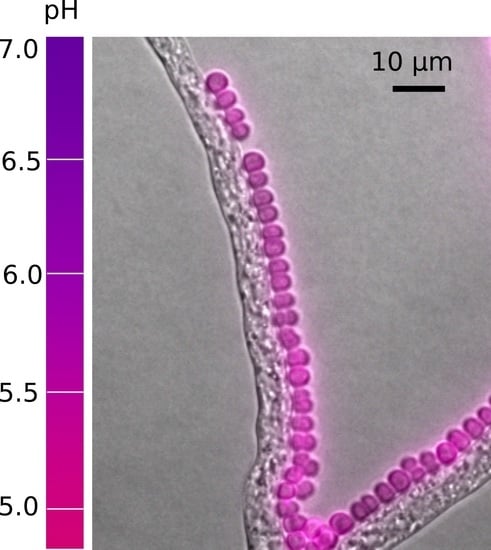
pH Distribution along Growing Fungal Hyphae at Microscale
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms, Growth Conditions and Analysis of Water Contact Angles
2.2. Time-Resolved In Vitro Hyphal Surface pH Sensing
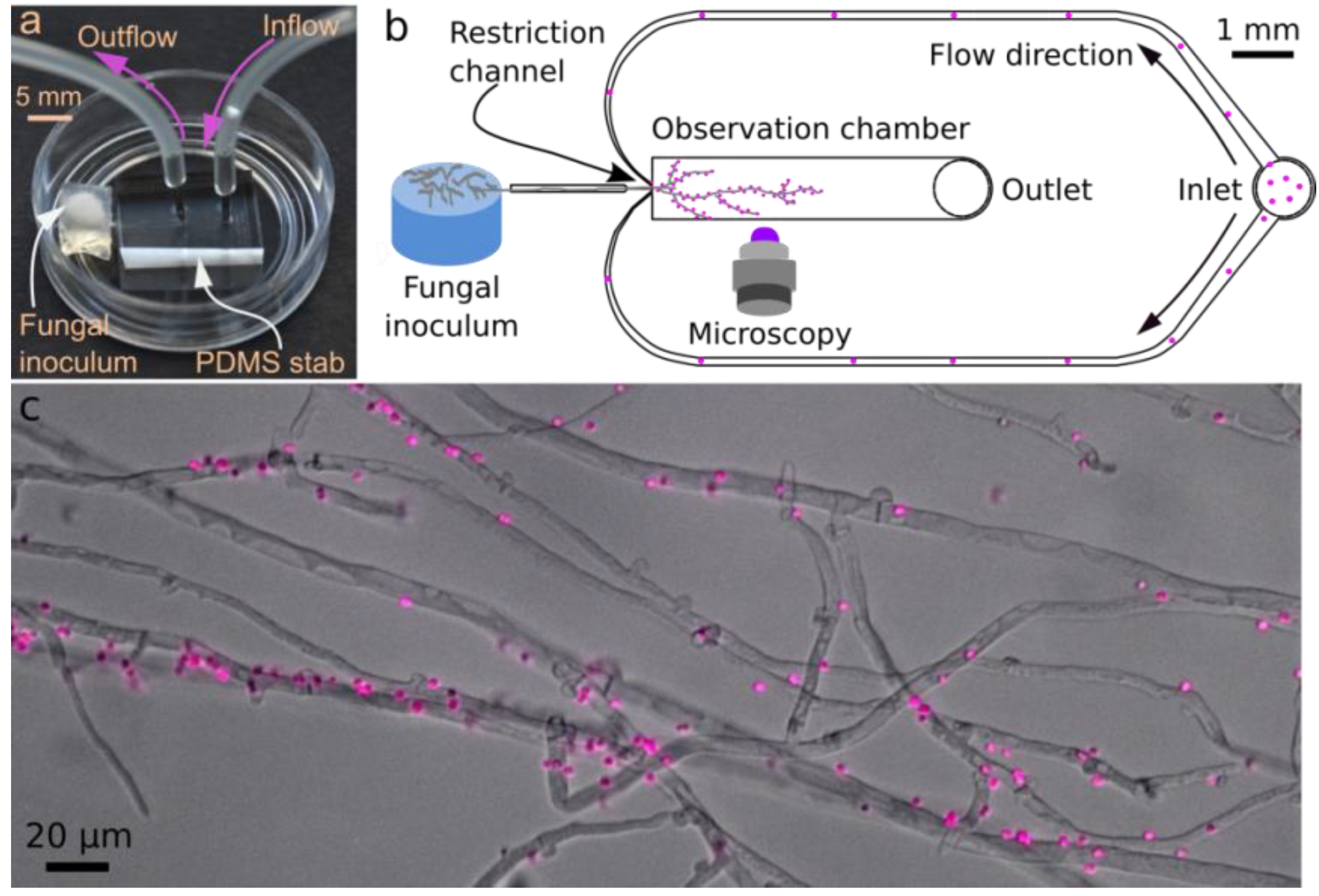
2.2.1. Microfluidic Device
2.2.2. Microfluidic Device: Fungal Inoculation and Loading of Bioreporters
2.2.3. Time-Resolved Microscopic Imaging
2.3. Image Analysis and Spatial Data Interpretation
3. Statistical Analysis
4. Results
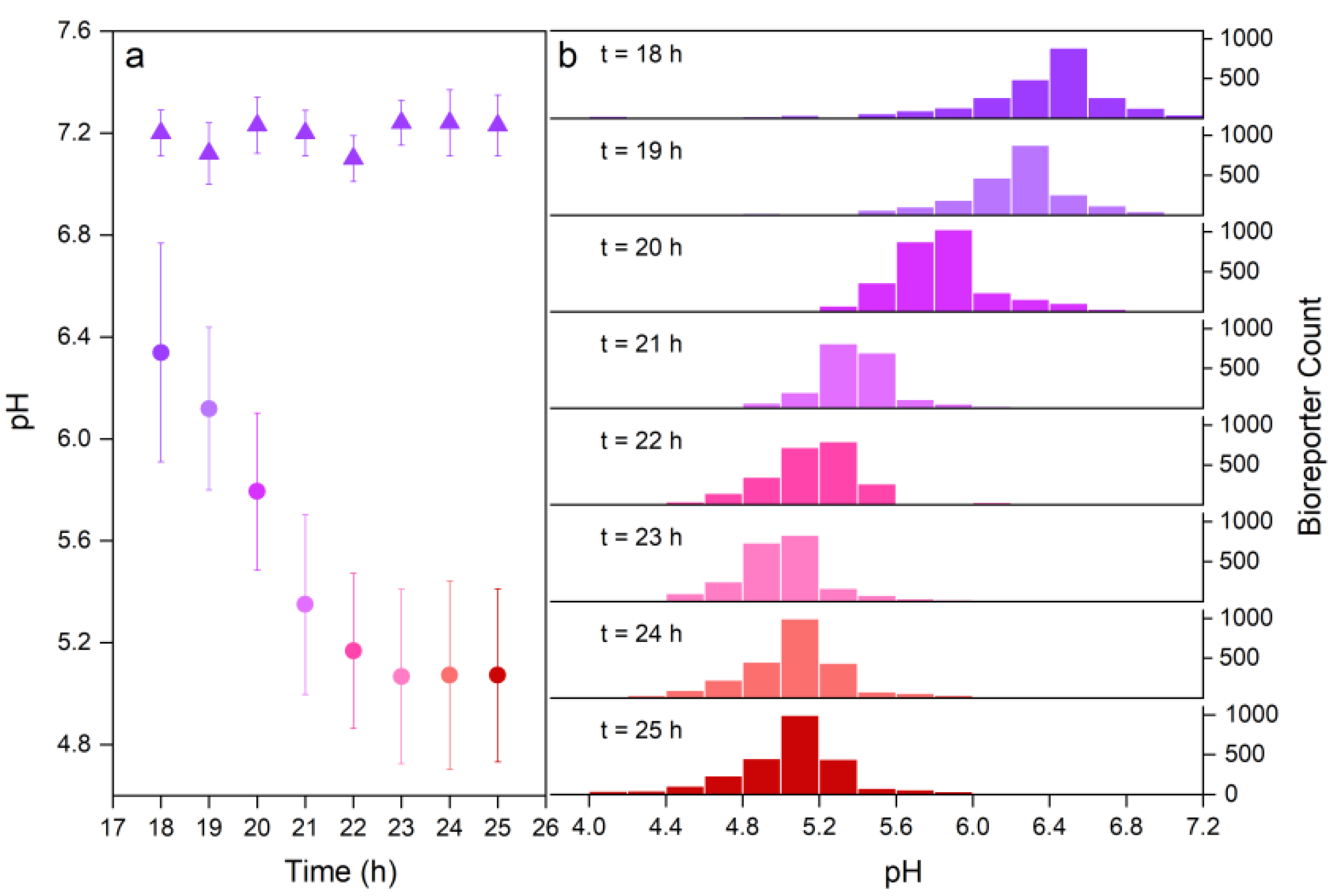
4.1. Time-Resolved Average Hyphal Surface pH
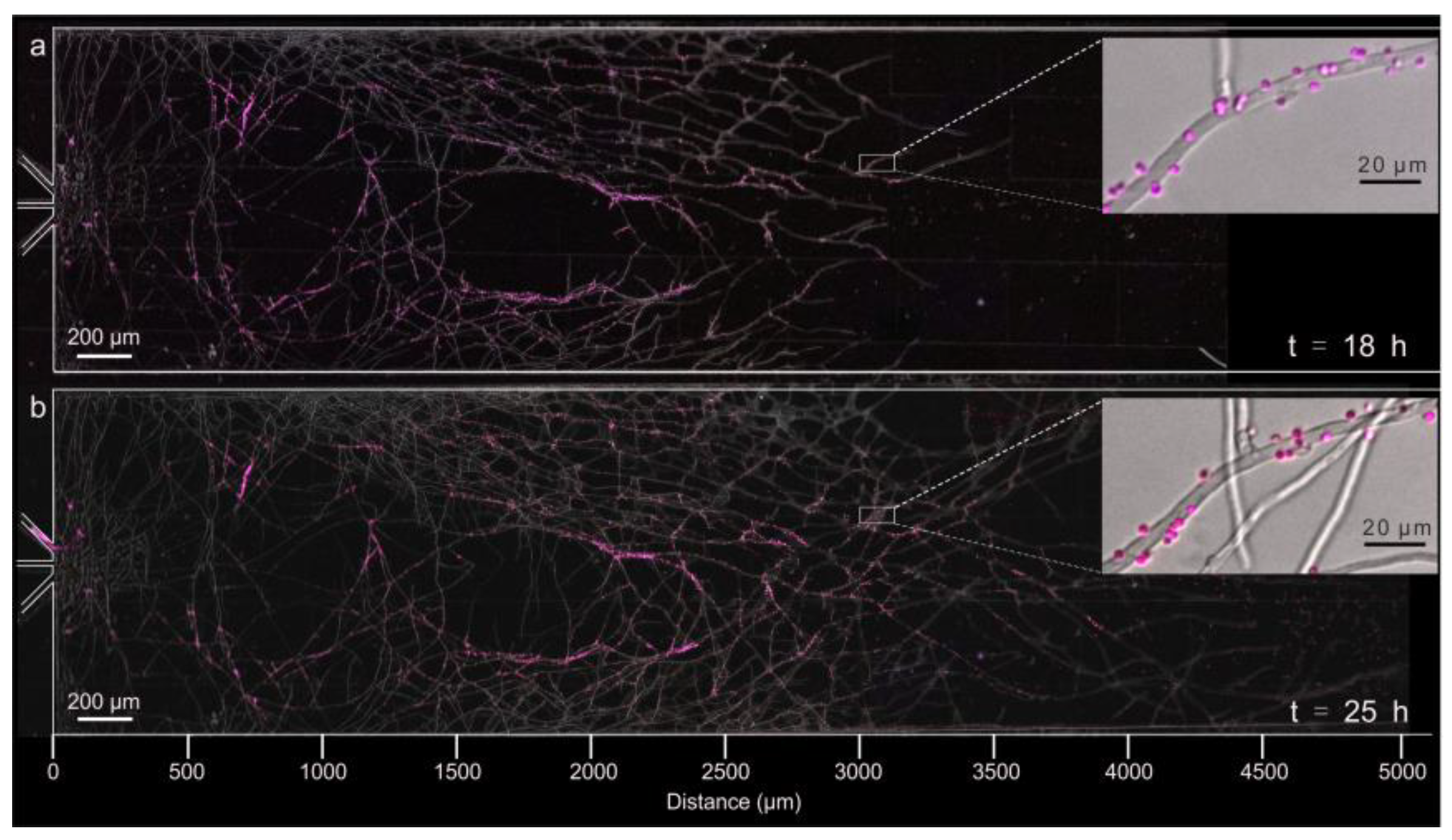
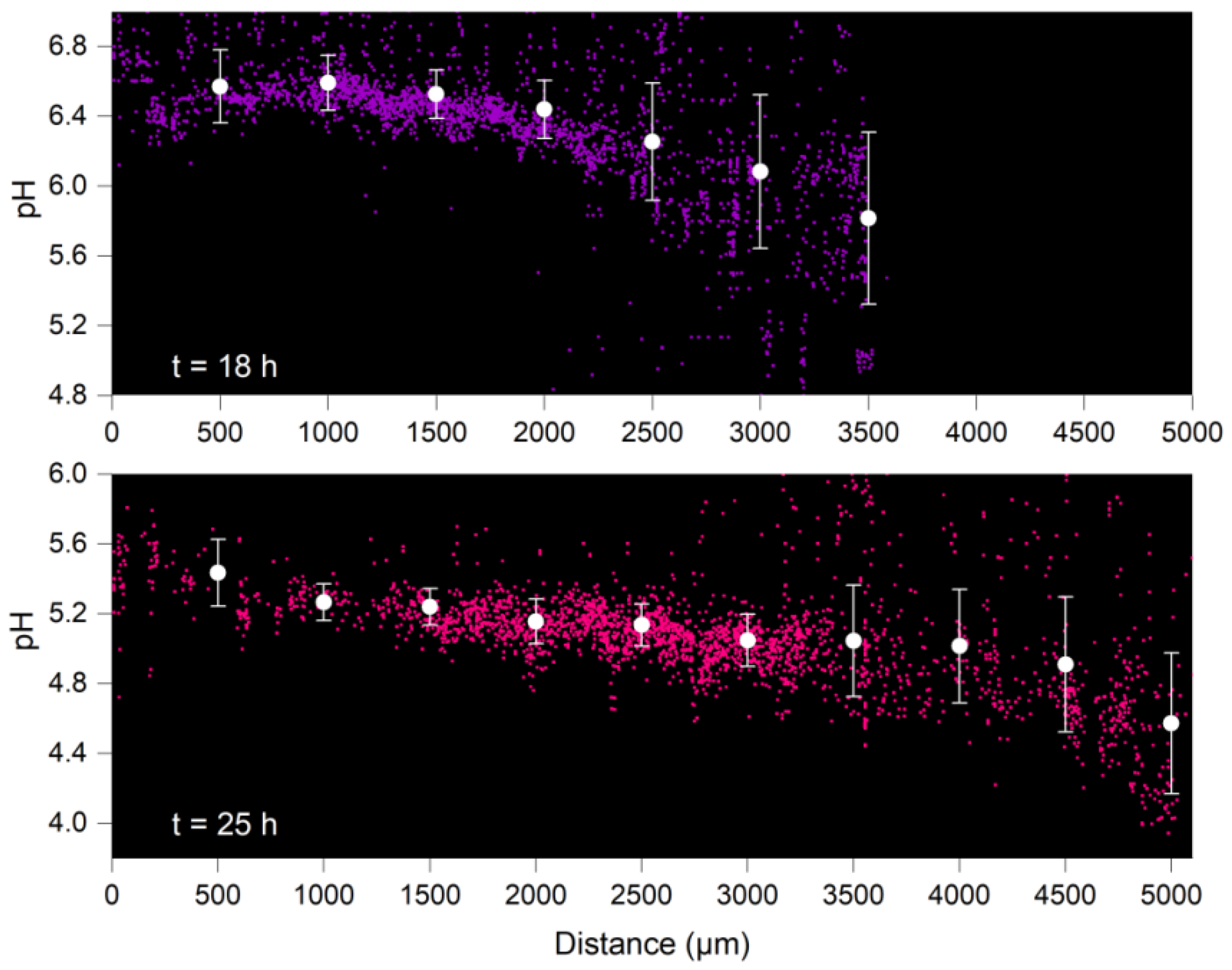
4.2. Distribution of Surface pH along Growing Hyphae
5. Discussion
5.1. Time-Resolved In Vitro Analysis of Hyphal Surface pH
5.2. pH on the Hyphal Surface and Its Ecological Significance
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Worrich, A.; Wick, L.Y.; Banitz, T. Ecology of contaminant biotransformation in the mycosphere: Role of transport processes. Adv. Appl. Microbiol. 2018, 104, 93–133. [Google Scholar] [PubMed]
- Liers, C.; Aranda, E.; Strittmatter, E.; Piontek, K.; Plattner, D.A.; Zorn, H.; Ullrich, R.; Hofrichter, M. Phenol oxidation by DyP-type peroxidases in comparison to fungal and plant peroxidases. J. Mol. Catal. B Enzym. 2014, 103, 41–46. [Google Scholar] [CrossRef]
- Boer, W.d.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- Kohlmeier, S.; Smits, T.H.M.; Ford, R.M.; Keel, C.; Harms, H.; Wick, L.Y. Taking the Fungal Highway: Mobilization of Pollutant-Degrading Bacteria by Fungi. Environ. Sci. Technol. 2005, 39, 4640–4646. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Kallies, R.; Kühn, I.; Schmidt, M.; Harms, H.; Chatzinotas, A.; Wick, L.Y. Phage co-transport with hyphal-riding bacteria fuels bacterial invasion in a water-unsaturated microbial model system. ISME J. 2021, 16, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.