A Special Phenotype of Aconidial Aspergillus niger SH2 and Its Mechanism of Formation via CRISPRi
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. The Formation of the Spore-like Propagule Assay in the Sporogenous Aspergillus niger SH2
2.3. The Purification of the Spore-like Propagule
2.4. The Routine and Fluorescent Staining of the Spore-like Propagules
2.5. Transmission Electron Microscopy
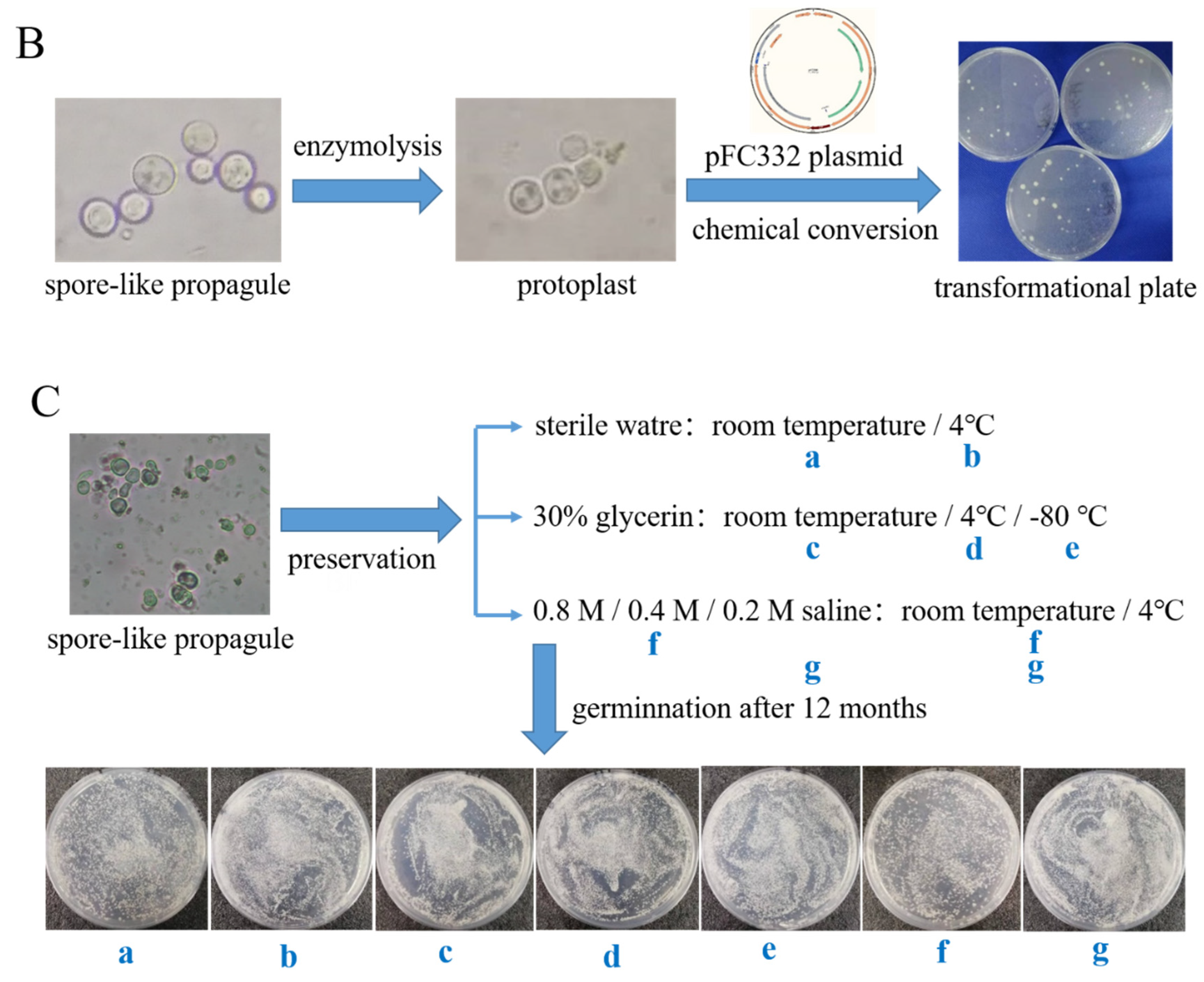
2.6. The Phenotypes and Genetic Transformation Assay of the Spore-like Propagule
2.7. RNA Purification, Quantitative RT-PCR Analysis, and RNA-seq
2.8. Construction of CRISPRi in Aspergillus niger SH2
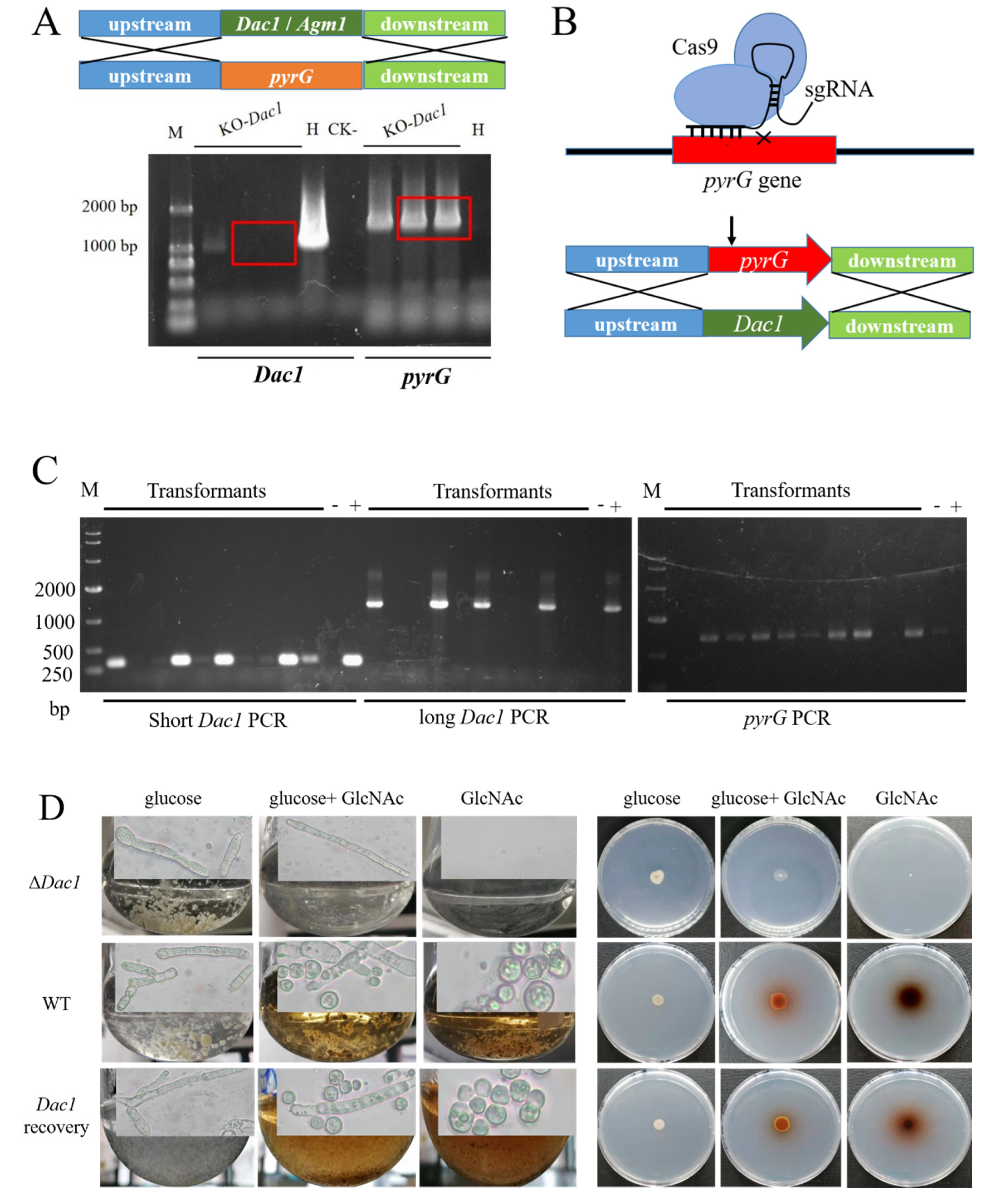
2.9. Construction of Gene Knockout or In-Situ Complementation Strains
2.10. The Phenotypes Assay of Gene Knockout or In-Situ Complementation Strains
2.11. Statistical Analyses
3. Results
3.1. The Induction, Morphology, and Subcellular Structure Assay of the Spore-like Propagule in Sporogenous Aspergillus niger SH2
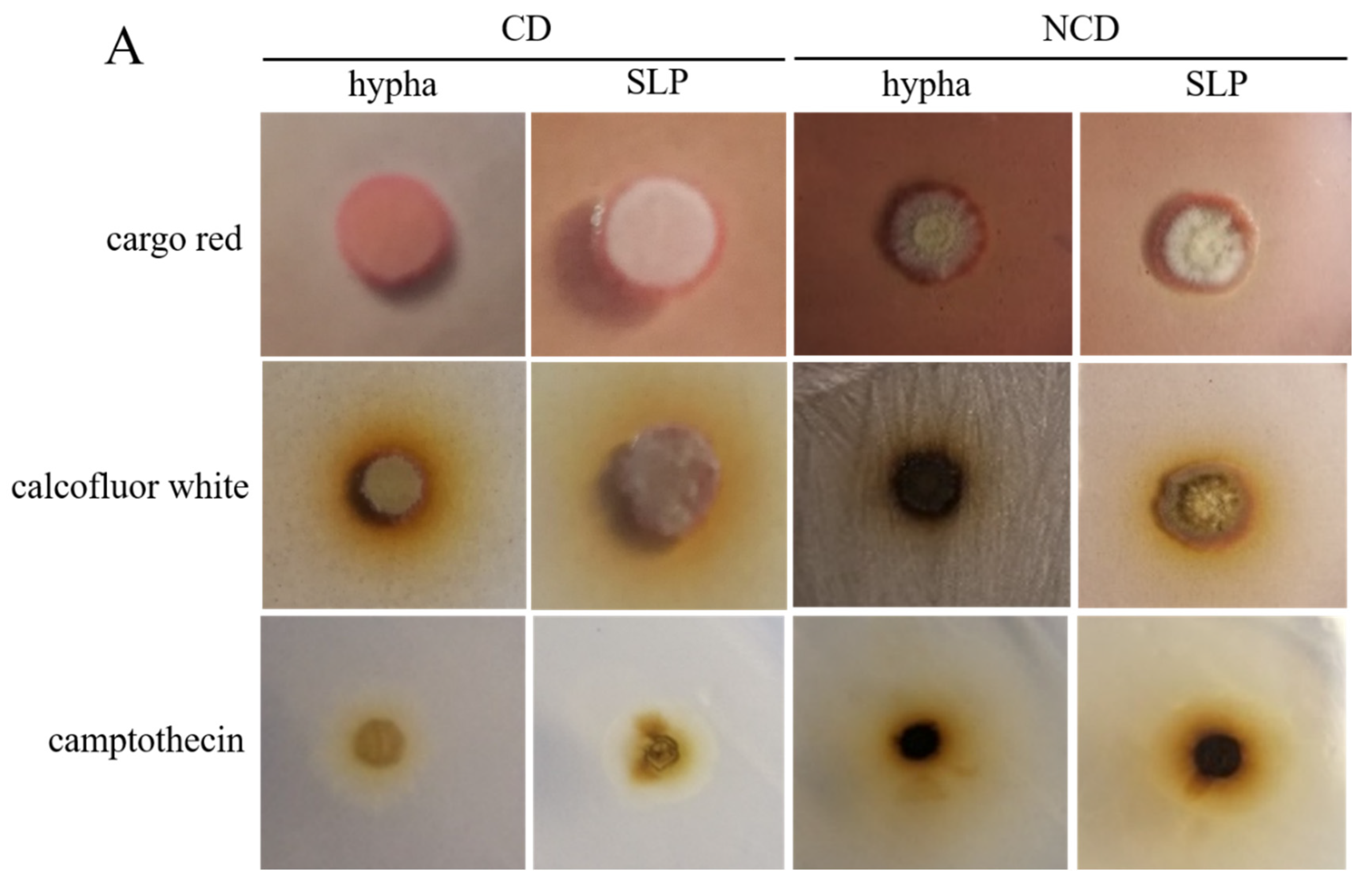
3.2. The Morphological Phenotype, Genetic Transformation, and Germination Assay of the Spore-like Propagule
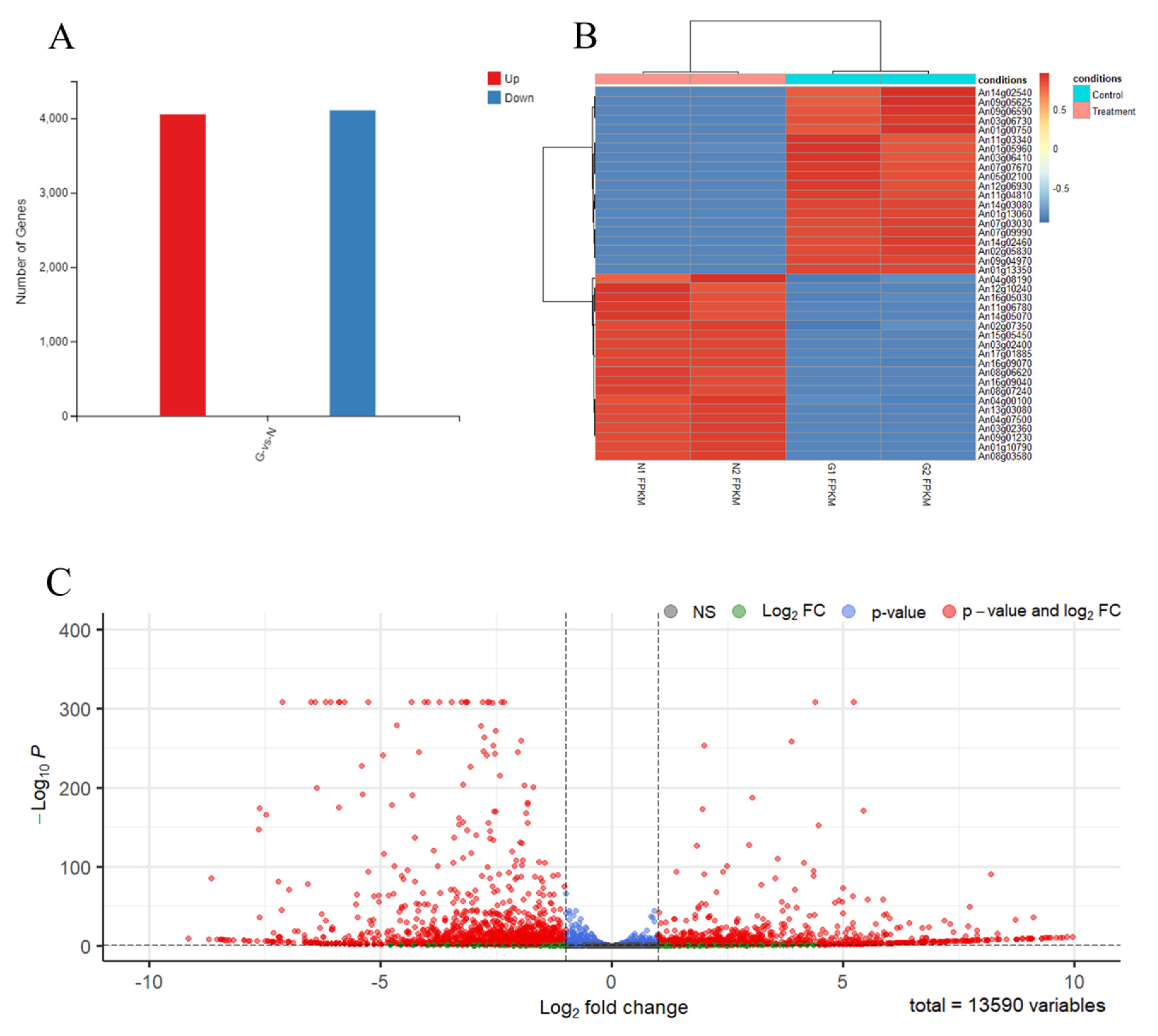
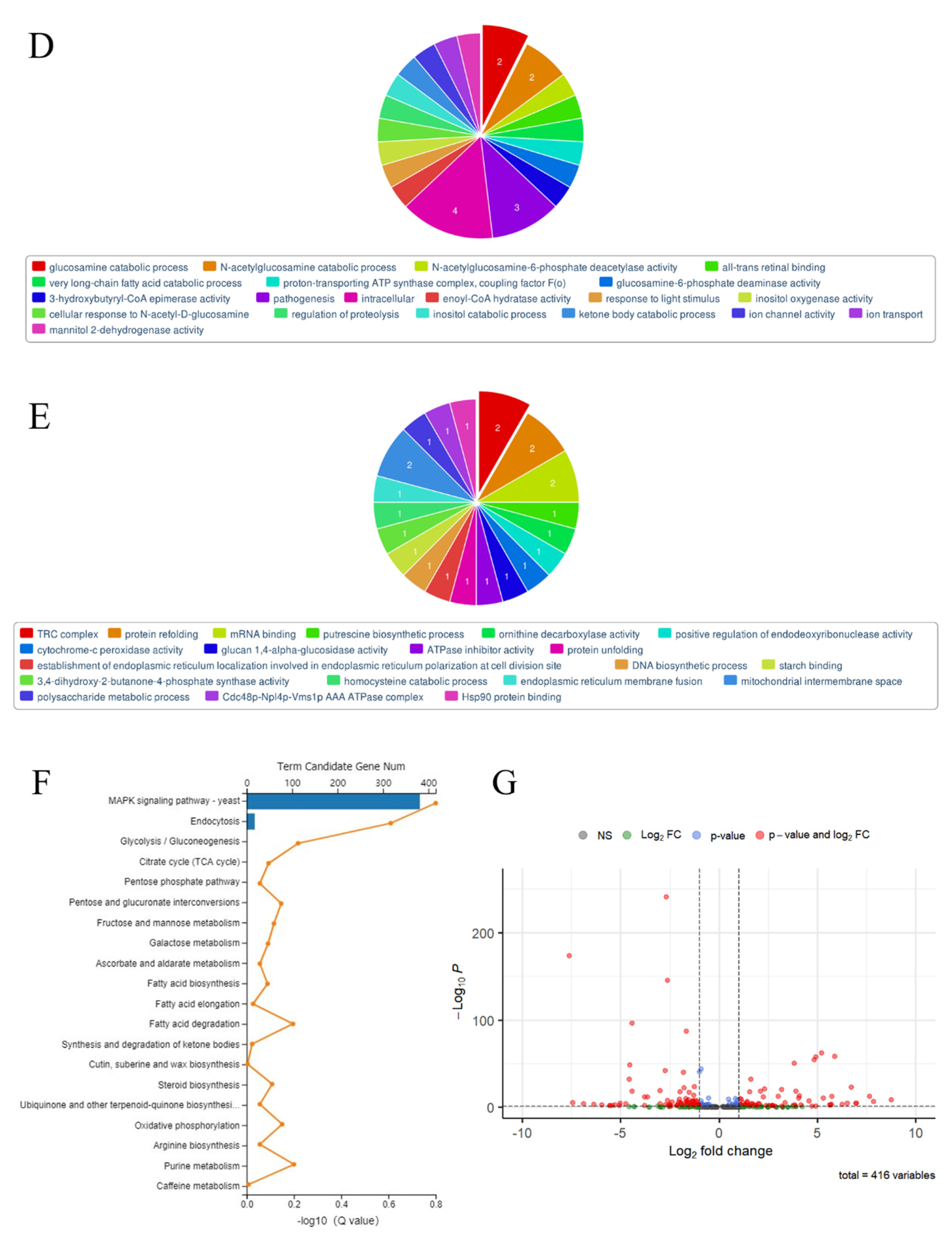
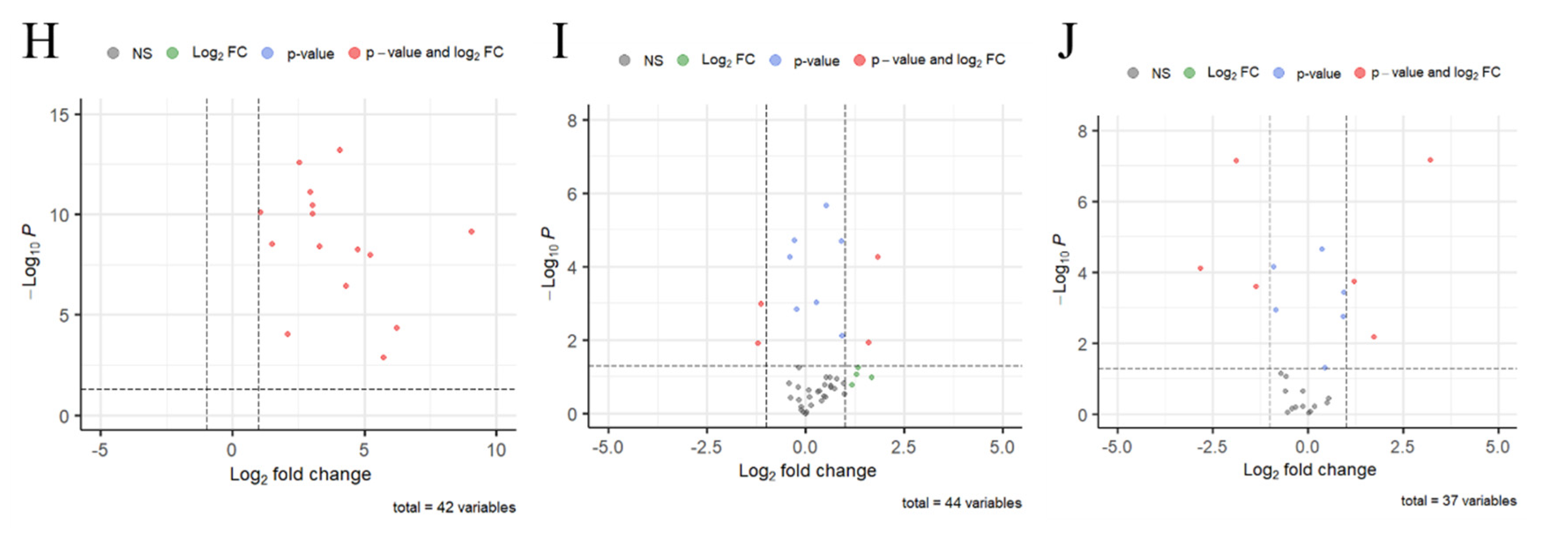
3.3. The RNA Sequence of Spore-Like Propagule
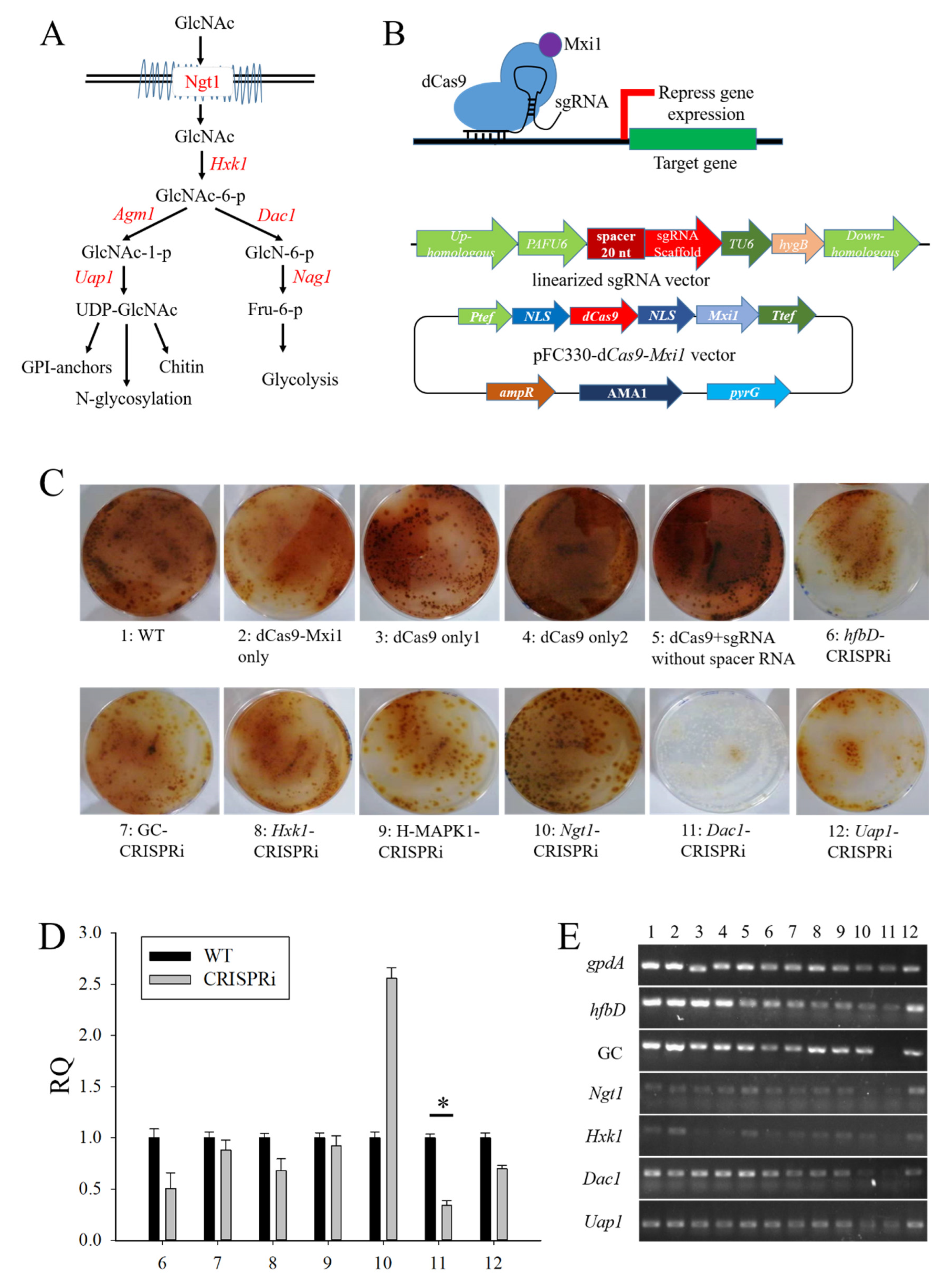
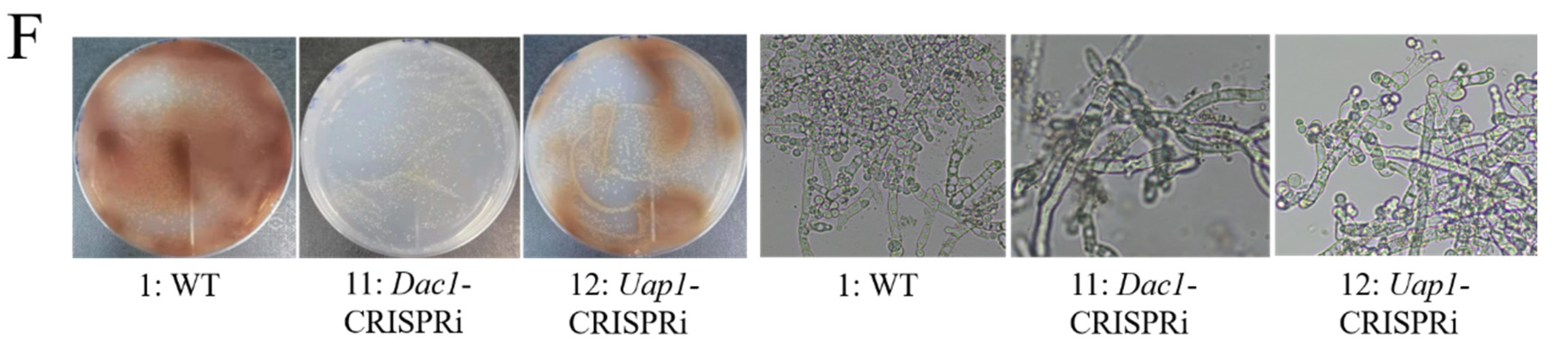
3.4. The Construction of CRISPRi on Conidia and GlcNAc Metabolic Pathway Genes in Aspergillus niger SH2
3.5. The Phenotypes Assay of Dac1 Gene Knockout and In-Situ Complementation Mutants in Aspergillus niger SH2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Demirci, E.; Arentshorst, M.; Yilmaz, B.; Swinkels, A.; Reid, I.D.; Visser, J.; Tsang, A.; Ram, A.F.J. Genetic Characterization of Mutations Related to Conidiophore Stalk Length Development in Aspergillus niger Laboratory Strain N402. Front. Genet. 2021, 12, 666684. [Google Scholar] [CrossRef] [PubMed]
