 46, 69–76. [Google Scholar] [CrossRef]
- Schilling, J.S.; Jellison, J. Oxalate regulation by two brown rot fungi decaying oxalate-amended and non-amended wood. Holzforschung 2005, 59, 681–688. [Google Scholar] [CrossRef]
- Arantes, V.; Goodell, B. Current understanding of brown-rot fungal biodegradation mechanisms: A review. ACS Symp. Ser. 2014, 1158, 3–21. [Google Scholar] [CrossRef]
- Hattori, T.; Hisamori, H.; Suzuki, S.; Umezawa, T.; Yoshimura, T.; Sakai, H. Rapid copper transfer and precipitation by wood-rotting fungi can effect copper removal from copper sulfate-treated wood blocks during solid-state fungal treatment. Int. Biodeterior. Biodegrad. 2015, 97, 195–201. [Google Scholar] [CrossRef]
- Mäkelä, M.R.; Hildén, K.; Lundell, T.K. Oxalate decarboxylase: Biotechnological update and prevalence of the enzyme in filamentous fungi. Appl. Microbiol. Biotechnol. 2010, 87, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, M.R.; Sietiö, O.M.; De Vries, R.P.; Timonen, S.; Hildén, K. Oxalate-metabolising genes of the white-rot fungus Dichomitus squalens are differentially induced on wood and at high proton concentration. PLoS ONE 2014, 9, e87959. [Google Scholar] [CrossRef] [PubMed]
- Dutton, M.V.; Evans, C.S.; Atkey, P.T.; Wood, D.A. Oxalate production by Basidiomycetes, including the white-rot species Coriolus versicolor and Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 1993, 39, 5–10. [Google Scholar] [CrossRef]
- Kuan, I.C.; Tien, M. Stimulation of Mn peroxidase activity: A possible role for oxalate in lignin biodegradation. Proc. Natl. Acad. Sci. USA 1993, 90, 1242–1246. [Google Scholar] [CrossRef] [PubMed]
- Köse, C.; Kartal, S.N. Tolerance of brown-rot and dry-rot fungi to CCA and ACQ wood preservatives. Turkish J. Agric. For. 2010, 34, 181–190. [Google Scholar] [CrossRef]
- Iqbal, M.; Edyvean, R.G.J. Biosorption of lead, copper and zinc ions on loofa sponge immobilized biomass of Phanerochaete chrysosporium. Miner. Eng. 2004, 17, 217–223. [Google Scholar] [CrossRef]
- Rudakiya, D.M.; Iyer, V.; Shah, D.; Gupte, A.; Nath, K. Biosorption Potential of Phanerochaete chrysosporium for Arsenic, Cadmium, and Chromium Removal from Aqueous Solutions. Glob. Challenges 2018, 2, 1800064. [Google Scholar] [CrossRef]
- Noormohamadi, H.R.; Fat’hi, M.R.; Ghaedi, M.; Ghezelbash, G.R. Potentiality of white-rot fungi in biosorption of nickel and cadmium: Modeling optimization and kinetics study. Chemosphere 2019, 216, 124–130. [Google Scholar] [CrossRef]
- Civardi, C.; Grolimund, D.; Schubert, M.; Wick, P.; Schwarze, F.W.M.R. Micronized copper-treated wood: Copper remobilization into spores from the copper-tolerant wood-destroying fungus Rhodonia placenta. Environ. Sci. Nano 2019, 6, 425–431. [Google Scholar] [CrossRef]
- Dusengemungu, L.; Kasali, G.; Gwanama, C.; Ouma, K.O. Recent Advances in Biosorption of Copper and Cobalt by Filamentous Fungi. Front. Microbiol. 2020, 11, 3285. [Google Scholar] [CrossRef] [PubMed]
- da Costa, L.G.; Brocco, V.F.; Paes, J.B.; Kirker, G.T.; Bishell, A.B. Biological and chemical remediation of CCA treated eucalypt poles after 30 years in service. Chemosphere 2022, 286, 131629. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, X.; Wang, D.; Cheng, Y. Contribution characteristics of the in situ extracellular polymeric substances ( EPS ) in Phanerochaete chrysosporium to Pb immobilization. Bioprocess Biosyst. Eng. 2017, 40, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, J.; Yang, R.; Wu, L. Distribution, characteristics of extracellular polymeric substances of Phanerochaete chrysosporium under lead ion stress and the influence on Pb removal. Sci. Rep. 2020, 10, 17633. [Google Scholar] [CrossRef] [PubMed]