-J.; Kleinsteuber, S.; Sträuber, H.; Dusny, C.; Harms, H.; Wick, L.Y. Impact of fungal hyphae on growth and dispersal of obligate anaerobic bacteria in aerated habitats. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.; Cowley, E.; Nobile, C.; Hartooni, N.; Newman, D.; Johnson, A. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef]
- Minamino, T.; Imae, Y.; Oosawa, F.; Kobayashi, Y.; Oosawa, K. Effect of intracellular pH on rotational speed of bacterial flagellar motors. J. Bacteriol. 2003, 185, 1190–1194. [Google Scholar] [CrossRef]
- Kim, M.; Or, D. Microscale pH variations during drying of soils and desert biocrusts affect HONO and NH3 emissions. Nat. Commun. 2019, 10, 3944. [Google Scholar] [CrossRef] [PubMed]
- Gadd, G.M.; Rhee, Y.J.; Stephenson, K.; Wei, Z. Geomycology: Metals, actinides and biominerals. Environ. Microbiol. Rep. 2012, 4, 270–296. [Google Scholar] [CrossRef] [PubMed]
- Schmalenberger, A.; Duran, A.; Bray, A.W.; Bridge, J.; Bonneville, S.; Benning, L.G.; Romero-Gonzalez, M.E.; Leake, J.R.; Banwart, S.A. Oxalate secretion by ectomycorrhizal Paxillus involutus is mineral-specific and controls calcium weathering from minerals. Sci. Rep. 2015, 5, 12187. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.-J.; Dusny, C.; Wang, L.; Appel, J.; Lindstaedt, K.; Schlosser, D.; Harms, H.; Wick, L.Y. Illuminate the hidden: In vivo mapping of microscale pH in the mycosphere using a novel whole-cell biosensor. ISME Commun. 2021, 1, 75. [Google Scholar] [CrossRef]
