A Perilipin Affects Lipid Droplet Homeostasis and Aerial Hyphal Growth, but Has Only Small Effects on Virulence in the Insect Pathogenic Fungus Beauveria bassiana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Insect Larvae
2.2. Molecular Manipulations
2.3. Expression Analysis
2.4. Lipid Profiles Analysis
2.5. Effect of BbPlin1 Gene Deletion on B. bassiana Growth
2.6. Phenotypic Characterizations and Insect Bioassays
2.7. Transcriptome Analysis of Mutant Strains
3. Results
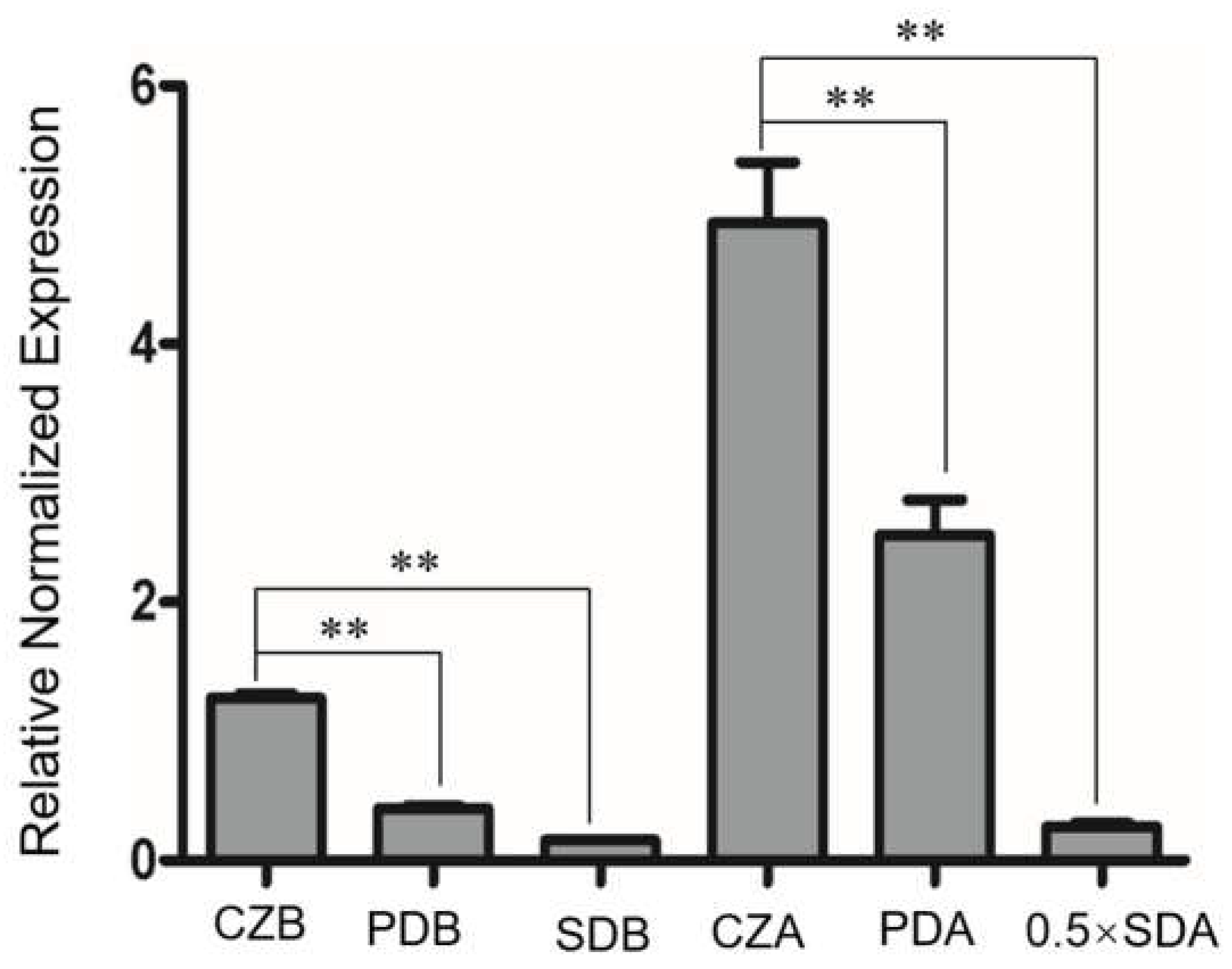
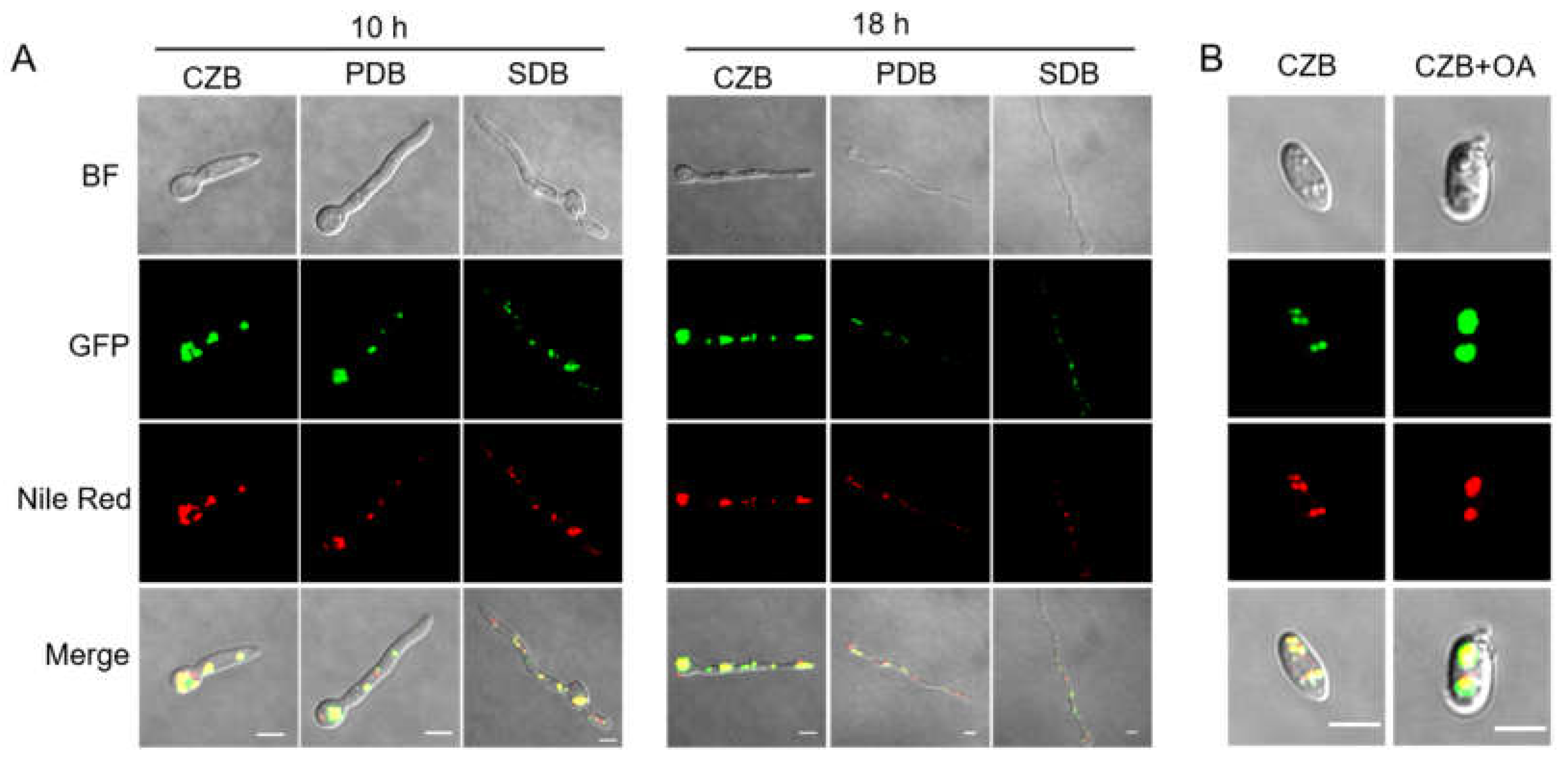
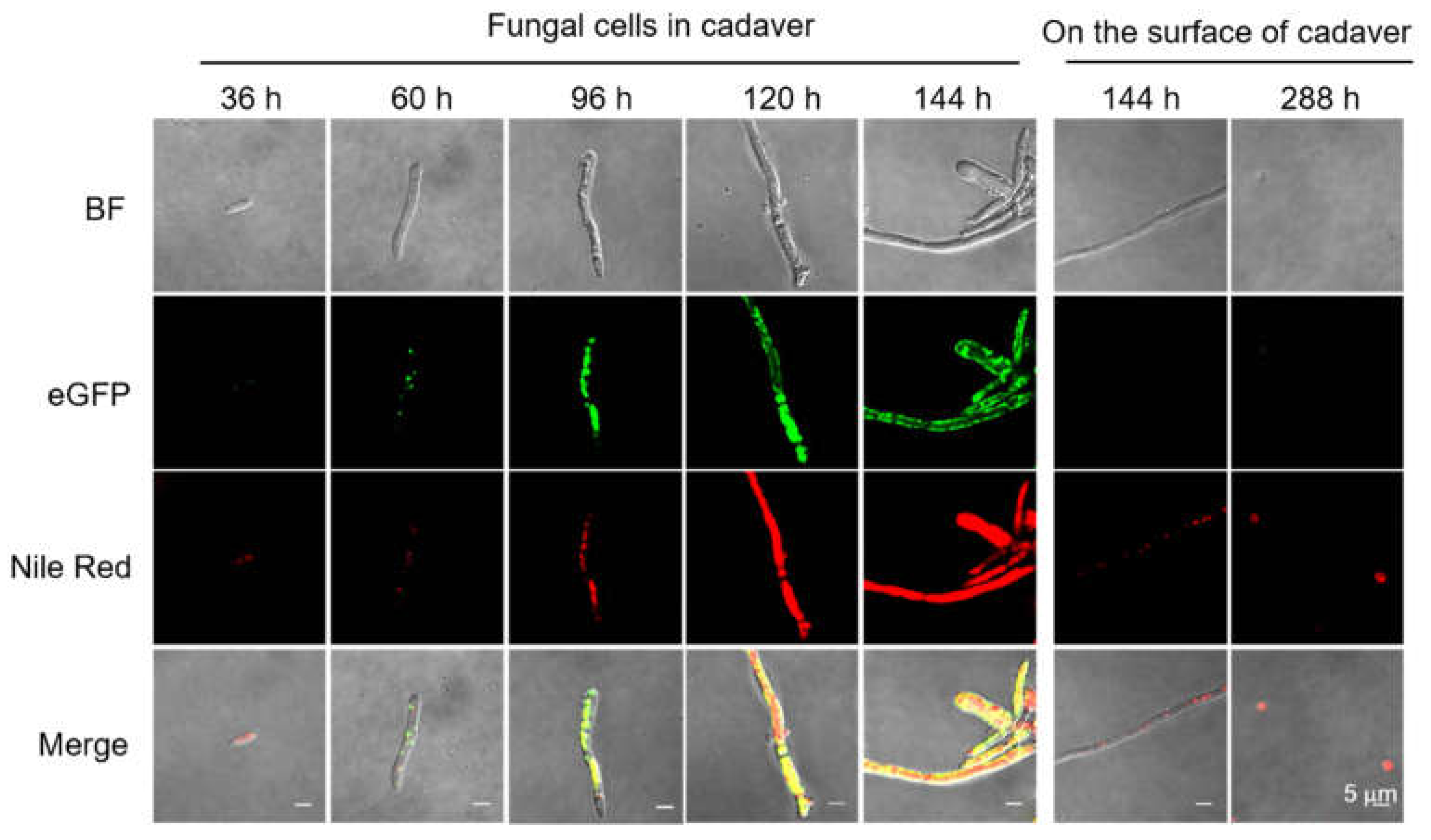
3.1. Sequence Analysis, Expression Patterns, and Cellular Localization of BbPlin1
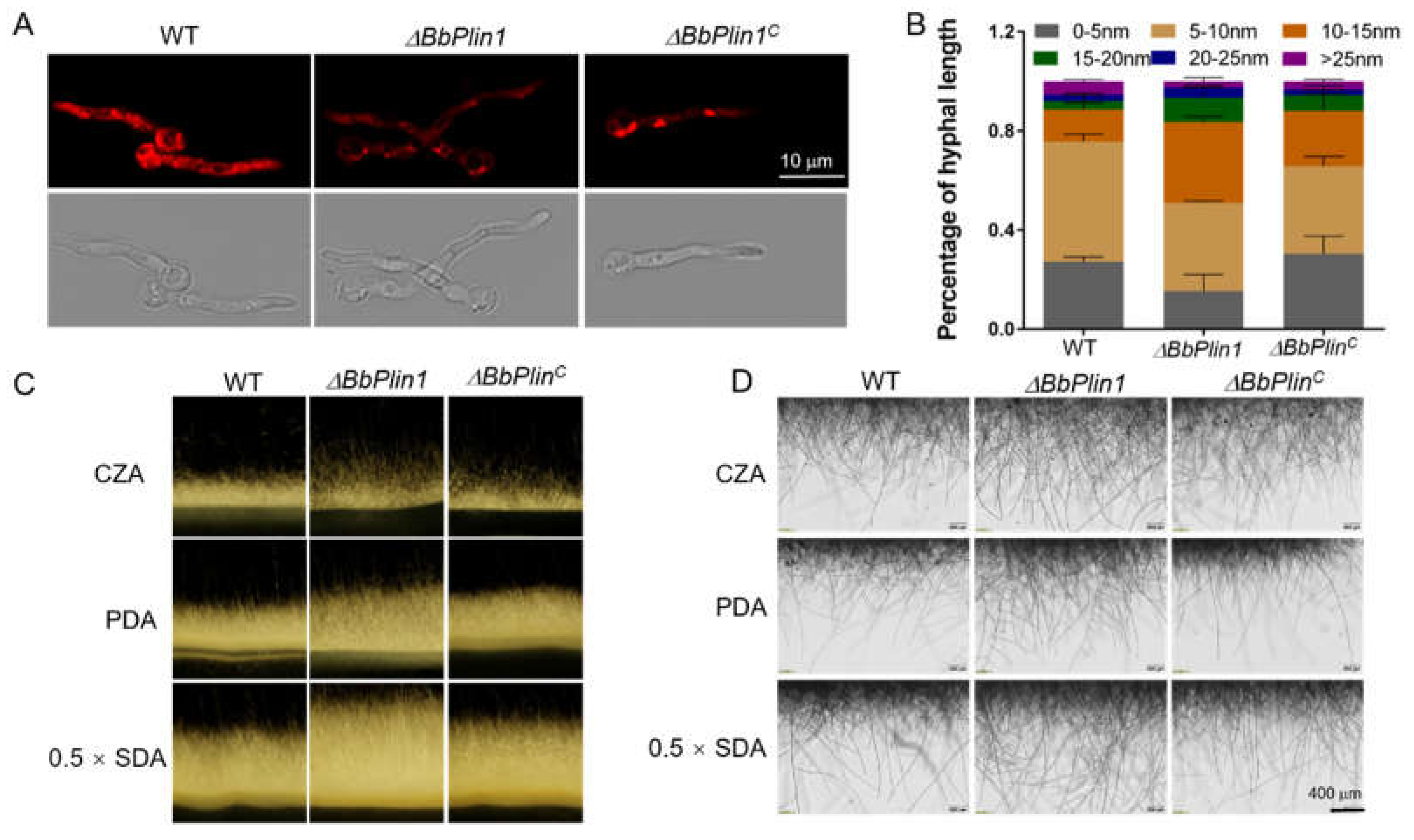
3.2. Generation of ΔBbPlin1 Mutants and Phenotypic Effects on Hyphal Growth and Lipid Droplet Accumulation
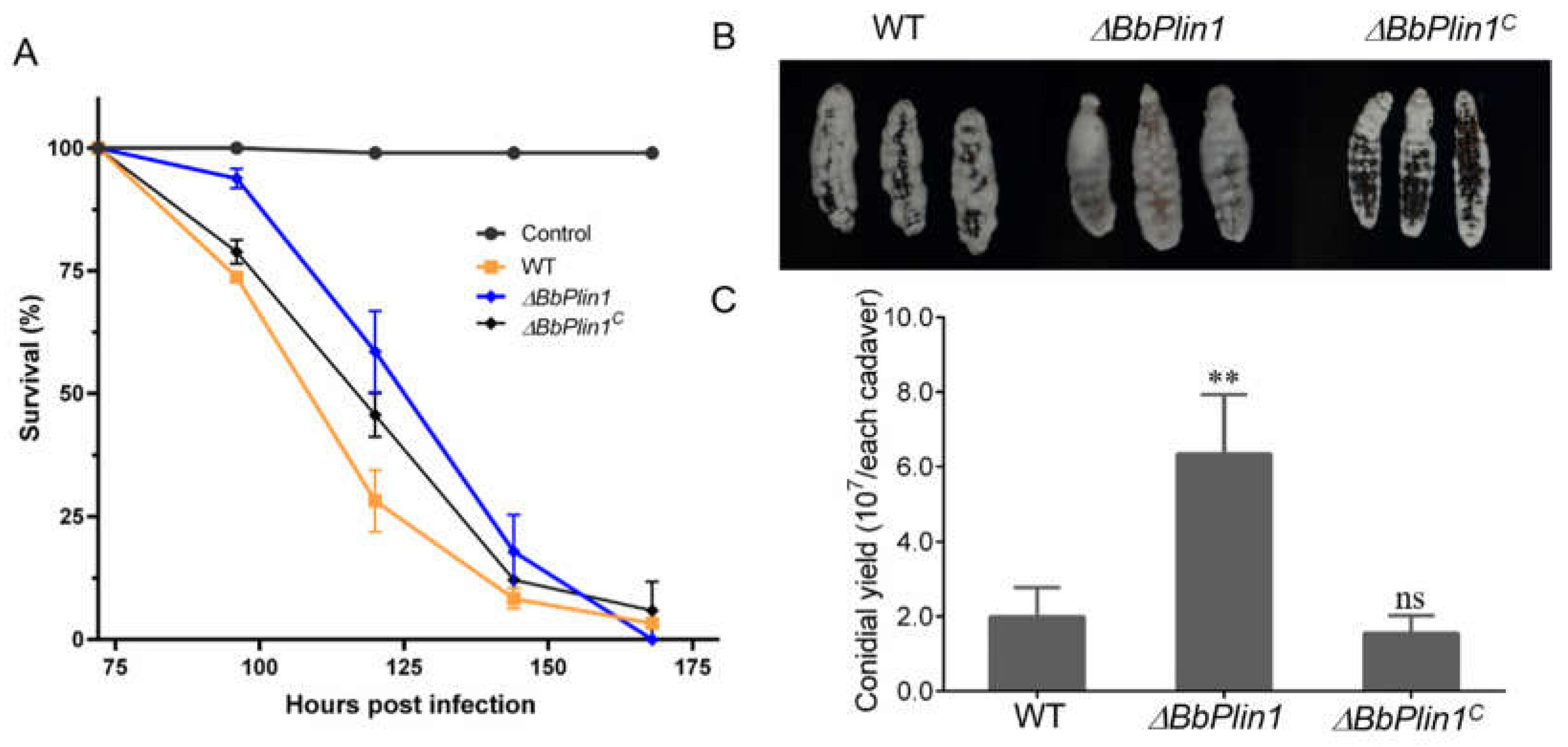
3.3. Loss of BbPlin1 Has a Minor Effect on Fungal Virulence
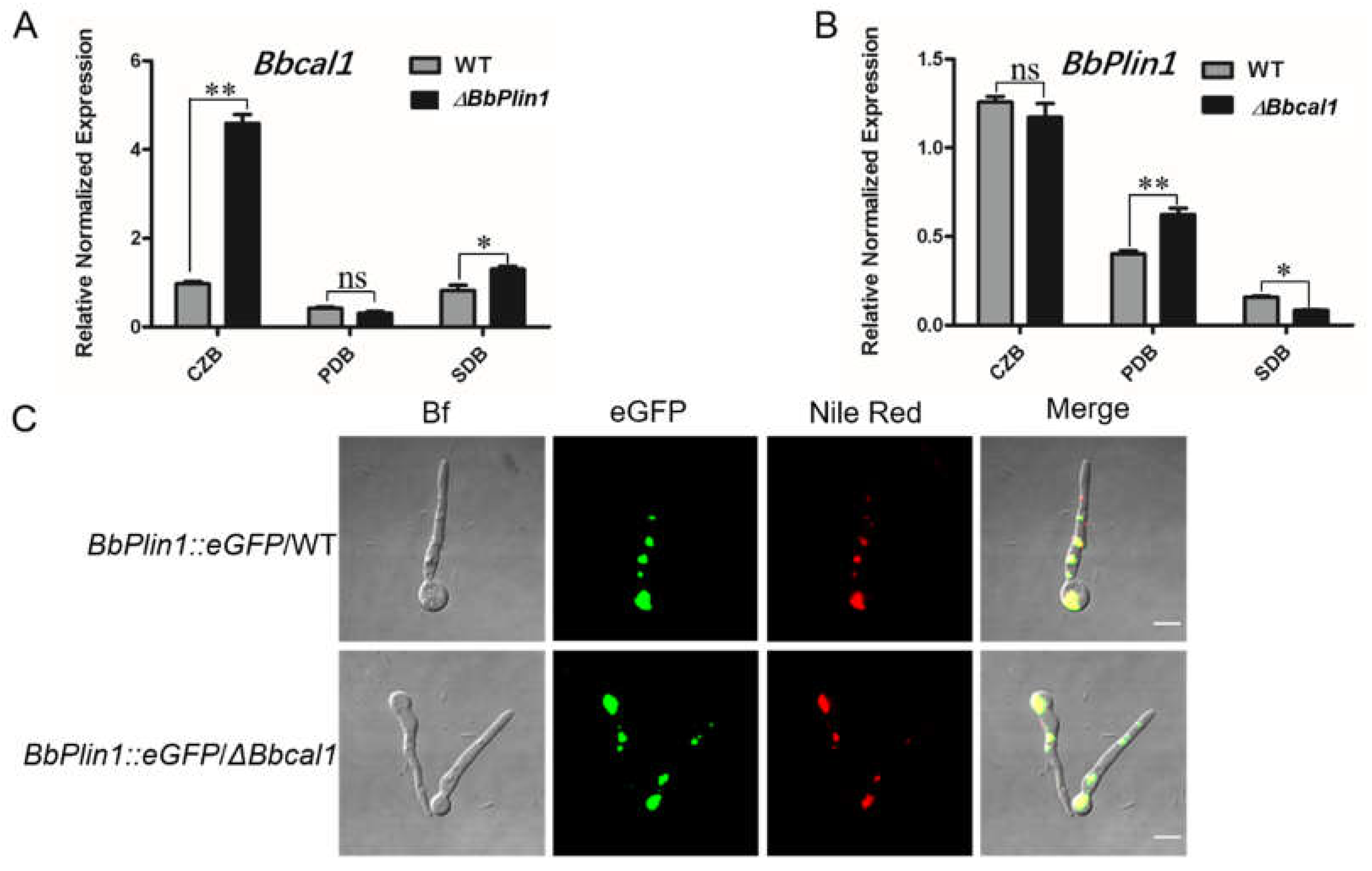
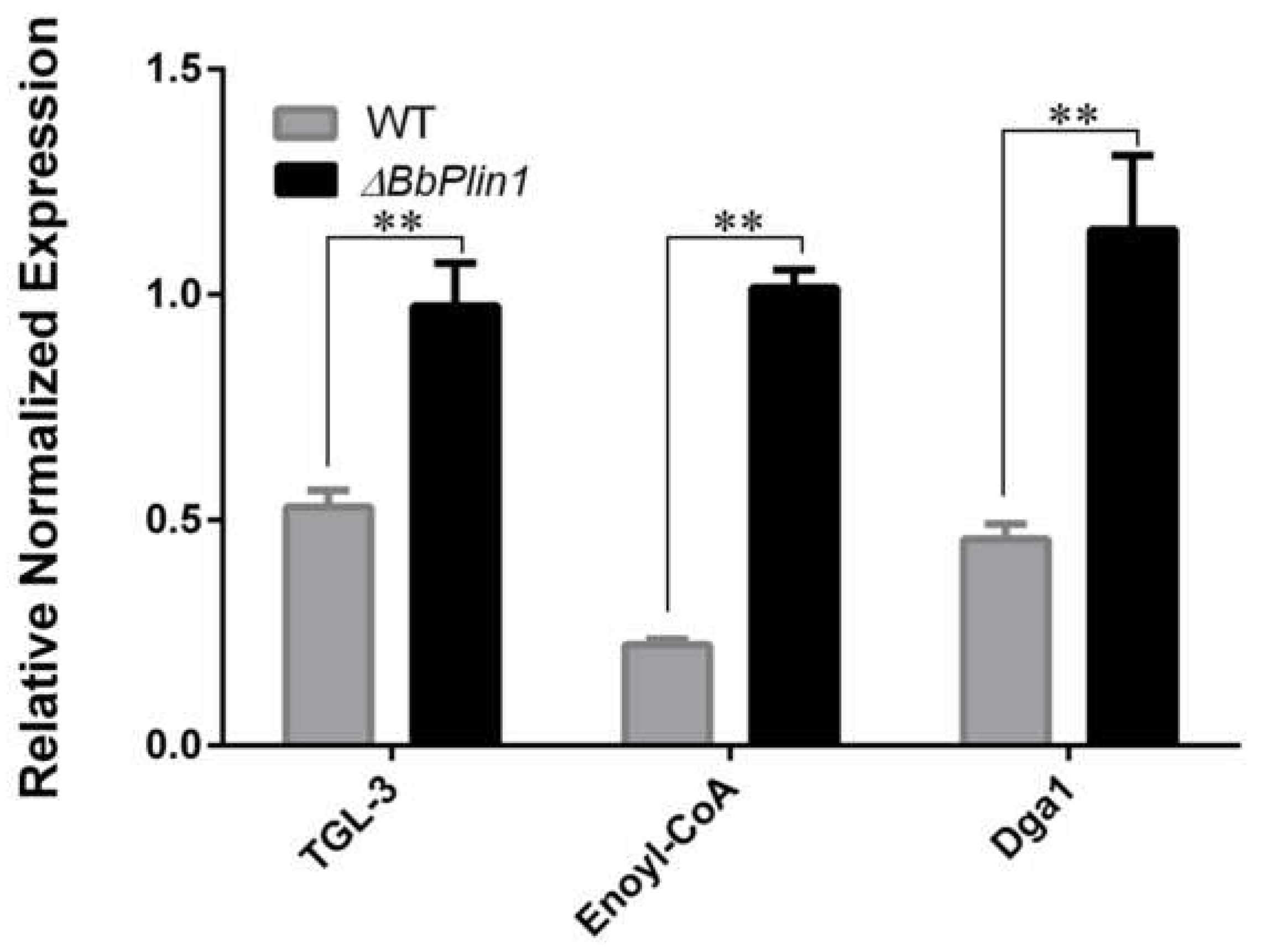
3.4. Compensatory Expression of Bbcal1 and Transcriptomic Analyses of the ΔbbPlin1 Mutant
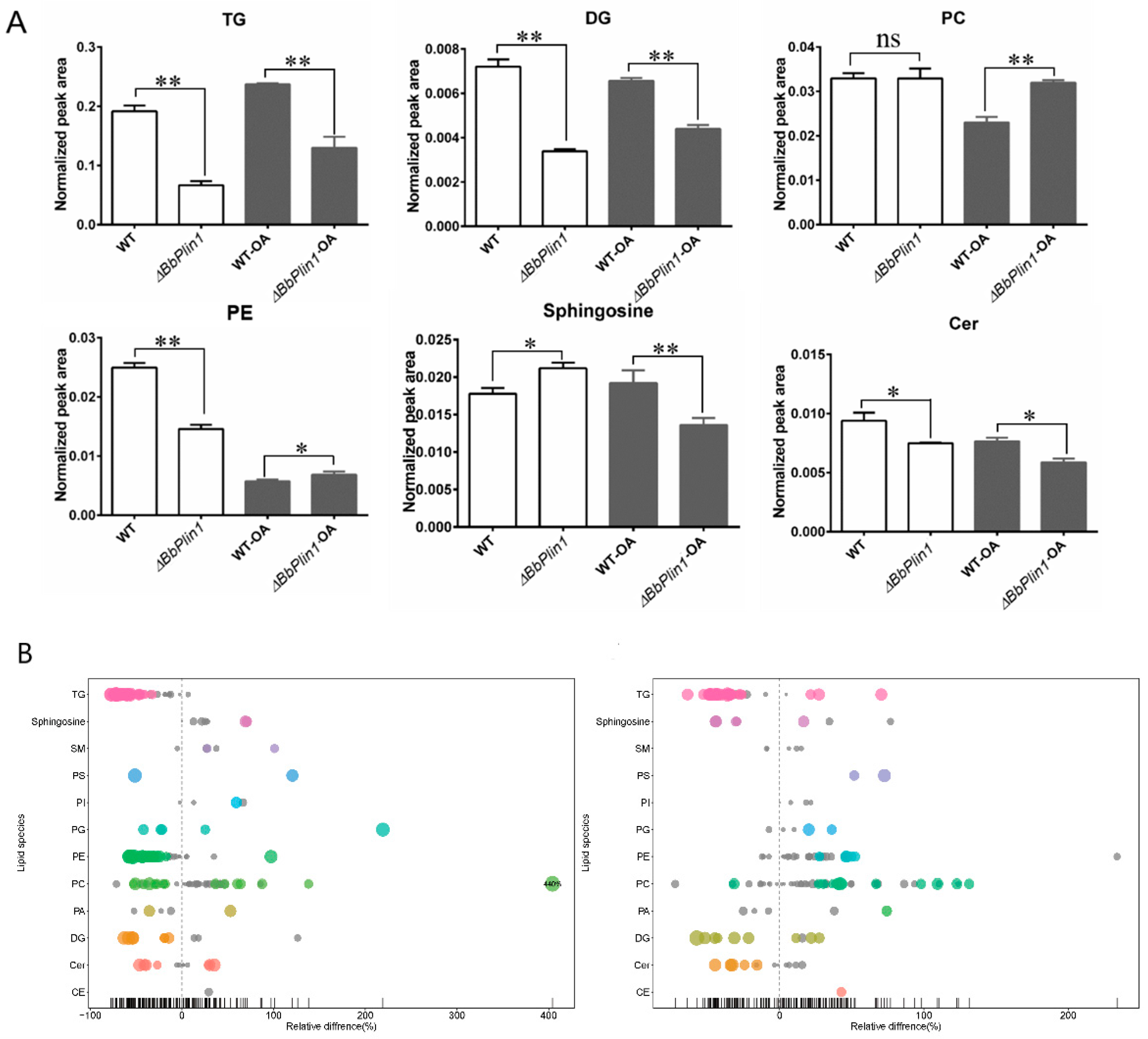
3.5. Loss of BbPlin1 Alters Cellular Lipid Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, W.; Zhang, M.; Zheng, S.; Li, Y.; Li, X.; Li, W.; Li, G.; Lin, Z.; Xie, Z.; Zhao, Z.; et al. Trapping toxins within lipid droplets is a resistance mechanism in fungi. Sci. Rep. 2015, 5, 15133. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Cordes, K.R.; Farese, R.V., Jr.; Walther, T.C. Lipid droplets at a glance. J. Cell Sci. 2009, 122, 749–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krahmer, N.; Farese, R.V., Jr.; Walther, T.C. Balancing the fat: Lipid droplets and human disease. EMBO Mol. Med. 2013, 5, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Ding, Y.; Gong, Z.; Yang, L.; Zhang, S.; Zhang, C.; Lin, X.; Shen, H.; Zou, H.; Xie, Z.; et al. Dynamics of the lipid droplet proteome of the Oleaginous yeast Rhodosporidium toruloides. Eukaryotic. Cell. 2015, 14, 252–264. [Google Scholar] [CrossRef] [Green Version]
- Bickel, P.E.; Tansey, J.T.; Welte, M.A. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta 2009, 1791, 419–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itabe, H.; Yamaguchi, T.; Nimura, S.; Sasabe, N. Perilipins: A diversity of intracellular lipid droplet proteins. Lipids. Health Dis. 2017, 16, 83. [Google Scholar] [CrossRef] [Green Version]
- Beller, M.; Bulankina, A.V.; Hsiao, H.H.; Urlaub, H.; Jäckle, H.; Kühnlein, R.P. PERILIPIN–dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 2010, 12, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef]
- Hsieh, K.; Lee, Y.K.; Londos, C.; Raaka, B.M.; Dalen, K.T.; Kimmel, A.R. Perilipin family members preferentially sequester to either triacylglycerol–specific or cholesteryl–ester–specific intracellular lipid storage droplets. J. Cell Sci. 2012, 125, 4067–4076. [Google Scholar] [CrossRef] [Green Version]
- Gao, Q.; Binns, D.D.; Kinch, L.N.; Grishin, N.V.; Ortiz, N.; Chen, X.; Goodman, J.M. Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J. Cell Biol. 2017, 216, 3199–3217. [Google Scholar] [CrossRef]