- Martău, G.-A.; Unger, P.; Schneider, R.; Venus, J.; Vodnar, D.C.; López-Gómez, J.P. Integration of Solid State and Submerged Fermentations for the Valorization of Organic Municipal Solid Waste. J. Fungi 2021, 7, 766. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.-F.; Ouyang, L.-M.; Schütze, T.; Cheng, S.; Meyer, V.; Zhuang, Y.-P. Comparative genomics of the aconidial Aspergillus niger strain LDM3 predicts genes associated with its high protein secretion capacity. Appl. Microbiol. Biotechnol. 2020, 104, 2623–2637. [Google Scholar] [CrossRef] [PubMed]
- Krull, R.; Wucherpfennig, T.; Esfandabadi, M.E.; Walisko, R.; Melzer, G.; Hempel, D.C.; Kampen, I.; Kwade, A.; Wittmann, C. Characterization and control of fungal morphology for improved production performance in biotechnology. J. Biotechnol. 2013, 163, 112–123. [Google Scholar] [CrossRef]
- Shukla, P. Synthetic Biology Perspectives of Microbial Enzymes and Their Innovative Applications. Indian J. Microbiol. 2019, 59, 401–409. [Google Scholar] [CrossRef]
- Kaup, B.-A.; Ehrich, K.; Pescheck, M.; Schrader, J. Microparticle-enhanced cultivation of filamentous microorganisms: Increased chloroperoxidase formation by Caldariomyces fumago as an example. Biotechnol. Bioeng. 2007, 99, 491–498. [Google Scholar] [CrossRef]
- Krull, R.; Cordes, C.; Horn, H.; Kampen, I.; Kwade, A.; Neu, T.R.; Nörtemann, B. Morphology of Filamentous Fungi: Linking Cellular Biology to Process Engineering Using Aspergillus niger. Biosyst. Eng. 2010, 121, 1–21. [Google Scholar] [CrossRef]
- Driouch, H.; Sommer, B.; Wittmann, C. Morphology engineering of Aspergillus niger for improved enzyme production. Biotechnol. Bioeng. 2010, 105, 1058–1068. [Google Scholar] [CrossRef]
- Papagianni, M.; Mattey, M. Morphological development of Aspergillus niger in submerged citric acid fermentation as a function of the spore inoculum level. Application of neural network and cluster analysis for characterization of mycelial morphology. Microb. Cell Factories 2006, 5, 3. [Google Scholar] [CrossRef]
- Teng, Y.; Xu, Y.; Wang, N. Changes in morphology of Rhizopus chinensis in submerged fermentation and their effect on production of mycelium-bound lipase. Bioprocess Biosyst. Eng. 2008, 32, 397–405. [Google Scholar] [CrossRef]