- Fomina, M.; Hillier, S.; Charnock, J.M.; Melville, K.; Alexander, I.J.; Gadd, G.M. Role of oxalic acid overexcretion in transformations of toxic metal minerals by Beauveria caledonica. Appl. Environ. Microbiol. 2005, 71, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Ahalya, N.; Ramachandra, T.V.; Kanamadi, R.D. Biosorption of heavy metals. Res. J. Chem. Environ. 2003, 7, 71–79. [Google Scholar] [CrossRef]
- Ramesh, G.; Podila, G.K.; Gay, G.; Marmeisse, R.; Reddy, M.S. Different Patterns of Regulation for the Copper and Cadmium Metallothioneins of the Ectomycorrhizal Fungus Hebeloma cylindrosporum. Appl. Environ. Microbiol. 2009, 75, 2266–2274. [Google Scholar] [CrossRef]
- Sácký, J.; Černý, J.; Šantrůček, J.; Borovička, J.; Leonhardt, T.; Kotrba, P. Cadmium hyperaccumulating mushroom Cystoderma carcharias has two metallothionein isoforms usable for cadmium and copper storage. Fungal Genet. Biol. 2021, 153, 103574. [Google Scholar] [CrossRef]
- Liu, B.; Dong, P.; Zhang, X.; Feng, Z.; Wen, Z.; Shi, L.; Xia, Y.; Chen, C.; Shen, Z.; Lian, C.; et al. Identification and characterization of eight metallothionein genes involved in heavy metal tolerance from the ectomycorrhizal fungus Laccaria bicolor. Environ. Sci. Pollut. Res. 2022, 29, 14430–14442. [Google Scholar] [CrossRef]
- Nguyen, H.; Rineau, F.; Vangronsveld, J.; Cuypers, A.; Colpaert, J.V.; Ruytinx, J. A novel, highly conserved metallothionein family in basidiomycete fungi and characterization of two representative SlMTa and SlMTb genes in the ectomycorrhizal fungus Suillus luteus. Environ. Microbiol. 2017, 19, 2577–2587. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandharikar, G.; Claudien, K.; Rose, C.; Billet, D.; Pollier, B.; Deveau, A.; Besserer, A.; Morel-Rouhier, M. Comparative Copper Resistance Strategies of Rhodonia placenta and Phanerochaete chrysosporium in a Copper/Azole-Treated Wood Microcosm. J. Fungi 2022, 8, 706. https://doi.org/10.3390/jof8070706
Pandharikar G, Claudien K, Rose C, Billet D, Pollier B, Deveau A, Besserer A, Morel-Rouhier M. Comparative Copper Resistance Strategies of Rhodonia placenta and Phanerochaete chrysosporium in a Copper/Azole-Treated Wood Microcosm. Journal of Fungi. 2022; 8(7):706. https://doi.org/10.3390/jof8070706
Chicago/Turabian StylePandharikar, Gaurav, Kévin Claudien, Christophe Rose, David Billet, Benoit Pollier, Aurélie Deveau, Arnaud Besserer, and Mélanie Morel-Rouhier. 2022. "Comparative Copper Resistance Strategies of Rhodonia placenta and Phanerochaete chrysosporium in a Copper/Azole-Treated Wood Microcosm" Journal of Fungi 8, no. 7: 706. https://doi.org/10.3390/jof8070706
APA StylePandharikar, G., Claudien, K., Rose, C., Billet, D., Pollier, B., Deveau, A., Besserer, A., & Morel-Rouhier, M. (2022). Comparative Copper Resistance Strategies of Rhodonia placenta and Phanerochaete chrysosporium in a Copper/Azole-Treated Wood Microcosm. Journal of Fungi, 8(7), 706. https://doi.org/10.3390/jof8070706






