Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions
Abstract
1. Introduction
2. Global and National Significance of Rice Blast in Present and Future Context
3. Molecular Interplay between Rice and M. oryzae
3.1. The Pathogen: M. oryzae
3.1.1. Magnaporthe oryzae Genomics
3.1.2. Pathogenicity Related Factors of M. oryzae
3.1.3. Editing Pathogenicity Genes in M. oryzae
3.1.4. Magnaporthe Host-Shifting
3.2. The Host: Rice
3.2.1. Genomes Sequenced
3.2.2. Resistance Genes as Solo Protectors
| Sl. No. | Gene ID | Chr. No. | Position (cM) | Source Cultivar | Reference |
|---|---|---|---|---|---|
| 1 | Pit | 1 | 9.08–12.17 | Tjahaja | [107] |
| 2 | Pi27(t) | 1 | 24.29–27.90 | IR64 (Indica) | [108] |
| 3 | Pi24(t) | 1 | 20.97–22.22 | Azuenca (Japonica) | [109] |
| 4 | Pitp(t) | 1 | 100.54–108.43 | Tetep | [110] |
| 5 | Pi35(t) | 1 | 132.0–136.6 | Hokkai 188 (Japonica) | [111] |
| 6 | Pi37 | 1 | 132.44–133.95 | St. No. 1 (Japonica) | [112] |
| 7 | Pish | 1 | 135.3-138.7 | Shin2 (Japonica) | [113] |
| 8 | Pid1(t) | 2 | 87.5–89.9 | Digu | [114] |
| 9 | Pig(t) | 2 | 137.38–140.54 | Guangchangzhan (Indica) | [115] |
| 10 | Pitq5 | 2 | 150.5–157.9 | Teqing | [116] |
| 11 | Piy1(t) | 2 | 153.2–154.1 | Yanxian No. 1 | [117] |
| 12 | Piy2(t) | 2 | 153.2–154.1 | Yanxian No. 1 | [117] |
| 13 | Pib | 2 | 153.2–154.1 | Tohoku IL9 | [118] |
| 14 | Pi25(t) | 2 | 137.44–150.90 | IR64 (Indica) | [119] |
| 15 | Pi14(t) | 2 | 1–26.90 | Maowangu | [120] |
| 16 | Pir-2-3(t) | 2 | 96.8–99.3 | IR64 (Indica) | [121] |
| 17 | Pitq2 | 2 | Teqing (Indica) | [122] | |
| 18 | Pirf2-1(t) | 2 | 109.6–112.2 | O. rufipogon (W) | [121] |
| 19 | Pi16(t) | 2 | 1–26.91 | Aus373 (Indica) | [123] |
| 20 | Pitq3 | 3 | Teqing (Indica) | [122] | |
| 21 | Pi68 | 3 | 6.8-9.7 | O. glumaepatula (W) | [124] |
| 22 | pi21 | 4 | 20.97–22.22 | Owarihatamochi | [125] |
| 23 | Pikur1 | 4 | 98.44–134.23 | Kuroka (Japonica) | [126] |
| 24 | Pi39(t) | 4 | 107.4–108.2 | Chubu 111 (Japonica) | [127] |
| 25 | Pitq4 | 4 | Teqing (Indica) | [122] | |
| 26 | Pi(t) | 4 | 9.08–12.17 | Tjahaja | [128] |
| 27 | Pi26(t) | 5 | 35.00–46.70 | Gumei 2 (Indica) | [119] |
| 28 | Pi23(t) | 5 | 43.02–76.70 | Sweon 365 | [125] |
| 29 | Pi10 | 5 | 58.08–75.41 | Tongil | [129] |
| 30 | Pi22(t) | 6 | 19.5–24.09 | Suweon365 (Japonica) | [125] |
| 31 | Pi26(t) | 6 | 35.00–46.70 | Azucena (Japonica) | [130] |
| 32 | Pi27(t) | 6 | 22.22–2.97 | IR64 (Indica) | [108] |
| 33 | Pi40(t) | 6 | 65.09–70.12 | O. australiensis (W) | [131] |
| 34 | Piz | 6 | 40.6–42.07 | Zenith (Japonica) | [132] |
| 35 | Piz-t | 6 | 58.7 | Toride 1 | [107] |
| 36 | Pi9 | 6 | 41.5–41.55 | O. minuta (W) | [133] |
| 37 | Pi25(t) | 6 | 72.32–77.03 | Gumei 2 | [119] |
| 38 | Pi8 | 6 | 19.5–24.09 | Kasalath (Indica) | [120] |
| 39 | Pi3(t) | 6 | Pai-kan-tao (Japonica) | [134] | |
| 40 | Pitq1 | 6 | 92.6-98.2 | Tequing (Indica) | [135] |
| 41 | Pi13(t) | 6 | 56.8-60.5 | Kasalath (Indica) | [136] |
| 42 | Pii1 | 6 | 88.8-90.6 | Fujisaka 5 (Indica) | [120] |
| 43 | Pid2 | 6 | 68.63–68.65 | Digu | [137] |
| 44 | Pigm(t) | 6 | 41.47–41.68 | Gumei 4 | [138] |
| 45 | Pi17(t) | 7 | 89.00–99.9 | DJ 123 | [120] |
| 46 | Pi36 | 8 | 11.48–11.53 | Q61 (Indica) | [127] |
| 47 | Pi33 | 8 | 23.66–24.61 | IR64 (Indica) | [134] |
| 48 | Pizh | 8 | 17.48–84.04 | Zhai-Ya-Quing8 (Indica) | [108] |
| 49 | Pi11 | 8 | Zhai-Ya-Quing8 (Indica) | [128] | |
| 50 | Pi29(t) | 8 | 38.65–64.96 | IR64 (Indica) | [108] |