- Takeshita, N. Control of Actin and Calcium for Chitin Synthase Delivery to the Hyphal Tip of Aspergillus. In The Fungal Cell Wall: An Armour and a Weapon for Human Fungal Pathogens; Part of the Current Topics in Microbiology and Immunology book series; Springer: Cham, Switzerland, 2020; Volume 425, pp. 113–129. [Google Scholar] [CrossRef]
- Riquelme, M. Tip Growth in Filamentous Fungi: A Road Trip to the Apex. Annu. Rev. Microbiol. 2013, 67, 587–609. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lu, J.; Zhang, G.; Liu, S.; Zhou, J.; Du, G.; Chen, J. Recent advances in the development of Aspergillus for protein production. Bioresour. Technol. 2022, 348, 126768. [Google Scholar] [CrossRef] [PubMed]
- Swift, R.J.; Wiebe, M.; Robson, G.D.; Trinci, A.P. Recombinant Glucoamylase Production byAspergillus nigerB1 in Chemostat and pH Auxostat Cultures. Fungal Genet. Biol. 1998, 25, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Wang, B.; He, P.; Lin, Y.; Pan, L. Genomic analysis of the aconidial and high-performance protein producer, industrially relevant Aspergillus niger SH2 strain. Gene 2014, 541, 107–114. [Google Scholar] [CrossRef]
- Yu, R.; Liu, J.; Wang, Y.; Wang, H.; Zhang, H. Aspergillus niger as a Secondary Metabolite Factory. Front. Chem. 2021, 9, 701022. [Google Scholar] [CrossRef]
- Geistlinger, T.; Darst, B.; Schuerg, T.; Ramesh, B.; Yoder, W. Methods and compositions for producing homokaryotic filamentous fungal cells. U.S. Patent Application 17/278,427, 2021. [Google Scholar]
- Kurt, T.; Marbà-Ardébol, A.-M.; Turan, Z.; Neubauer, P.; Junne, S.; Meyer, V. Rocking Aspergillus: Morphology-controlled cultivation of Aspergillus niger in a wave-mixed bioreactor for the production of secondary metabolites. Microb. Cell Factories 2018, 17, 128. [Google Scholar] [CrossRef]
- Driouch, H.; Hänsch, R.; Wucherpfennig, T.; Krull, R.; Wittmann, C. Improved enzyme production by bio-pellets of Aspergillus niger: Targeted morphology engineering using titanate microparticles. Biotechnol. Bioeng. 2011, 109, 462–471. [Google Scholar] [CrossRef]