- Hyde, S.M.; Wood, P.M.J.M. A mechanism for production of hydroxyl radicals by the brown-rot fungus Coniophora puteana: Fe (III) reduction by cellobiose dehydrogenase and Fe (II) oxidation at a distance from the hyphae. Microbiology 1997, 143, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.d.; Meier, R.J.; Wolfbeis, O.S. Fluorescent pH-sensitive nanoparticles in an agarose matrix for imaging of bacterial growth and metabolism. Angew. Chem. Int. Ed. 2013, 52, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Wencel, D.; Abel, T.; McDonagh, C. Optical chemical pH sensors. Anal. Chem. 2014, 86, 15–29. [Google Scholar] [CrossRef]
- Marzocchi, U.; Bonaglia, S.; van de Velde, S.; Hall, P.O.; Schramm, A.; Risgaard-Petersen, N.; Meysman, F.J. Transient bottom water oxygenation creates a niche for cable bacteria in long-term anoxic sediments of the Eastern Gotland Basin. Environ. Microbiol. 2018, 20, 3031–3041. [Google Scholar] [CrossRef] [PubMed]
- Agethen, S.; Sander, M.; Waldemer, C.; Knorr, K.-H. Plant rhizosphere oxidation reduces methane production and emission in rewetted peatlands. Soil Biol. Biochem. 2018, 125, 125–135. [Google Scholar] [CrossRef]
- Dal Co, A.; Van Vliet, S.; Ackermann, M. Emergent microscale gradients give rise to metabolic cross-feeding and antibiotic tolerance in clonal bacterial populations. Philos. Trans. R. Soc. B 2019, 374, 20190080. [Google Scholar] [CrossRef] [PubMed]
- Rendler, T.; Neburkova, J.; Zemek, O.; Kotek, J.; Zappe, A.; Chu, Z.; Cigler, P.; Wrachtrup, J. Optical imaging of localized chemical events using programmable diamond quantum nanosensors. Nat. Commun. 2017, 8, 14701. [Google Scholar] [CrossRef] [PubMed]
- Neil, A.G.; Darryl, L.K.; Franklin, M.H. Growing hyphae of Achlya bisexualis generate a longtitudinal pH gradient in the surrounding medium. Microbiology 1984, 130, 2967–2974. [Google Scholar]
- Zacchetti, B.; Wösten, H.A.; Claessen, D. Multiscale heterogeneity in filamentous microbes. Biotechnol. Adv. 2018, 36, 2138–2149. [Google Scholar] [CrossRef]
