Agrobacterium tumefaciens-Mediated Transformation of Candida glabrata
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Media
2.3. Growth Conditions
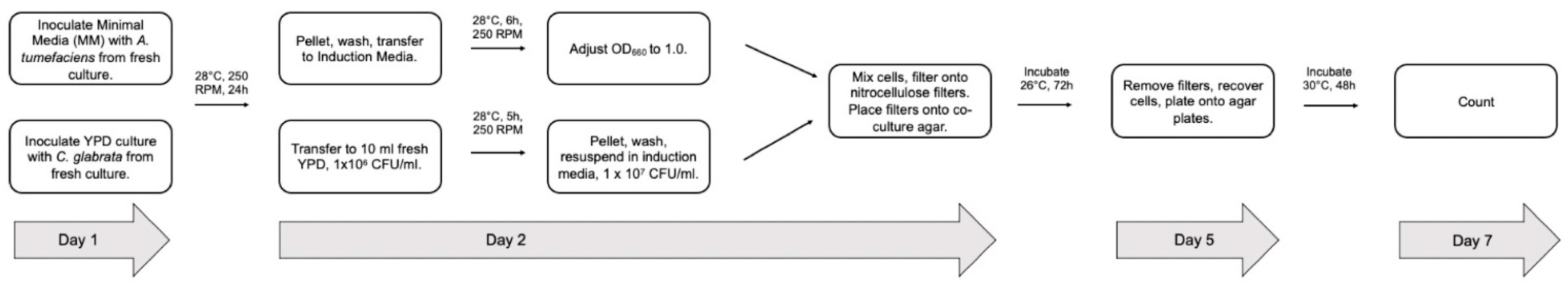
2.4. Standard Transformation Protocol
2.5. Optimization of the Agrobacterium Mediated Transformation System for C. glabrata
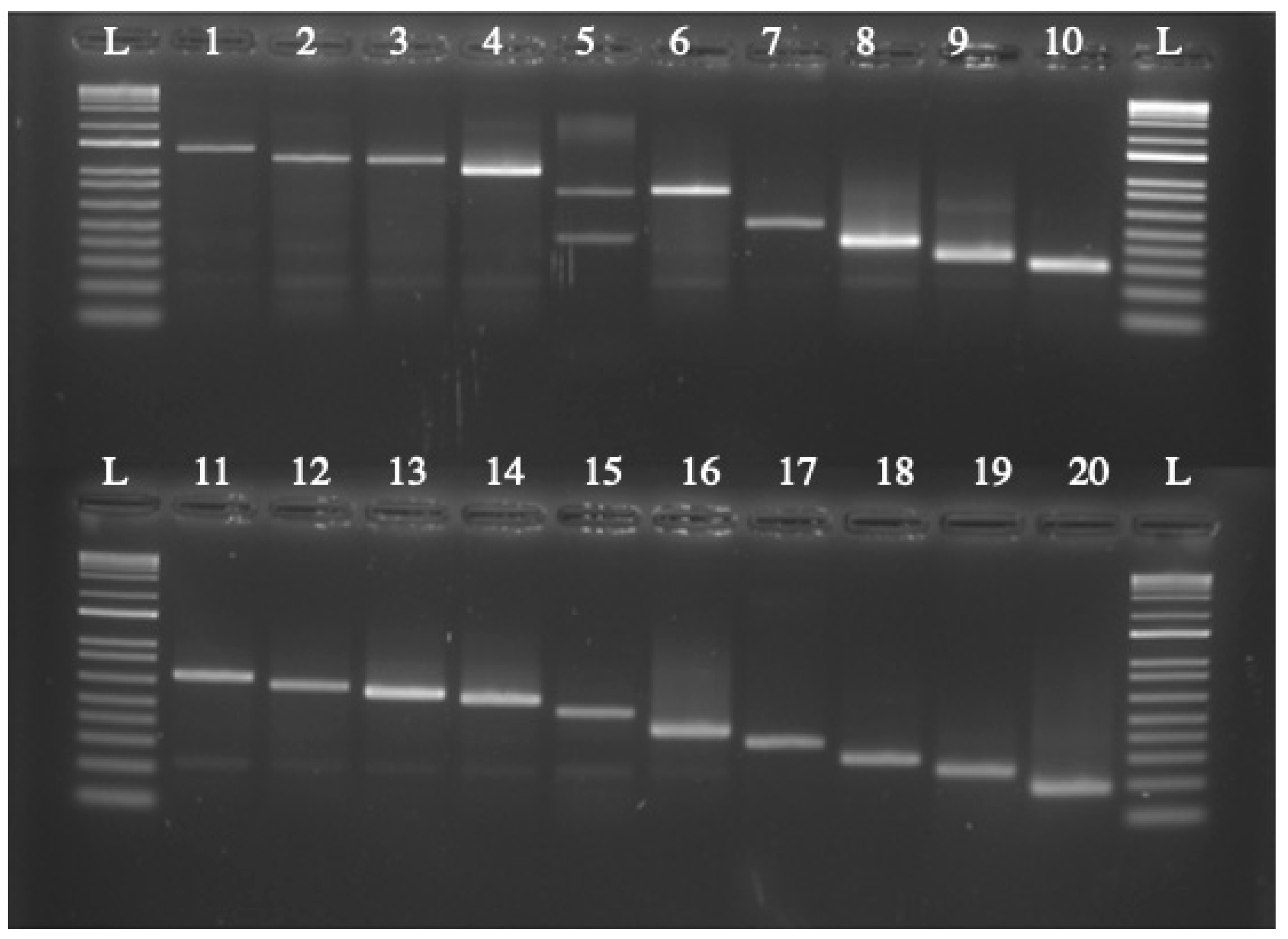
2.6. Identification of Insertion Site by Vectorette PCR
3. Results
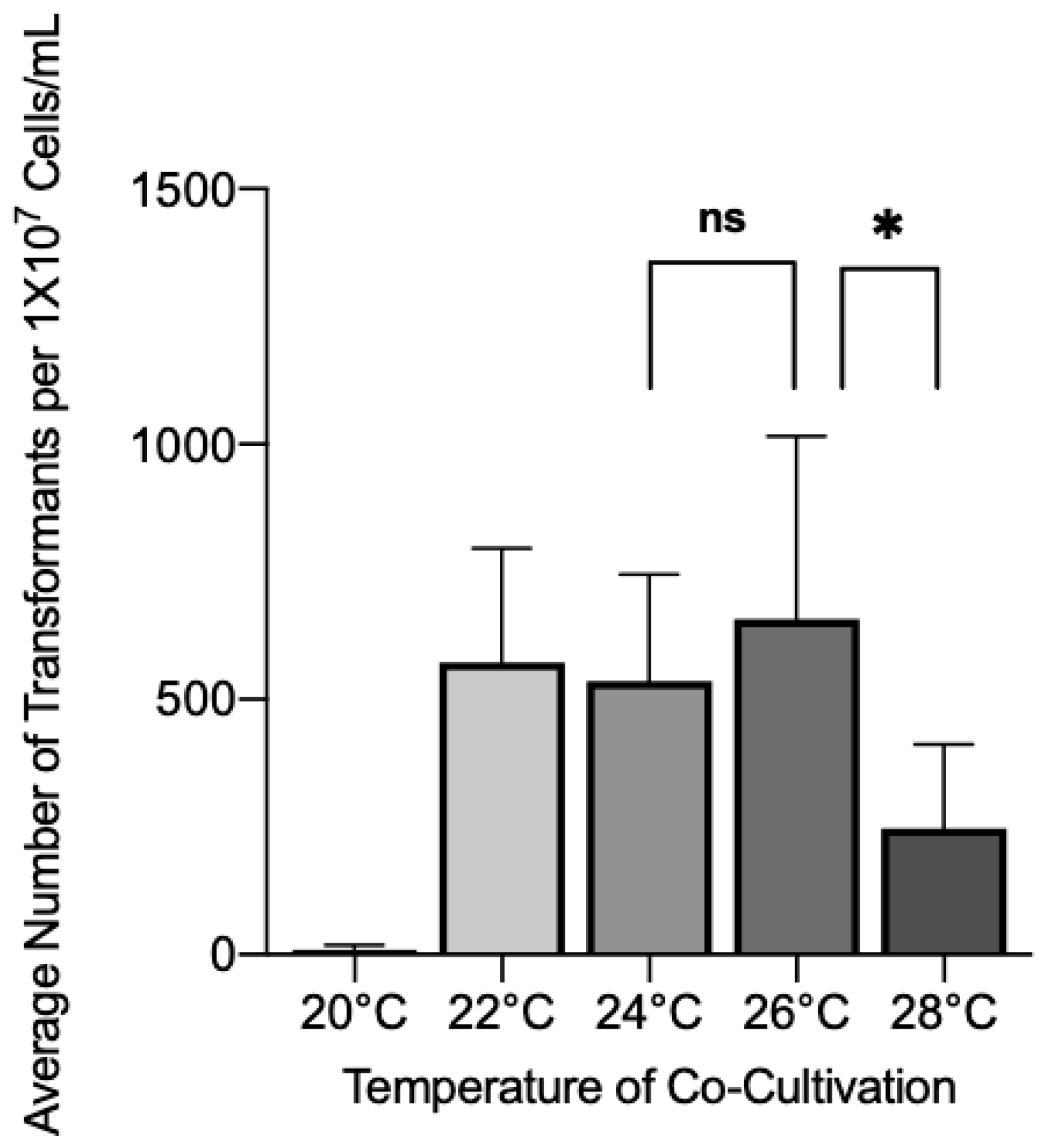
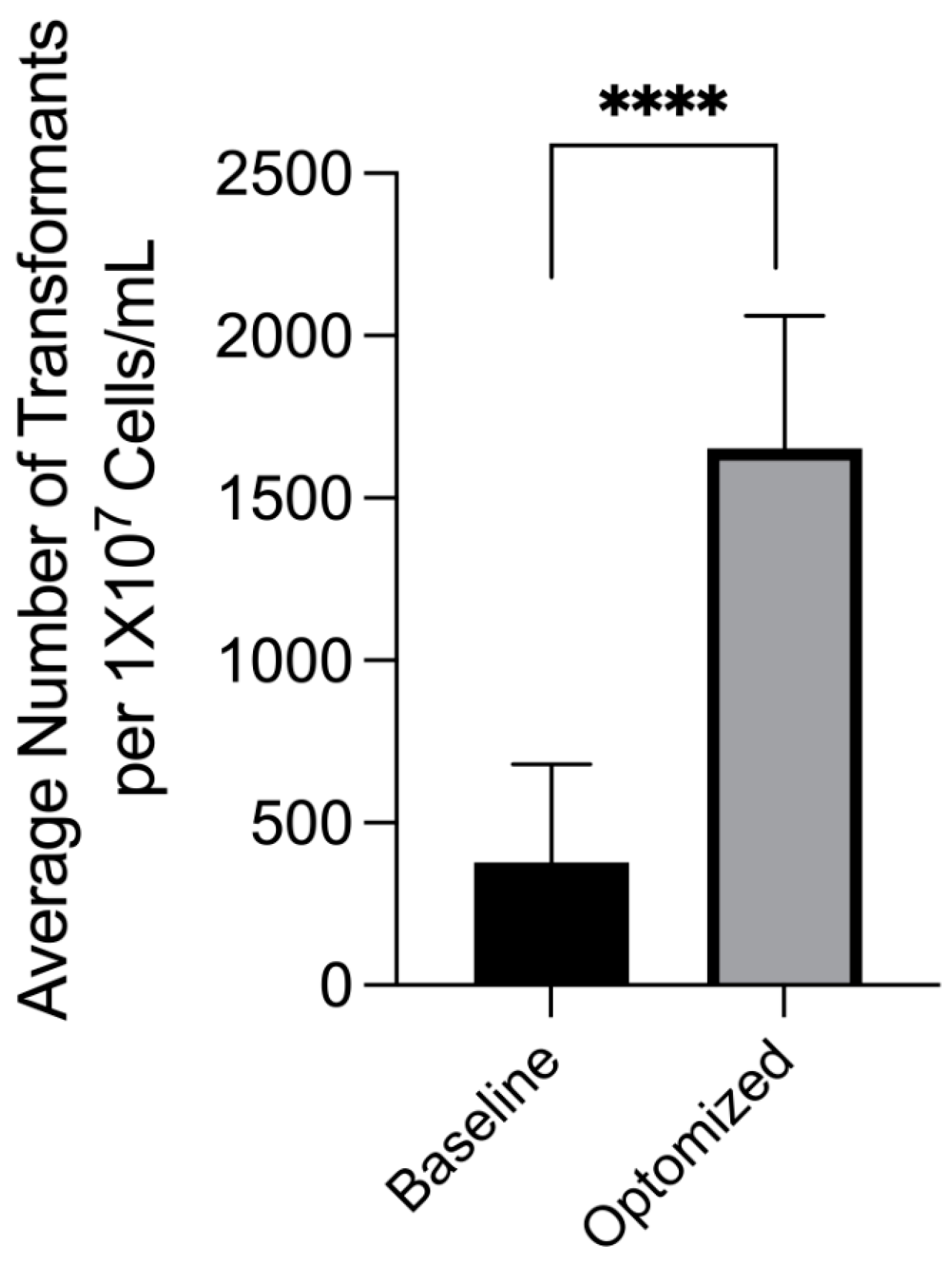
3.1. Effect of Co-Cultivation Parameters on Transformation Efficiency
3.2. Effect of Acetosyringone on Transformation Efficiency
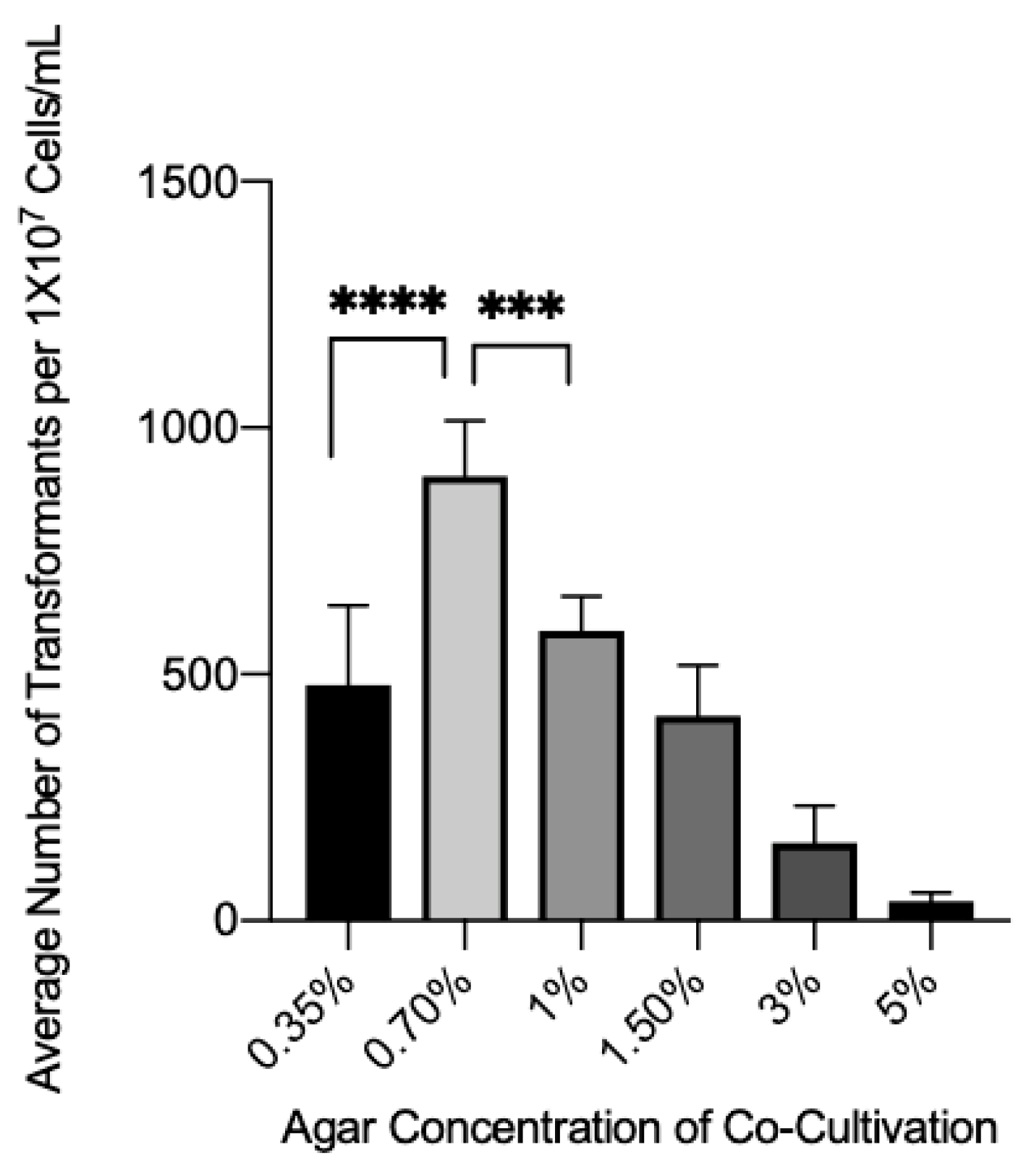
3.3. Effect of Agar Concentration on Transformation Efficiency
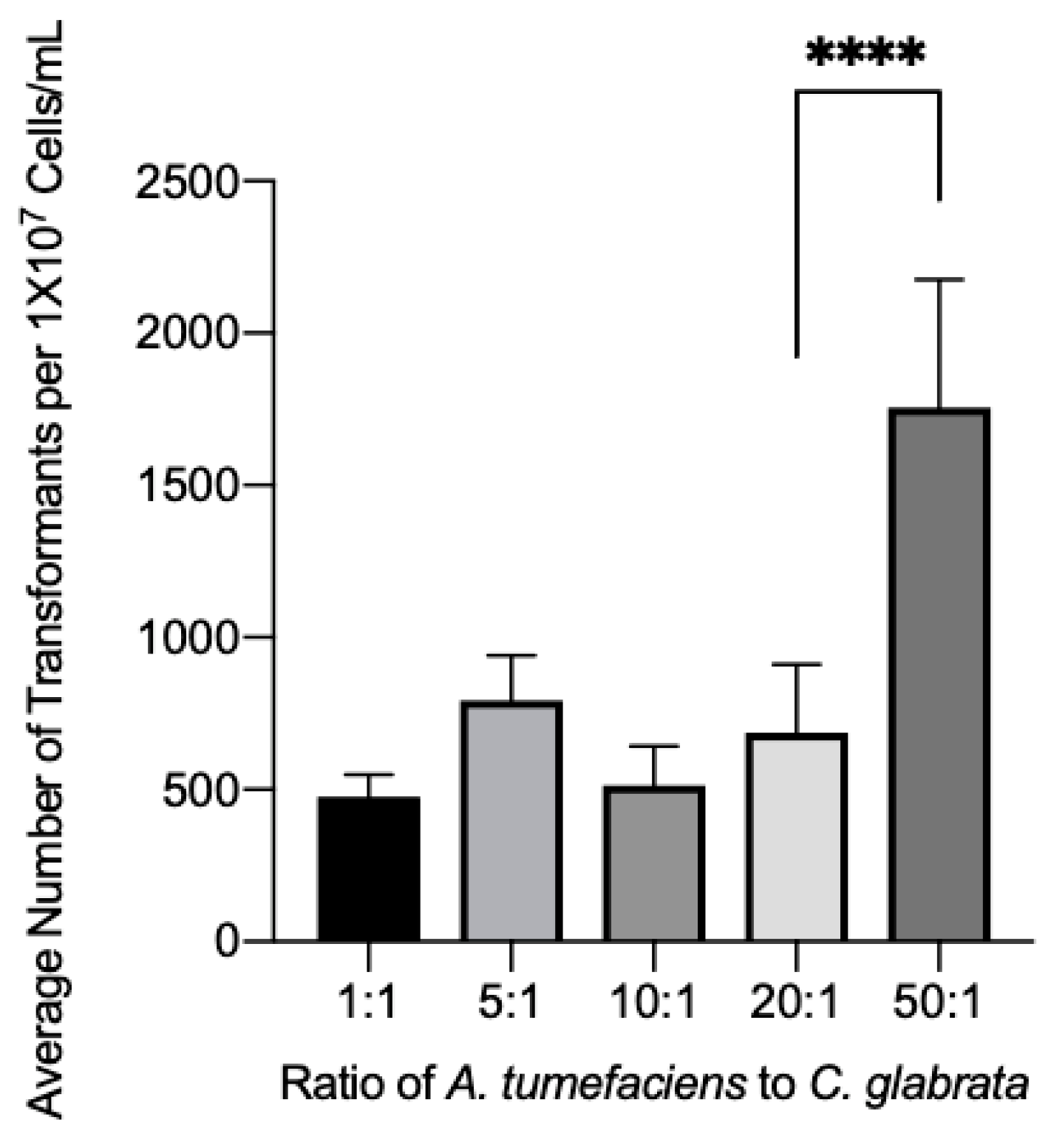
3.4. Effect of the Bacterial:Fungal Cell Ratio on Transformation Efficiency
3.5. Optimized Transformation Protocol