- De Queiroz, M. Characterization of an Aspergillus nidulans mutant with abnormal distribution of nuclei in hyphae, metulae, phialides and conidia. FEMS Microbiol. Lett. 1998, 166, 49–55. [Google Scholar] [CrossRef][Green Version]
- Hara, S.; Tsuji, R.F.; Hatamoto, O.; Masuda, T. A Simple Method for Enrichment of Uninucleate Conidia of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2002, 66, 693–696. [Google Scholar] [CrossRef][Green Version]
- Strippoli, V.; Simonetti, N. Specific induction of chlamydospore formation inCandida albicans by N-acetyl-d-glucosamine. Experientia 1975, 31, 130–131. [Google Scholar] [CrossRef]
- Simonetti, N.S.; Strippoli, V.; Cassone, A. Yeast-mycelial conversion induced by N-acetyi-D-glucosamine in Candida alhicans. Nature 1974, 250, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, F.J.; Konopka, J.B. Identification of an N-Acetylglucosamine Transporter That Mediates Hyphal Induction in Candida albicans. Mol. Biol. Cell 2007, 18, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Oikonomou, P.; Tavazoie, S. Comprehensive Genome-wide Perturbations via CRISPR Adaptation Reveal Complex Genetics of Antibiotic Sensitivity. Cell 2020, 180, 1002–1017.e31. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Fang, H.; Zhang, D. Expanding application of CRISPR-Cas9 system in microorganisms. Synth. Syst. Biotechnol. 2020, 5, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Ostrov, N.; Wong, B.G.; Gold, M.A.; Khalil, A.S.; Church, G.M. Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nat. Microbiol. 2019, 4, 1105–1113. [Google Scholar] [CrossRef]
- Sanson, K.R.; Hanna, R.E.; Hegde, M.; Donovan, K.F.; Strand, C.; Sullender, M.E.; Vaimberg, E.W.; Goodale, A.; Root, D.E.; Piccioni, F.; et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 2018, 9, 5416. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Gilbert, L.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Román, E.; Coman, I.; Prieto, D.; Alonso-Monge, R.; Pla, J. Implementation of a CRISPR-Based System for Gene Regulation in Candida albicans. mSphere 2019, 4, e00001-19. [Google Scholar] [CrossRef]