- Lin, C.; Liu, X.; Shi, T.; Li, C.; Huang, G. The Colletotrichum gloeosporioides perilipin homologue CAP 20 regulates functional appressorial formation and fungal virulence. J. Phytopathol. 2018, 166, 216–225. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; St Leger, R.J. The Metarhizium anisopliae Perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J. Biol. Chem. 2007, 282, 21110–21115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Yan, J.; Liu, C.; Xing, J.; Ren, Z.; Hendy, A.; Zheng, L.; Huang, J.; Chen, X.L. Perilipin LDP1 coordinates lipid droplets formation and utilization for appressorium–mediated infection in Magnaporthe oryzae. Environ. Microbiol. 2020, 22, 2843–2857. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Li, S.; Zhong, Z.; Sun, M. A Perilipin Gene from Clonostachys rosea f. Catenulata HL–1–1 Is Related to Sclerotial Parasitism. Int. J. Mol. Sci. 2015, 16, 5347–5362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keyhani, N.O. Lipid biology in fungal stress and virulence: Entomopathogenic fungi. Fungal Biol. 2018, 122, 420–429. [Google Scholar] [CrossRef]
- Pedrini, N.; Zhang, S.; Juárez, M.P.; Keyhani, N.O. Molecular characterization and expression analysis of a suite of cytochrome P450 enzymes implicated in insect hydrocarbon degradation in the entomopathogenic fungus Beauveria bassiana. Microbiol. Read. Engl. 2010, 156, 2549–2557. [Google Scholar] [CrossRef] [Green Version]