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vazquez, J.A.; Dembry, L.M.; Sanchez, V.; Vazquez, M.A.; Sobel, J.D.; Dmuchowski, C.; Zervos, M.J. Nosocomial Candida glabrata colonization: An epidemiologic study. J. Clin. Microbiol. 1998, 36, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Gabaldon, T.; Carrete, L. The birth of a deadly yeast: Tracing the evolutionary emergence of virulence traits in Candida glabrata. FEMS Yeast Res. 2016, 16, fov110. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Domergue, R.; Castano, I.; De Las Penas, A.; Zupancic, M.; Lockatell, V.; Hebel, J.R.; Johnson, D.; Cormack, B.P. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 2005, 308, 866–870. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention, Office of Infectious Disease. Antibiotic resistance threats in the United States, 2013. April 2013. Available online: http://www.cdc.gov/drugresistance/threat-report-2013 (accessed on 28 June 2017).
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance 2014; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Li, L.; Redding, S.; Dongari-Bagtzoglou, A. Candida glabrata: An emerging oral opportunistic pathogen. J. Dent. Res. 2007, 86, 204–215. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and mechanisms of antifungal resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]
- Wiederhold, N.P.; Lewis, R.E. The echinocandin antifungals: An overview of the pharmacology, spectrum and clinical efficacy. Expert Opin. Investig. Drugs 2003, 12, 1313–1333. [Google Scholar] [CrossRef]
- Rodrigues, C.F.; Silva, S.; Henriques, M. Candida glabrata: A review of its features and resistance. Eur J. Clin. Microbiol. Infect. Dis. 2014, 33, 673–688. [Google Scholar] [CrossRef]
- Singh-Babak, S.D.; Babak, T.; Diezmann, S.; Hill, J.A.; Xie, J.L.; Chen, Y.L.; Poutanen, S.M.; Rennie, R.P.; Heitman, J.; Cowen, L.E. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 2012, 8, e1002718. [Google Scholar] [CrossRef]
- Beyda, N.D.; John, J.; Kilic, A.; Alam, M.J.; Lasco, T.M.; Garey, K.W. FKS mutant Candida glabrata: Risk factors and outcomes in patients with candidemia. Clin. Infect. Dis. 2014, 59, 819–825. [Google Scholar] [CrossRef]