| 51 | Pii2(t) | 9 | 4.09–28.89 | Azucena | [139] |
| 52 | Pi5(t) | 9 | 31.3–33.0 | RIL125, RIL249 and RIL260(Moroberekan) | [140] |
| 53 | Pi3(t) | 9 | 31.3–33.1 | Kan-Tao | [128] |
| 54 | Pi15 | 9 | 38.56–38.74 | GA25 (Japonica) | [120] |
| 55 | Pii | 9 | 9.16–113.72 | Ishikari Shiroke (Japonica) | [141] |
| 56 | Pi28(t) | 10 | 78.26–90.67 | IR64 (Indica) | [108] |
| 57 | Pia | 11 | 1.01–2.09 | Aichi Asahi (Japonica) | [126] |
| 58 | PiCO39(t) | 11 | 25.21–27.55 | CO39 (Indica) | [142] |
| 59 | Pilm2 | 11 | 54.54–113.5 | Lemont | [116] |
| 60 | Pi30(t) | 11 | 1.76–26.31 | IR64 (Indica) | [108] |
| 61 | Pi7(t) | 11 | 71.4–84.3 | RIL29 (Japonica) | [143] |
| 62 | Pi34 | 11 | 77.69–77.96 | Chubu32 (Japonica) | [144] |
| 63 | Pi38 | 11 | 76.55–87.91 | Tadukan (Indica) | [145] |
| 64 | PBR | 11 | 80.5–120.3 | St. No. 1 | [146] |
| 65 | Pb1 | 11 | 85.7–91.4 | Modan | [147] |
| 66 | Pi44(t) | 11 | 91.4–117.9 | RIL29 (Japonica) | [148] |
| 67 | Pik-h (Pi54) | 11 | 99.0–99.05 | Tetep | [95] |
| 68 | Pi1 | 11 | 105.99–113.49 | LAC23 (Japonica) | [149] |
| 69 | Pik-m | 11 | 109.25–110.13 | Tsuyuake (Japonica) | [150] |
| 70 | Pi18(t) | 11 | 107.18–113.50 | Suweon365 (Japonica) | [132] |
| 71 | Pik | 11 | 109.25–110.13 | Kusabue (Indica) | [151] |
| 72 | Pik-p | 11 | 109.25–110.14 | HR22 (Indica) | [107] |
| 73 | Pik-s | 11 | 109.25–110.15 | Shin 2 (Japonica) | [152] |
| 74 | Pik-g | 11 | 109.25–110.16 | GA20 (Japonica) | [120] |
| 75 | Pise1 | 11 | 22.96–66.92 | Sensho | [153] |
| 76 | Pi f | 11 | 98.78–113.84 | Chugoku 31-1 (St. No. 1) | [154] |
| 77 | Mpiz | 11 | 16.29–66.92 | Zenith (Japonica) | [155] |
| 78 | Pikur2 | 11 | 11.36–73.49 | Kuroka (Japonica) | [125] |
| 79 | Pish | 11 | 110.3–111.8 | Nipponbare (Japonica) | [113] |
| 80 | Pib2 | 11 | 105.99–113.49 | Lemont (Japonica) | [122] |
| 81 | Pi44 | 11 | 85.7–89.7 | Moroberekan (Japonica) | [148] |
| 82 | Pi47 | 11 | Xiangzi (Indica) | [116] | |
| 83 | Pise | 11 | 22.96–66.92 | Sensho | [153] |
| 84 | Piis1 | 11 | 11.36–76.11 | Imochi Shirazu (Japonica) | [153] |
| 85 | Pi24(t) | 12 | 20.97–22.22 | Azuenca (Japonica) | [156] |
| 86 | Pi62(t) | 12 | 9.7–77 | Tsuyuake (Japonica) | [157] |
| 87 | Pitq6 | 12 | 23.0–30.92 | Tequing (Indica) | [116] |
| 88 | Pi6(t) | 12 | 1–1.68 | Apura (Indica) | [158] |
| 89 | Pi12 | 12 | 27.95–60.48 | Moroberekan (Japonica) | [159] |
| 90 | Pi21(t) | 12 | 20.94–22.22 | Owarihatamochi (Japonica) | [125] |
| 91 | Pi31(t) | 12 | 30.92–47.66 | IR64 (Indica) | [108] |
| 92 | Pi32(t) | 12 | 52.41–75.46 | IR64 (Indica) | [108] |
| 93 | Pi157 | 12 | 49.5–62.2 | Moroberekan (Japonica) | [123] |
| 94 | Pita | 12 | 42.41–42.43 | Tadukan (Indica) | [107] |
| 95 | Pita-2 | 12 | 40.31–52.84 | Shimokita (Japonica) | [160] |
| 96 | Pi19(t) | 12 | 35.30–53.67 | Aichi Asahi (Japonica) | [161] |
| 97 | Pi39(t) | 12 | - | Chubu 111 (Japonica) | [127,162] |
| 98 | Pi20(t) | 12 | 51.5–51.8 | IR24 (Indica) | [163] |
| 99 | Pi20 | 12 | 49.6-50.4 | IR24 (Indica) | [164] |
| 100 | Pi42(t) | 12 | 58.9-56-7 | DHR9 (Indica) | [151] |
| 101 | Pi48 | 12 | Xiangzi 3150 (Indica) | [116] | |
| 102 | PiGD-3(t) | 12 | 55.8 | Sanhuangzhan 2 | [138] |
3.2.3. Chemical Modulators
3.2.4. Modulation of Coding RNA (mRNA) of Rice upon M. oryzae Infection
3.2.5. Small and Long Non-Coding RNA Play a Regulatory Role in Rice upon M. oryzae Infection