- Krijgsheld, P.; Altelaar, A.M.; Post, H.; Ringrose, J.H.; Müller, W.H.; Heck, A.J.; Wösten, H.A. Spatially resolving the secretome within the mycelium of the cell factory Aspergillus niger. J. Proteome Res. 2012, 11, 2807–2818. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E.; Wilke, S.K.; Ahrén, D.; Au, C.H.; Birren, B.W.; Borodovsky, M.; Burns, C.; Canbäck, B.; Casselton, L.A.; Cheng, C. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. USA 2010, 107, 11889–11894. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.E.; Stöckli, M.; van Swaay, D.; Sabotič, J.; Kallio, P.T.; Künzler, M.; deMello, A.J.; Aebi, M. Probing bacterial–fungal interactions at the single cell level. Integr. Biol. 2014, 6, 935–945. [Google Scholar] [CrossRef]
- Smits, T.H.; Wick, L.Y.; Harms, H.; Keel, C. Characterization of the surface hydrophobicity of filamentous fungi. Environ. Microbiol. 2003, 5, 85–91. [Google Scholar] [CrossRef]
- Smits, M.M.; Herrmann, A.M.; Duane, M.; Duckworth, O.W.; Bonneville, S.; Benning, L.G.; Lundström, U. The fungal–mineral interface: Challenges and considerations of micro-analytical developments. Fungal Biol. Rev. 2009, 23, 122–131. [Google Scholar] [CrossRef]
- Stanley, C.E.; Shrivastava, J.; Brugman, R.; Heinzelmann, E.; van Swaay, D.; Grossmann, G. Dual-flow-RootChip reveals local adaptations of roots towards environmental asymmetry at the physiological and genetic levels. New Phytol. 2018, 217, 1357–1369. [Google Scholar] [CrossRef]
- Miesenböck, G.; De Angelis, D.A.; Rothman, J.E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998, 394, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Mahon, M.J. pHluorin2: An enhanced, ratiometric, pH-sensitive green florescent protein. Adv. Biosci. Biotechnol. 2011, 2, 132. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Benoit, I.; van den Esker, M.H.; Patyshakuliyeva, A.; Mattern, D.J.; Blei, F.; Zhou, M.; Dijksterhuis, J.; Brakhage, A.A.; Kuipers, O.P.; de Vries, R.P. Bacillus acillus subtilis attachment to Aspergillus spergillus niger hyphae results in mutually altered metabolism. Environ. Microbiol. 2015, 17, 2099–2113. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, N.; Stanley, C.E.; Harms, H.; Chatzinotas, A.; Wick, L.Y. Mycelial effects on phage retention during transport in a microfluidic platform. Environ. Sci. Technol. 2019, 53, 11755–11763. [Google Scholar] [CrossRef] [PubMed]
- Miquel Guennoc, C.; Rose, C.; Labbé, J.; Deveau, A. Bacterial biofilm formation on the hyphae of ectomycorrhizal fungi: A widespread ability under controls? FEMS Microbiol. Ecol. 2018, 94, fiy093. [Google Scholar] [CrossRef] [PubMed]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Herve, V.; Labbe, J.; Lastovetsky, O.A. Bacterial–fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef]
- Toljander, J.F.; Artursson, V.; Paul, L.R.; Jansson, J.K.; Finlay, R.D. Attachment of different soil bacteria to arbuscular mycorrhizal fungal extraradical hyphae is determined by hyphal vitality and fungal species. FEMS Microbiol. Lett. 2006, 254, 34–40. [Google Scholar] [CrossRef]
- Hermansson, M. The DLVO theory in microbial adhesion. Colloids Surf. B Biointerfaces 1999, 14, 105–119. [Google Scholar] [CrossRef]
- Chrysikopoulos, C.V.; Syngouna, V.I. Attachment of bacteriophages MS2 and ΦX174 onto kaolinite and montmorillonite: Extended-DLVO interactions. Colloids Surf. B Biointerfaces 2012, 92, 74–83. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, S.; Cai, L.; Liu, X.; Wu, H.; Xin, F.; Zhang, M.; Jiang, M. Improved treatment and utilization of rice straw by Coprinopsis cinerea. Appl. Biochem. Biotechnol. 2018, 184, 616–629. [Google Scholar] [CrossRef]