- Xiao, G.; Ying, S.H.; Zheng, P.; Wang, Z.L.; Zhang, S.; Xie, X.Q.; Feng, M.G. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012, 2, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ortiz–Urquiza, A.; Keyhani, N.O. Action on the surface: Entomopathogenic fungi versus the insect cuticle. Insects 2013, 4, 357–374. [Google Scholar] [CrossRef]
- Ortiz–Urquiza, A.; Keyhani, N.O. Molecular genetics of Beauveria bassiana infection of insects. Adv. Genet. 2016, 94, 165–249. [Google Scholar] [CrossRef]
- Fan, Y.; Borovsky, D.; Hawkings, C.; Ortiz–Urquiza, A.; Keyhani, N.O. Exploiting host molecules to augment mycoinsecticide virulence. Nat. Biotechnol. 2012, 30, 35–37. [Google Scholar] [CrossRef]
- Ortiz–Urquiza, A.; Luo, Z.; Keyhani, N.O. Improving mycoinsecticides for insect biological control. Appl. Microbiol. Biot. 2015, 99, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.W.; Robalino, I.V.; Keyhani, N.O. Uptake of the fluorescent probe FM4–64 by hyphae and haemolymph–derived in vivo hyphal bodies of the entomopathogenic fungus Beauveria bassiana. Microbiol. Read. Engl. 2009, 155, 3110–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanchoo, A.; Lewis, M.W.; Keyhani, N.O. Lectin mapping reveals stage–specific display of surface carbohydrates in in vitro and haemolymph–derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiol. Read. Engl. 2009, 155, 3121–3133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedrini, N.; Ortiz–Urquiza, A.; Huarte–Bonnet, C.; Zhang, S.; Keyhani, N.O. Targeting of insect epicuticular lipids by the entomopathogenic fungus Beauveria bassiana: Hydrocarbon oxidation within the context of a host–pathogen interaction. Front. Microbiol. 