- Farmakiotis, D.; Tarrand, J.J.; Kontoyiannis, D.P. Drug-resistant Candida glabrata infection in cancer patients. Emerg. Infect. Dis. 2014, 20, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Kummasook, A.; Cooper, C.R., Jr.; Vanittanakom, N. An improved Agrobacterium-mediated transformation system for the functional genetic analysis of Penicillium marneffei. Med. Mycol. 2010, 48, 1066–1074. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McClelland, C.M.; Wickes, B.L. Agrobacterium tumefaciens as a molecular tool for the study of fungal pathogens. In Applied Mycology; Rai, M., Bridge, P.D., Eds.; CABI: Wallingford, UK, 2009; pp. 239–257. [Google Scholar]
- Hood, E.E.; Gelvin, S.B.; Melchers, L.S.; Hoekema, A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993, 2, 208–218. [Google Scholar] [CrossRef]
- Shen, J.; Guo, W.; Kohler, J.R. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect. Immun. 2005, 73, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Khang, C.H.; Park, S.Y.; Lee, Y.H.; Kang, S. A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum. Fungal Genet. Biol. 2005, 42, 483–492. [Google Scholar] [CrossRef]
- Bundock, P.; den Dulk-Ras, A.; Beijersbergen, A.; Hooykaas, P.J. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995, 14, 3206–3214. [Google Scholar] [CrossRef]
- Xiao, X.; Feng, J.; Li, Y.; Chen, Z.; Shi, M.; Xi, L.; Mylonakis, E.; Zhang, J. Agrobacterium tumefaciens-mediated transformation: An efficient tool for targeted gene disruption in Talaromyces marneffei. World J. Microbiol. Biotechnol. 2017, 33, 183. [Google Scholar] [CrossRef]
- Rho, H.S.; Kang, S.; Lee, Y.H. Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea. Mol. Cells 2001, 12, 407–411. [Google Scholar]
- Clarke, D.L.; Woodlee, G.L.; McClelland, C.M.; Seymour, T.S.; Wickes, B.L. The Cryptococcus neoformans STE11alpha gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol. Microbiol. 2001, 40, 200–213. [Google Scholar] [CrossRef]