- Liaud, N.; Giniés, C.; Navarro, D.; Fabre, N.; Crapart, S.; Gimbert, I.H.; Levasseur, A.; Raouche, S.; Sigoillot, J.-C. Exploring fungal biodiversity: Organic acid production by 66 strains of filamentous fungi. Fungal Biol. Biotechnol. 2014, 1, 1. [Google Scholar] [CrossRef]
- Mäkelä, M.R.; Hildén, K.; Lundell, T.K. Oxalate decarboxylase: Biotechnological update and prevalence of the enzyme in filamentous fungi. Appl. Microbiol. Biotechnol. 2010, 87, 801–814. [Google Scholar] [CrossRef]
- Jahangeer, S.; Khan, N.; Jahangeer, S.; Sohail, M.; Shahzad, S.; Ahmad, A.; Khan, S.A. Screening and characterization of fungal cellulases isolated from the native environmental source. Pak. J. Bot. 2005, 37, 739. [Google Scholar]
- Gummadi, S.N.; Panda, T. Purification and biochemical properties of microbial pectinases—A review. Process Biochem. 2003, 38, 987–996. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 2010, 42, 391–404. [Google Scholar] [CrossRef]
- Kilaru, S.; Hoegger, P.J.; Kües, U. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 2006, 50, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Floudas, D.; Bentzer, J.; Ahrén, D.; Johansson, T.; Persson, P.; Tunlid, A. Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi. ISME J. 2020, 14, 2046–2059. [Google Scholar] [CrossRef] [PubMed]
- Schulze, M.; Geisler, L.; Majcherczyk, A.; Rühl, M. Signal peptide replacement resulted in recombinant homologous expression of laccase Lcc8 in Coprinopsis cinerea. AMB Express 2019, 9, 36. [Google Scholar] [CrossRef]
- Schneider, P.; Caspersen, M.B.; Mondorf, K.; Halkier, T.; Skov, L.K.; Østergaard, P.R.; Brown, K.M.; Brown, S.H.; Xu, F. Characterization of a Coprinus cinereus laccase. Enzym. Microb. Technol. 1999, 25, 502–508. [Google Scholar] [CrossRef]
- Pan, K.; Zhao, N.; Yin, Q.; Zhang, T.; Xu, X.; Fang, W.; Hong, Y.; Fang, Z.; Xiao, Y. Induction of a laccase Lcc9 from Coprinopsis cinerea by fungal coculture and its application on indigo dye decolorization. Bioresour. Technol. 2014, 162, 45–52. [Google Scholar] [CrossRef]