2013, 4, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Widemann, E.; Bernard, G.; Lesot, A.; Pinot, F.; Pedrini, N.; Keyhani, N.O. CYP52X1, representing new cytochrome P450 subfamily, displays fatty acid hydroxylase activity and contributes to virulence and growth on insect cuticular substrates in entomopathogenic fungus Beauveria bassiana. J. Biol. Chem. 2012, 287, 13477–13486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Biogenesis and functions of lipid droplets in plants: Thematic review series: Lipid droplet synthesis and metabolism: From yeast to man. J. Lipid. Res. 2012, 53, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Ortiz–Urquiza, A.; Garrett, T.; Pei, Y.; Keyhani, N.O. Involvement of a caleosin in lipid storage, spore dispersal, and virulence in the entomopathogenic filamentous fungus, Beauveria bassiana. Environ. Microbiol. 2015, 17, 4600–4614. [Google Scholar] [CrossRef] [Green Version]
- Ortiz–Urquiza, A.; Fan, Y.; Garrett, T.; Keyhani, N.O. Growth substrates and caleosin–mediated functions affect conidial virulence in the insect pathogenic fungus Beauveria bassiana. Microbiol. Read. Engl. 2016, 162, 1913–1921. [Google Scholar] [CrossRef]
- Fang, W.; Zhang, Y.; Yang, X.; Zheng, X.; Duan, H.; Li, Y.; Pei, Y. Agrobacterium tumefaciens–mediated transformation of Beauveria bassiana using an herbicide resistance gene as a selection marker. J. Invertebr. Pathol. 2004, 85, 18–24. [Google Scholar] [CrossRef]