3.3. Interplay between Rice-M. oryzae: A Classical Example for Plant-Pathogen Interactions
3.3.1. Gene-for-Gene Model
3.3.2. Guard Model
3.3.3. Decoy Model
4. Resistance Response of Rice to Blast Disease
4.1. Resistance Response Based on Quantitative Trait Loci (QTL)
4.2. Resistance Gene Mediated Resistance
4.2.1. The Blast Resistance Genes Identified, Mapped, and Cloned in Rice
| Genes & Alleles | Encoded Protein | Chr. No | Cognate AVR Gene | Chromosomal Location | Donor | Reference |
|---|---|---|---|---|---|---|
| Pish | NLR | 1 | - | 33,136,846–33,145,541 | Nipponbare | [257] |
| Pi35 | NLR | 1 | - | 33,838,140–35,206,760 | Hokkai 188 | [111] |
| Pi37 | NLR | 1 | - | 33,116,117–33,124,371 | St. No. 1 | [112] |
| Pi64 | NLR | 1 | - | 33,098,072–33,104,550 | Yangmaogu | [254] |
| Pit | NLR | 1 | - | 2,686,729–2,687,700 | K59 | [258,259] |
| Pi-b | NLR | 2 | AVR-Pib | 35,979,234 | Tohoku IL9 | [38,118] |
| pi21 | Proline-rich metal binding protein | 4 | - | 19,836,301–19,835,131 | Owarihatamochi | [96] |
| Pi63 | NLR | 4 | - | 31,553,065–31,558,406 | Kahei | [260] |
| PiPR1 | NLR | 4 | - | 316,00,121–31,604,201 | [261] | |
| Pi9 | NLR | 6 | AVR-Pi9 | 2,410,176–2,418,568 | 75-1-127 | [37,133] |
| Pi2 | NLR | 6 | 1,043,5816–10,441,907 | Jefferson | [262] | |
| Piz-t | NLR | 6 | Avr-Pizt | 10,387,509–10,390,465 | Zenith | [36,262] |
| Pi50 | NLR | 6 | - | 10,375,846–10,380,263 | Er-Ba-zhan (EBZ) | [97] |
| Pizh | NLR | 6 | - | 10,087,244–10,478,622 | [263] | |
| Pigm | NLR | 6 | - | Near to 10,435,816–10,441,907 | Gumei4 | [100,264] |
| Pi-d2 | B-lectin receptor kinase | 6 | - | 17,164,851–17,160,330 | Digu | [137] |
| Pi-d3 | NLR | 6 | - | 13,058,027–13,055,162 | Digu | [265,266] |
| Pi25 | NLR | 6 | - | 13,058,027–13,055,162 (Pid3 allele) | Gumei2 | [253] |
| Pid3-A4 | NLR | 6 | - | 13,058,027–13,055,162 (Pid3 allele) | A4 (Oryza rufipogon) | [267] |
| Pi36 | NLR | 8 | - | 2,878,953–2,890,634 | Kasalath | [268] |
| Pi5 | NLR | 9 | - | 9,674,695–9,674,000 | RIL260 | [236] |
| Pii | NLR | 9 | AVR-Pii | 9,674,695–9,674,000 | Hitomebore | [16,269] |
| Pi56 | NLR | 9 | - | 9,777,527–9,780,698 | Sanhuangzhan No. 2 | [270] |
| Pb1 | NLR | 11 | - | 14,705,215–14,714,572 | Modan | [252,271] |
| Pik | NLR | 11 | AVR-Pik | 27,984,697–27,989,134 | Kusabue | [16,272] |
| Pik-p | NLR | 11 | AVR-Pikp | 27,978,568–27,980,621 | K60 | [237] |
| Pikm | NLR | 11 | AVR-Pikm | 27,984,697–27,989,134 | Tsuyuake | [235] |
| Pike | NLR | 11 | - | 27,984,697–27,989,134 (Pik allele) | Xiangzao143 | [273] |
| Pik-h | NLR | 11 | - | 27,984,697–27,989,134 (Pik allele) | K3 | [238] |
| Pi1 | NLR | 11 | - | 27,984,697–27,989,134 (Pik allele) | C101LAC | [274] |
| Pi54 | NLR | 11 | AVR-Pi54 | 25,262,834–25,264,520 | Tetep | [39,275] |
| Pi54rh | NLR | 11 | Avr-Pi54 | 25,262,834–25,264,520 (Pi54 allele) | Oryza rhizomatis (nrcpb 002) | [171] |
| Pi54of | NLR | 11 | AVR-Pi54 | 25,262,834–25,264,520 (Pi54 allele) | Oryza officinalis (nrcpb004) | [89] |
| Pia | NLR | 11 | AVR-Pia | 6,546,026–6,541,924 | Sasanishiki | [16,44,276] |
| Pi-CO39 | NLR | 11 | AVR-CO39 | 6,888,057-6,291,466 | CO39 | [142,277] |
| Pi-ta | NLR | 12 | AVR-Pita | 10,612,068–10,606,359 | Yashiro-mochi | [32,162] |
| Pi65 | LRR-RLK | 12 | 28,376,327–28,379,731 | GangYu129 | [278] | |
| Ptr | ARM repeat domain protein | 12 | - | 10822534–10833768 | M2354 | [101] |
4.2.2. Resistance Response Mediated by Alleles of Known R Genes
5. Molecular Mechanisms of Leaf and Panicle Blast