- Wensing, L.; Sharma, J.; Uthayakumar, D.; Proteau, Y.; Chavez, A.; Shapiro, R.S. A CRISPR Interference Platform for Efficient Genetic Repression in Candida albicans. mSphere 2019, 4, e00002-19. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Suresh, S.; Schlecht, U.; Wu, M.; Wagih, O.; Peltz, G.; Davis, R.W.; Steinmetz, L.M.; Parts, L.; St.Onge, R.P. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Ding, H.; Shen, L.; He, G.-J.; Liu, H.; Tian, X.; Tao, C.; Bai, X.; Liang, J.; Jin, C.; et al. A unique cell wall synthetic response evoked by glucosamine determines pathogenicity-associated fungal cellular differentiation. PLoS Genet. 2021, 17, e1009817. [Google Scholar] [CrossRef] [PubMed]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- van Hartingsveldt, W.; Mattern, I.E.; van Zeijl, C.M.; Pouwels, P.H.; van den Hondel, C.A. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Gen. Genet. 1987, 206, 71–75. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [CrossRef]
- Semighini, C.P.; Savoldi, M.; Goldman, G.H.; Harris, S.D. Functional Characterization of the Putative Aspergillus nidulans Poly(ADP-Ribose) Polymerase Homolog PrpA. Genetics 2006, 173, 87–98. [Google Scholar] [CrossRef]
- Herrero, A.B.; López, M.C.; Fernández-Lago, L.; Domínguez, A. Candida albicans and Yarrowia lipolytica as alternative models for analysing budding patterns and germ tube formation in dimorphic fungi. Microbiology 1999, 145, 2727–2737. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, S.A.; Naseem, S.; Konopka, J.B.; Sil, A. N-acetylglucosamine (GlcNAc) Triggers a Rapid, Temperature-Responsive Morphogenetic Program in Thermally Dimorphic Fungi. PLoS Genet. 2013, 9, e1003799. [Google Scholar] [CrossRef] [PubMed]
- Kopecká, M.; Gabriel, M. The influence of congo red on the cell wall and (1–3)-beta-D-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch. Microbiol. 1992, 158, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Mora-Montes, H.M.; Netea, M.G.; Ferwerda, G.; Lenardon, M.D.; Brown, G.D.; Mistry, A.R.; Kullberg, B.J.; O’Callaghan, C.A.; Sheth, C.C.; Odds, F.C.; et al. Recognition and Blocking of Innate Immunity Cells by Candida albicans Chitin. Infect. Immun. 2011, 79, 1961–1970. [Google Scholar] [CrossRef]