- Welte, M.A.; Gould, A.P. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 2017, 1862, 1260–1272. [Google Scholar] [CrossRef]
- Hashemi, H.F.; Goodman, J.M. The life cycle of lipid droplets. Curr. Opin. Cell Biol. 2015, 33, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar, L.R.; Pardo, J.P.; Lomelí, M.M.; Bocardo, O.I.L.; Juárez Oropeza, M.A.; Guerra Sánchez, G. Lipid droplets accumulation and other biochemical changes induced in the fungal pathogen Ustilago maydis under nitrogen–starvation. Arch. Microbiol. 2017, 199, 1195–1209. [Google Scholar] [CrossRef] [PubMed]
- Sestric, R.; Munch, G.; Cicek, N.; Sparling, R.; Levin, D.B. Growth and neutral lipid synthesis by Yarrowia lipolytica on various carbon substrates under nutrient–sufficient and nutrient–limited conditions. Bioresour. Technol. 2014, 164, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Pedrini, N.; Ortiz–Urquiza, A.; Huarte–Bonnet, C.; Fan, Y.; Juárez, M.P.; Keyhani, N.O. Tenebrionid secretions and a fungal benzoquinone oxidoreductase form competing components of an arms race between a host and pathogen. Proc. Natl. Acad. Sci. USA 2015, 112, E3651–E3660. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Liu, X.; Keyhani, N.O.; Tang, G.; Pei, Y.; Zhang, W.; Tong, S. Regulatory cascade and biological activity of Beauveria bassiana oosporein that limits bacterial growth after host death. Proc. Natl. Acad. Sci. USA 2017, 114, E1578–E1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecuona, R.; Clement, J.-L.; Riba, G.; Joulie, C.; Juárez, P. Spore Germination and Hyphal Growth of Beauveria sp. on Insect Lipids. J. Econ. Entomol. 1997, 90, 119–123. [Google Scholar] [CrossRef]