6. Management of Blast Disease Using Host Resistance
6.1. Introgression of QTLs for Blast Resistance
6.2. Introgression of R-Genes for Blast Resistance
6.3. Transgenic Approach for Blast Management
6.4. Genome Editing of Immunity Regulators
7. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Thirze, H. Modelling Grain Surplus and Deficit in Cameroon for 2030. Master’s Thesis, Lund University, Lund, Sweeden, 2016; p. 393. [Google Scholar]
- Asibi, A.E.; Chai, Q.; Coulter, J.A. Rice blast: A disease with implications for global food security. Agronomy 2019, 9, 451. [Google Scholar] [CrossRef]
- Samal, P.; Babu, S. The shape of rice agriculture towards 2050. In Proceedings of the Conference IAAE 30th International Conference of Agricultural Economists, Vancouver, BC, Canada, 28 July–2 August 2018. [Google Scholar]
- Sharma, T.R.; Rai, A.K.; Gupta, S.K.; Vijayan, J.; Devanna, B.N.; Ray, S. Rice blast management through host-plant resistance: Retrospect and prospects. Agric. Res. 2012, 1, 37–52. [Google Scholar] [CrossRef]
- Kaundal, R.; Kapoor, A.S.; Raghava, G.P. Machine learning techniques in disease forecasting: A case study on rice blast prediction. BMC Bioinform. 2006, 7, 485. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.P.; Lee, S.; Wang, J.; Ma, L.; Bianco, T.; Jia, Y. Current advances on genetic resistance to rice blast disease. Rice-Germplasm Genet. Improv. 2014, 23, 195–217. [Google Scholar]
- Kato, H. Rice blast disease. Pestic. Outlook 2001, 12, 23–25. [Google Scholar] [CrossRef]
- Gnanamanickam, S.S. Rice and its importance to human life. In Biological Control of Rice Diseases; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–11. [Google Scholar]
- Variar, M. Pathogenic variation in M. grisea and breeding for blast resistance in India. In JIRCAS Working Report No. 53; Japan International Center for Agricultural Sciences: Tsukuba, Japan, 2006; pp. 87–95. [Google Scholar]
- Wilson, R.A.; Talbot, N.J. Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 2009, 7, 185–195. [Google Scholar] [CrossRef]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef]
- Singh, P.K.; Mahato, A.K.; Jain, P.; Rathour, R.; Sharma, V.; Sharma, T.R. Comparative genomics reveals the high copy number variation of a retro transposon in different Magnaporthe isolates. Front. Microbiol. 2019, 10, 966. [Google Scholar] [CrossRef]
- Xue, M.; Yang, J.; Li, Z.; Hu, S.; Yao, N.; Dean, R.A.; Zhao, W.; Shen, M.; Zhang, H.; Li, C.; et al. Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2012, 8, e1002869. [Google Scholar] [CrossRef]
- Chen, C.; Lian, B.; Hu, J.; Zhai, H.; Wang, X.; Venu, R.; Liu, E.; Wang, Z.; Chen, M.; Wang, B.; et al. Genome comparison of two Magnaporthe oryzae field isolates reveals genome variations and potential virulence effectors. BMC Genom. 2013, 14, 887. [Google Scholar] [CrossRef]
- Gowda, M.; Shirke, M.D.; Mahesh, H.B.; Chandarana, P.; Rajamani, A.; Chattoo, B.B. Genome analysis of rice-blast fungus Magnaporthe oryzae field isolates from southern India. Genome Data 2015, 5, 284–291. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoshida, K.; Saitoh, H.; Fujisawa, S.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Tosa, Y.; Chuma, I.; Takano, Y.; Win, J.; et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 2009, 21, 1573–1591. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, Y.; Zhao, M.; Jing, M.; Liu, X.; Liu, M.; Guo, X.; Zhang, X.; Chen, Y.; Liu, Y.; et al. Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PLoS Pathog. 2015, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, B.; Lu, G.; Zhong, Z.; Wang, Z. Genome sequence resource of Magnaporthe oryzae laboratory strain 2539. Mol. Plant Microbe Interact. 2020, 33, 1029–1031. [Google Scholar] [CrossRef]