- Arnold, C.; Hodgson, I.J. Vectorette PCR: A novel approach to genomic walking. PCR Methods Appl. 1991, 1, 39–42. [Google Scholar] [CrossRef]
- Veluthambi, K.; Krishnan, M.; Gould, J.H.; Smith, R.H.; Gelvin, S.B. Opines stimulate induction of the vir genes of the Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 1989, 171, 3696–3703. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Brockman, N.E.; Wickes, B.L. Optimizing Transformation Frequency of Cryptococcus neoformans and Cryptococcus gattii Using Agrobacterium tumefaciens. J. Fungi 2021, 7, 520. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.L.; Metz, M.; Monks, D.E.; Mullaney, M.L.; Hall, T.; Nester, E.W. Purine synthesis and increased Agrobacterium tumefaciens transformation of yeast and plants. Proc. Natl. Acad. Sci. USA 2003, 100, 6634–6639. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.J.; Carmona, J.A.; Cunha, C.; Carvalho, A.; Rappleye, C.A.; Goldman, W.E.; Hooykaas, P.J.; Leao, C.; Ludovico, P.; Rodrigues, F. Towards a molecular genetic system for the pathogenic fungus Paracoccidioides brasiliensis. Fungal Genet. Biol. 2007, 44, 1387–1398. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Spain, S.; Andrade, P.I.; Brockman, N.E.; Fu, J.; Wickes, B.L. Agrobacterium tumefaciens-Mediated Transformation of Candida glabrata. J. Fungi 2022, 8, 596. https://doi.org/10.3390/jof8060596
D’Spain S, Andrade PI, Brockman NE, Fu J, Wickes BL. Agrobacterium tumefaciens-Mediated Transformation of Candida glabrata. Journal of Fungi. 2022; 8(6):596. https://doi.org/10.3390/jof8060596
Chicago/Turabian StyleD’Spain, Samantha, Pilar I. Andrade, Nohelli E. Brockman, Jianmin Fu, and Brian L. Wickes. 2022. "Agrobacterium tumefaciens-Mediated Transformation of Candida glabrata" Journal of Fungi 8, no. 6: 596. https://doi.org/10.3390/jof8060596
APA StyleD’Spain, S., Andrade, P. I., Brockman, N. E., Fu, J., & Wickes, B. L. (2022). Agrobacterium tumefaciens-Mediated Transformation of Candida glabrata. Journal of Fungi, 8(6), 596. https://doi.org/10.3390/jof8060596