- Brenelli, L.; Squina, F.M.; Felby, C.; Cannella, D. Laccase-derived lignin compounds boost cellulose oxidative enzymes AA9. Biotechnol. Biofuels 2018, 11, 10. [Google Scholar] [CrossRef]
- Houtman, C.J.; Kitin, P.; Houtman, J.C.; Hammel, K.E.; Hunt, C.G. Acridine orange indicates early oxidation of wood cell walls by fungi. PLoS ONE 2016, 11, e0159715. [Google Scholar] [CrossRef]
- Weber, R.; Pitt, D. Filamentous fungi—Growth and physiology. Appl. Mycol. Biotechnol. 2001, 1, 13–54. [Google Scholar]
- Li, X.L.; George, E.; Marschner, H. Phosphorus depletion and pH decrease at the root–soil and hyphae–soil interfaces of VA mycorrhizal white clover fertilized with ammonium. New Phytol. 1991, 119, 397–404. [Google Scholar] [CrossRef]
- Roncal, T.; Ugalde, U.; Irastorza, A. Calcium-induced conidiation in Penicillium cyclopium: Calcium triggers cytosolic alkalinization at the hyphal tip. J. Bacteriol. 1993, 175, 879–886. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bittisnich, D.; Williamson, R. Tip-localised H+-fluxes and the applicability of the acid-growth hypothesis to tip-growing cells: Control of chloronemal extension in Funaria hygrometrica by auxin and light. Planta 1989, 178, 96–102. [Google Scholar] [CrossRef]
- Kutschera, U. The current status of the acid-growth hypothesis. New Phytol. 1994, 126, 549–569. [Google Scholar] [CrossRef]
- Marre, E. Fusicoccin: A tool in plant physiology. Annu. Rev. Plant Physiol. 1979, 30, 273–288. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.; Preston, G.M. Bacterial mycophagy: Definition and diagnosis of a unique bacterial–fungal interaction. New Phytol. 2008, 177, 859–876. [Google Scholar] [CrossRef]