- Reddy, B.; Kumar, A.; Mehta, S.; Sheoran, N.; Chinnusamy, V.; Prakash, G. Hybrid de novo genome-reassembly reveals new insights on pathways and pathogenicity determinants in rice blast pathogen Magnaporthe oryzae RMg_Dl. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Langner, T.; Harant, A.; Gomez-Luciano, L.B.; Shrestha, R.K.; Malmgren, A.; Latorre, S.M.; Burbano, H.A.; Win, J.; Kamoun, S. Genomic rearrangements generate hypervariable mini-chromosomes in host-specific isolates of the blast fungus. PLoS Genet. 2021, 17, e1009386. [Google Scholar] [CrossRef]
- Chen, K.; Feng, J.; Chen, S.; Su, J.; Yang, J.; Wang, C.; Feng, A.; Chen, B.; Zhu, X.; Wang, W. Comparative Analysis of the Genomes of Three Field Isolates of the Rice Blast Fungus Magnaporthe oryzae from Southern China. Agric. Sci. 2021, 12, 713–725. [Google Scholar]
- Kumar, A.; Sheoran, N.; Prakash, G.; Ghosh, A.; Chikara, S.K.; Rajashekara, H.; Singh, U.D.; Aggarwal, R.; Jain, R.K. Genome sequence of a unique Magnaporthe oryzae RMg-Dl isolate from India that causes blast disease in diverse cereal crops, obtained using PacBio single-molecule and Illumina HiSeq2500 sequencing. Genome Announc. 2017, 5, e01570-16. [Google Scholar] [CrossRef]
- Chiapello, H.; Mallet, L.; Guerin, C.; Aguileta, G.; Amselem, J.; Kroj, T.; Ortega-Abboud, E.; Lebrun, M.H.; Henrissat, B.; Gendrault, A.; et al. Deciphering genome content and evolutionary relationships of isolates from the fungus Magnaporthe oryzae attacking different host plants. Genome Biol. Evol. 2015, 7, 2896–2912. [Google Scholar] [CrossRef]
- Zhu, K.P.; Bao, J.D.; Zhang, L.H.; Xue, Y.; Yuan, L.; Zhu, M.H.; Lin, Q.Y.; Ao, Z.; Zhen, Z.; Bo, Z.; et al. Comparative analysis of the genome of the field isolate V86010 of the rice blast fungus Magnaporthe oryzae from Philippines. J. Integr. Agric. 2017, 16, 2222–2230. [Google Scholar] [CrossRef]
- Were, V.M.; Mwongera, D.T.; Soanes, D.M.; Shrestha, R.K.; Ryder, L.; Foster, A.J.; Mutiga, S.K.; Rotich, F.; Win, J.; Langer, T.; et al. Genome sequences of sixty Magnaporthe oryzae isolates from multiple host plant species. Zenedo 2021, 3, 1–9. [Google Scholar]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Tomma, B.P.J.; et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef]
- Yang, C.; Yu, Y.; Huang, J.; Meng, F.; Pang, J.; Zhao, Q.; Islam, A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 2019, 31, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, C.Y.; Park, S.Y.; Kim, K.T.; Jeon, J.; Chung, H.; Choi, G.; Kwon, S.; Choi, J.; Jeon, J.; et al. Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun. 2020, 11, 5845. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Sweigard, J.A.; Valent, B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 1995, 8, 939–948. [Google Scholar] [CrossRef]
- Sweigard, J.A.; Carroll, A.M.; Kang, S.; Farrall, L.; Chumley, F.G.; Valent, B. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 1995, 7, 1221–1233. [Google Scholar]