- Cairns, T.C.; Nai, C.; Meyer, V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 2018, 5, 13. [Google Scholar] [CrossRef]
- Monge, R.A.; Román, E.; Nombela, C.; Pla, J. The MAP kinase signal transduction network in Candida albicans. Microbiology 2006, 152, 905–912. [Google Scholar] [CrossRef]
- Xu, J.-R. MAP Kinases in Fungal Pathogens. Fungal Genet. Biol. 2000, 31, 137–152. [Google Scholar] [CrossRef]
- Williams, R.B.; Lorenz, M.C. Multiple Alternative Carbon Pathways Combine To Promote Candida albicans Stress Resistance, Immune Interactions, and Virulence. mBio 2020, 11, e03070-19. [Google Scholar] [CrossRef]
- Hai, T.; Curran, T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. PANS 1991, 88, 3720–3724. [Google Scholar] [CrossRef]
- Garcia-Gimeno, M.A.; Struhl, K. Aca1 and Aca2, ATF/CREB Activators in Saccharomyces cerevisiae, Are Important for Carbon Source Utilization but Not the Response to Stress. Mol. Cell. Biol. 2000, 20, 4340–4349. [Google Scholar] [CrossRef]
- Takeda, T.; Toda, T.; Kominami, K.; Kohnosu, A.; Yanagida, M.; Jones, N. Schizosaccharomyces pombe atfl encodes a transcription factor required for sexual development and entry into stationary phase. EMBO 1995, 14, 6193–6208. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Arima, T.-H.; Iwashita, K.; Yamada, O.; Gomi, K.; Akita, O. Aspergillus oryzae atfB encodes a transcription factor required for stress tolerance in conidia. Fungal Genet. Biol. 2008, 45, 922–932. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, M.; Krijgsheld, P.; Bleichrodt, R.; Menke, H.; Stam, H.; Stark, J.; Wösten, H.; Dijksterhuis, J. Germination of conidia of Aspergillus niger is accompanied by major changes in RNA profiles. Stud. Mycol. 2013, 74, 59–70. [Google Scholar] [CrossRef]