- Stanley, C.E.; van der Heijden, M.G. Microbiome-on-a-chip: New frontiers in plant–microbiota research. Trends Microbiol. 2017, 25, 610–613. [Google Scholar] [CrossRef]
- Yang, P.; Oliveira da Rocha Calixto, R.; van Elsas, J.D. Migration of Paraburkholderia terrae BS001 along old fungal hyphae in soil at various pH levels. Microb. Ecol. 2018, 76, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zhang, M.; van Elsas, J.D. Role of flagella and type four pili in the co-migration of Burkholderia terrae BS001 with fungal hyphae through soil. Sci. Rep. 2017, 7, 2997. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.F. Flagellar movement driven by proton translocation. FEBS Lett. 2003, 545, 86–95. [Google Scholar] [CrossRef]
- Simon, A.; Bindschedler, S.; Job, D.; Wick, L.Y.; Filippidou, S.; Kooli, W.M.; Verrecchia, E.P.; Junier, P. Exploiting the fungal highway: Development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium. FEMS Microbiol. Ecol. 2015, 91, fiv116. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, B.-J.; Stanley, C.E.; Dusny, C.; Schlosser, D.; Harms, H.; Wick, L.Y. pH Distribution along Growing Fungal Hyphae at Microscale. J. Fungi 2022, 8, 599. https://doi.org/10.3390/jof8060599
Xiong B-J, Stanley CE, Dusny C, Schlosser D, Harms H, Wick LY. pH Distribution along Growing Fungal Hyphae at Microscale. Journal of Fungi. 2022; 8(6):599. https://doi.org/10.3390/jof8060599
Chicago/Turabian StyleXiong, Bi-Jing, Claire E. Stanley, Christian Dusny, Dietmar Schlosser, Hauke Harms, and Lukas Y. Wick. 2022. "pH Distribution along Growing Fungal Hyphae at Microscale" Journal of Fungi 8, no. 6: 599. https://doi.org/10.3390/jof8060599
APA StyleXiong, B.-J., Stanley, C. E., Dusny, C., Schlosser, D., Harms, H., & Wick, L. Y. (2022). pH Distribution along Growing Fungal Hyphae at Microscale. Journal of Fungi, 8(6), 599. https://doi.org/10.3390/jof8060599