- Farman, M.L.; Leong, S.A. Chromosome walking to the AVR1-CO39 avirulence gene of Magnaporthe grisea: Discrepancy between the physical and genetic maps. Genetics 1998, 150, 1049–1058. [Google Scholar] [CrossRef]
- Orbach, M.J.; Farrall, L.; Sweigard, J.A.; Chumley, F.G.; Valent, B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2019–2032. [Google Scholar] [CrossRef]
- Böhnert, H.U.; Fudal, I.; Dioh, W.; Tharreau, D.; Notteghem, J.L.; Lebrun, M.H. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 2004, 16, 2499–2513. [Google Scholar] [CrossRef]
- Miki, S.; Matsui, K.; Kito, H.; Otsuka, K.; Ashizawa, T.; Yasuda, N.; Fukiya, S.; Sato, J.; Hirayae, K.; Fujita, Y.; et al. Molecular cloning and characterization of the AVR-Pia locus from a Japanese field isolate of Magnaporthe oryzae. Mol. Plant Pathol. 2009, 10, 361–374. [Google Scholar] [CrossRef]
- Wu, W.; Wang, L.; Zhang, S.; Li, Z.; Zhang, Y.; Lin, F.; Pan, Q. Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant-Microbe Interact. 2014, 27, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, B.; Wu, J.; Lu, G.; Hu, Y.; Zhang, X.; Zhang, Z.; Zhao, Q.; Feng, Q.; Zhang, H.; et al. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 2009, 22, 411–420. [Google Scholar] [CrossRef]
- Wu, J.; Kou, Y.; Bao, J.; Li, Y.; Tang, M.; Zhu, X.; Ponaya, A.; Xiao, G.; Li, J.; Li, C.; et al. Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol. 2015, 206, 1463–1475. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Wu, W.; He, L.; Yang, X.; Pan, Q. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Singh, P.K.; Gupta, D.K.; Mahato, A.K.; Sarkar, C.; Rathour, R.; Singh, N.K.; Sharma, T.R. Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Front. Plant Sci. 2016, 8, 1140. [Google Scholar] [CrossRef] [PubMed]
- De Guillen, K.; Ortiz-Vallejo, D.; Gracy, J.; Fournier, E.; Kroj, T.; Padilla, A. Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog. 2015, 11, e1005228. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, N.; Tsujimoto Noguchi, M.; Fujita, Y. Partial mapping of avirulence genes AVR-Pii and AVR-Pia in the rice blast fungus Magnaporthe oryzae. Can. J. Plant Pathol. 2006, 28, 494–498. [Google Scholar] [CrossRef]
- Chen, Q.H.; Wang, Y.C.; Li, A.N.; Zhang, Z.G.; Zheng, X.B. Molecular mapping of two cultivar-specific avirulence genes in the rice blast fungus Magnaporthe grisea. Mol. Genet. Genom. 2007, 277, 139–148. [Google Scholar] [CrossRef]
- Wang, B.H.; Ebbole, D.J.; Wang, Z.H. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J. Integr. Agric. 2017, 16, 2746–2760. [Google Scholar] [CrossRef]
- Okuyama, Y.; Kanzaki, H.; Abe, A.; Yoshida, K.; Tamiru, M.; Saitoh, H.; Fujibe, T.; Matsumura, H.; Shenton, M.; Galam, D.C.; et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant 2011, 66, 467–479. [Google Scholar] [CrossRef]
- Césari, S.; Kanzaki, H.; Fujiwara, T.; Bernoux, M.; Chalvon, V.; Kawano, Y.; Shimamoto, K.; Dodds, P.; Terauchi, R.; Kroj, T. The NB-LRR proteins RGA 4 and RGA 5 interact functionally and physically to confer disease resistance. EMBO J. 2014, 33, 1941–1959. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.H.; Zhang, T.; Liu, Q.Y.; Guo, H.S. Trans-kingdom RNAs and their fates in recipient cells: Advances, utilization and perspectives. Plant Commun. 2021, 2, 100167. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Wang, H.; Hu, P.; Hamby, R.; Jin, H. Small RNAs—Big players in plant-microbe interactions. Cell Host Microbe 2019, 26, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- LaMonte, G.; Philip, N.; Reardon, J.; Lacsina, J.R.; Majoros, W.; Chapman, L.; Thornburg, C.D.; Telen, M.J.; Ohler, U.; Nicchitta, C.V.; et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 2012, 12, 187–199. [Google Scholar] [CrossRef]