- Hagiwara, D.; Takahashi, H.; Kusuya, Y.; Kawamoto, S.; Kamei, K.; Gonoi, T. Comparative transcriptome analysis revealing dormant conidia and germination associated genes in Aspergillus species: An essential role for AtfA in conidial dormancy. BMC Genom. 2016, 17, 358. [Google Scholar] [CrossRef] [PubMed]
- Lew, S.Q.; Lin, C.-H. N-acetylglucosamine-mediated morphological transition in Candida albicans and Candida tropicalis. Curr. Genet. 2021, 67, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Kamthan, M.; Kamthan, A.; Ruhela, D.; Maiti, P.; Bhavesh, N.S.; Datta, A. Upregulation of galactose metabolic pathway by N-acetylglucosamine induced endogenous synthesis of galactose in Candida albicans. Fungal Genet. Biol. 2013, 54, 15–24. [Google Scholar] [CrossRef]
- Li, Z.; Vizeacoumar, F.J.; Bahr, S.; Li, J.; Warringer, J.; Vizeacoumar, F.S.; Min, R.; VanderSluis, B.; Bellay, J.; DeVit, M.; et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 2011, 29, 361–367. [Google Scholar] [CrossRef]
- Breslow, D.; Cameron, D.M.; Collins, S.; Schuldiner, M.; Stewart-Ornstein, J.; Newman, H.W.; Braun, S.; Madhani, H.; Krogan, N.J.; Weissman, J.S. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 2008, 5, 711–718. [Google Scholar] [CrossRef]
- Yan, Z.; Costanzo, M.; Heisler, L.; Paw, J.; Kaper, F.; Andrews, B.J.; Boone, C.; Giaever, G.; Nislow, C. Yeast Barcoders: A chemogenomic application of a universal donor-strain collection carrying bar-code identifiers. Nat. Methods 2008, 5, 719–725. [Google Scholar] [CrossRef]
- Reynolds, A.; Leake, D.; Boese, Q.; Scaringe, S.; Marshall, W.S.; Khvorova, A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004, 22, 326–330. [Google Scholar] [CrossRef]
- Fellmann, C.; Zuber, J.; McJunkin, K.; Chang, K.; Malone, C.D.; Dickins, R.A.; Xu, Q.; Hengartner, M.O.; Elledge, S.J.; Hannon, G.J.; et al. Functional Identification of Optimized RNAi Triggers Using a Massively Parallel Sensor Assay. Mol. Cell 2011, 41, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Naseem, S.; Parrino, S.M.; Buenten, D.M.; Konopka, J.B. Novel roles for GlcNAc in cell signaling. Commun. Integr. Biol. 2012, 5, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Naseem, S.; Gunasekera, A.; Araya, E.; Konopka, J.B. N-Acetylglucosamine (GlcNAc) Induction of Hyphal Morphogenesis and Transcriptional Responses in Candida albicans Are Not Dependent on Its Metabolism. J. Biol. Chem. 2011, 286, 28671–28680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ghosh, S.; Bhatt, D.N.; Narula, A.; Datta, A. Magnaporthe oryzae aminosugar metabolism is essential for successful host colonization. Environ. Microbiol. 2016, 18, 2768. [Google Scholar] [CrossRef]
- Kappel, L.; Gaderer, R.; Flipphi, M.; Seidl-Seiboth, V. The N-acetylglucosamine catabolic gene cluster in Trichoderma reesei is controlled by the Ndt80-like transcription factor RON1. Mol. Microbiol. 2015, 99, 640–657. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, L.; Yuan, S.; Zhou, Q.; Wang, X.; Wang, L.; Hu, Z.; Yan, Y. NGT1 Is Essential for N-Acetylglucosamine-Mediated Filamentous Growth Inhibition and HXK1 Functions as a Positive Regulator of Filamentous Growth in Candida tropicalis. Int. J. Mol. Sci. 2020, 21, 4036. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.-Y.; Li, L.-X.; Yao, L.-L.; Zheng, J.-W.; Wang, B.; Pan, L. A Special Phenotype of Aconidial Aspergillus niger SH2 and Its Mechanism of Formation via CRISPRi. J. Fungi 2022, 8, 679. https://doi.org/10.3390/jof8070679
Yu L-Y, Li L-X, Yao L-L, Zheng J-W, Wang B, Pan L. A Special Phenotype of Aconidial Aspergillus niger SH2 and Its Mechanism of Formation via CRISPRi. Journal of Fungi. 2022; 8(7):679. https://doi.org/10.3390/jof8070679
Chicago/Turabian StyleYu, Le-Yi, Lin-Xiang Li, Lin-Lin Yao, Jun-Wei Zheng, Bin Wang, and Li Pan. 2022. "A Special Phenotype of Aconidial Aspergillus niger SH2 and Its Mechanism of Formation via CRISPRi" Journal of Fungi 8, no. 7: 679. https://doi.org/10.3390/jof8070679
APA StyleYu, L.-Y., Li, L.-X., Yao, L.-L., Zheng, J.-W., Wang, B., & Pan, L. (2022). A Special Phenotype of Aconidial Aspergillus niger SH2 and Its Mechanism of Formation via CRISPRi. Journal of Fungi, 8(7), 679. https://doi.org/10.3390/jof8070679