- James, R.R.; Buckner, J.S.; Freeman, T.P. Cuticular lipids and silverleaf whitefly stage affect conidial germination of Beauveria bassiana and Paecilomyces fumosoroseus. J. Invertebr. Pathol. 2003, 84, 67–74. [Google Scholar] [CrossRef]
- Kraševec, N.; Panevska, A.; Lemež, Š.; Razinger, J.; Sepčić, K.; Anderluh, G.; Podobnik, M. Lipid–Binding Aegerolysin from Biocontrol Fungus Beauveria bassiana. Toxins 2021, 13, 820. [Google Scholar] [CrossRef]
- Tansey, J.T.; Sztalryd, C.; Gruia–Gray, J.; Roush, D.L.; Zee, J.V.; Gavrilova, O.; Reitman, M.L.; Deng, C.X.; Li, C.; Kimmel, A.R.; et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet–induced obesity. Proc. Natl. Acad. Sci. USA 2001, 98, 6494–6499. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.Y.; Pang, M.Y.; Feng, M.G.; Ying, S.H. A peroxisomal sterol carrier protein 2 (Scp2) contributes to lipid trafficking in differentiation and virulence of the insect pathogenic fungus Beauveria bassiana. Fungal Genet. Biol. FG B 2022, 158, 103651. [Google Scholar] [CrossRef]
- Peng, Y.J.; Zhang, H.; Feng, M.G.; Ying, S.H. Steryl Acetyl Hydrolase 1 (BbSay1) Links Lipid Homeostasis to Conidiogenesis and Virulence in the Entomopathogenic Fungus Beauveria bassiana. J. Fungi 2022, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Lu, Z.; Wang, H.; Li, N.; Song, G.; Zhu, Q.; Sun, J.; Zhang, Y. A secretory phospholipase A2 of a fungal pathogen contributes to lipid droplet homeostasis, assimilation of insect–derived lipids, and repression of host immune responses. Insect. Sci. 2022. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Liu, Y.; Keyhani, N.O.; Zhu, S.; Wang, J.; Wang, J.; Jin, D.; Fan, Y. A Perilipin Affects Lipid Droplet Homeostasis and Aerial Hyphal Growth, but Has Only Small Effects on Virulence in the Insect Pathogenic Fungus Beauveria bassiana. J. Fungi 2022, 8, 634. https://doi.org/10.3390/jof8060634
Wang X, Liu Y, Keyhani NO, Zhu S, Wang J, Wang J, Jin D, Fan Y. A Perilipin Affects Lipid Droplet Homeostasis and Aerial Hyphal Growth, but Has Only Small Effects on Virulence in the Insect Pathogenic Fungus Beauveria bassiana. Journal of Fungi. 2022; 8(6):634. https://doi.org/10.3390/jof8060634
Chicago/Turabian StyleWang, Xiaoyun, Yu Liu, Nemat O. Keyhani, Shengan Zhu, Jing Wang, Junyao Wang, Dan Jin, and Yanhua Fan. 2022. "A Perilipin Affects Lipid Droplet Homeostasis and Aerial Hyphal Growth, but Has Only Small Effects on Virulence in the Insect Pathogenic Fungus Beauveria bassiana" Journal of Fungi 8, no. 6: 634. https://doi.org/10.3390/jof8060634
APA StyleWang, X., Liu, Y., Keyhani, N. O., Zhu, S., Wang, J., Wang, J., Jin, D., & Fan, Y. (2022). A Perilipin Affects Lipid Droplet Homeostasis and Aerial Hyphal Growth, but Has Only Small Effects on Virulence in the Insect Pathogenic Fungus Beauveria bassiana. Journal of Fungi, 8(6), 634. https://doi.org/10.3390/jof8060634