- Buck, A.H.; Coakley, G.; Simbari, F.; McSorley, H.J.; Quintana, J.F.; Le Bihan, T.; Kumar, S.; Abreu-Goodger, C.; Lear, M.; Harcus, Y.; et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014, 5, 5488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.L.; Zhao, J.H.; Wang, S.; Jin, Y.; Chen, Z.Q.; Fang, Y.Y.; Hua, C.L.; Ding, S.W.; Guo, H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Jiao, J.; Peng, D. Wheat microRNA1023 suppresses invasion of Fusarium graminearum via targeting and silencing FGSG_03101. J. Plant Interact. 2018, 13, 514–521. [Google Scholar] [CrossRef]
- Raman, V.; Simon, S.A.; Romag, A.; Demirci, F.; Mathioni, S.M.; Zhai, J.; Meyers, B.C.; Donofrio, N.M. Physiological stressors and invasive plant infections alter the small RNA transcriptome of the rice blast fungus, Magnaporthe oryzae. BMC Genom. 2013, 14, 326. [Google Scholar] [CrossRef]
- Raman, V.; Simon, S.A.; Demirci, F.; Nakano, M.; Meyers, B.C.; Donofrio, N.M. Small RNA functions are required for growth and development of Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2017, 30, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Nunes, C.C.; Gowda, M.; Sailsbery, J.; Xue, M.; Chen, F.; Brown, D.E.; Oh, Y.; Mitchell, T.K.; Dean, R.A. Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genom. 2011, 12, 288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, J.R. Effectors and effector delivery in Magnaporthe oryzae. PLoS Pathog. 2014, 10, e1003826. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia striiformis f. sp. tritici mi croRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. N. Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.M.; Mao, H.Y.; Li, S.J.; Feng, T.; Zhang, Z.Y.; Cheng, L.; Luo, S.J.; Borkovich, K.A.; Ouyang, S.Q. Fol-milR1, a pathogenicity factor of Fusarium oxysporum, confers tomato wilt disease resistance by impairing host immune responses. N. Phytol. 2021, 232, 705–718. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, S.; Chang, H.; Zhan, M.; Qin, Q.M.; Zhang, B.; Li, Z.; Liu, Y. Mining Magnaporthe oryzae sRNAs with potential Transboundary regulation of Rice genes associated with growth and defense through expression profile analysis of the pathogen-infected Rice. Front. Genet. 2019, 10, 296. [Google Scholar] [CrossRef]
- Schuster, M.; Kahmann, R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet. Biol. 2019, 130, 43–53. [Google Scholar] [CrossRef]
- Molla, K.A.; Karmakar, S.; Islam, M.T. Wide horizons of CRISPR-cas-derived technologies for basic biology, agriculture, and medicine. In CRISPR—Cas Methods; Islam, M.T., Bhowmik, P.K., Molla, K.A., Eds.; Springer Protocols Handbooks; Humana: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Foster, A.J.; Martin-Urdiroz, M.; Yan, X.; Wright, H.S.; Soanes, D.M.; Talbot, N.J. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 2018, 8, 14355. [Google Scholar] [CrossRef]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef]
- Pohl, C.; Kiel, J.A.; Driessen, A.J.; Bovenberg, R.A.; Nygard, Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 2016, 5, 754–764. [Google Scholar] [CrossRef]
- Shi, T.Q.; Liu, G.N.; Ji, R.Y.; Shi, K.; Song, P.; Ren, L.J.; Huang, H.; Ji, X.J. CRISPR/Cas9-based genome editing of the filamentous fungi: The state of the art. Appl. Microbiol. Biotechnol. 2017, 101, 7435–7443. [Google Scholar] [CrossRef] [PubMed]
- Naduthodi, M.I.; Barbosa, M.J.; van der Oost, J. Progress of CRISPR-Cas based genome editing in photosynthetic microbes. Biotechnol. J. 2018, 13, e1700591. [Google Scholar] [CrossRef]
- Huang, J.; Cook, D.E. CRISPR-Cas12a ribonucleoprotein-mediated gene editing in the plant pathogenic fungus Magnaporthe oryzae. STAR Protoc. 2022, 3, 101072. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Malaker, P.K.; Barma, N.C.; Tiwary, T.P.; Collis, W.J.; Duveiller, E.P.; Singh, K.; Joshi, A.K.; Singh, R.P.; Braun, H.J.; Peterson, G.L.; et al. First report of wheat blast caused by Magnaporthe oryzae pathotype triticum in Bangladesh. Plant Dis. 2016, 100, 2330. [Google Scholar] [CrossRef]
- Tembo, B.; Mulenga, R.M.; Sichilima, S.; M’siska, K.K.; Mwale, M.; Chikoti, P.C.; Singh, P.K.; He, X.; Pedley, K.F.; Peterson, G.L.; et al. Detection and characterization of fungus (Magnaporthe oryzae pathotype Triticum) causing wheat blast disease on rain-fed grown wheat (Triticum aestivum L.) in Zambia. PLoS ONE 2020, 15, e0238724. [Google Scholar] [CrossRef]
- Devanna, B.N.; Sharma, T.R. Wheat blast disease management: Cues from the advancements in molecular biology of rice-Magnaporthe pathosystem. J. Plant Biochem. Biotechnol. 2018, 27, 249–259. [Google Scholar] [CrossRef]
- Kim, K.H.; Choi, E.D. Retrospective study on the seasonal forecast-based disease intervention of the wheat blast outbreaks in Bangladesh. Front. Plant Sci. 2020, 11, 570381. [Google Scholar] [CrossRef]
- Islam, M.T.; Croll, D.; Gladieux, P.; Soanes, D.M.; Persoons, A.; Bhattacharjee, P.; Hossain, M.; Gupta, D.R.; Rahman, M.; Mahboob, M.G.; et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 2016, 14, 84. [Google Scholar] [CrossRef]
- Prabhu, A.S.; Filippi, M.C.; Castro, N. Pathogenic variation among isolates of Pyricularia oryzae affecting rice, wheat, and grasses in Brazil. Int. J. Pest Manag. 1992, 38, 367–371. [Google Scholar]
- Urashima, A.S.; Igarashi, S.; Kato, H. Host range, mating type, and fertility of Pyricularia grisea from wheat in Brazil. Plant Dis. 1993, 77, 1211–1216. [Google Scholar] [CrossRef]
- Urashima, A.S.; Hashimoto, Y.; Don, L.D.; Kusaba, M.; Tosa, Y.; Nakayashiki, H.; Mayama, S. Molecular analysis of the wheat blast population in Brazil with a homolog of retrotransposon MGR583. Jpn. J. Phytopathol. 1999, 65, 429–436. [Google Scholar] [CrossRef]
- Urashima, A.S.; Galbieri, R.; Stabili, A. DNA fingerprinting and sexual characterization revealed two distinct populations of Magnaporthe grisea in wheat blast from Brazil. Czech J. Genet. Plant Breed. 2005, 41, 238–245. [Google Scholar] [CrossRef]
- Roy, K.K.; Reza, M.M.A.; Mustarin, K.E.; Malaker, P.K.; Barma, N.C.D.; He, X.; Singh, P.K. First report of barley blast caused by Magnaporthe oryzae pathotype Triticum (MoT) in Bangladesh. J. Gen. Plant Pathol. 2021, 87, 184–191. [Google Scholar] [CrossRef]
- Roy, K.K.; Rahman, M.E.; Reza, M.A.; Mustarin, K.E.; Malaker, P.K.; Barma, N.C.D.; Hossain, I.; He, X.; Singh, P.K. First report of triticale blast caused by the fungus Magnaporthe oryzae pathotype Triticum in Bangladesh. Can. J. Plant Pathol. 2021, 43, 288–295. [Google Scholar] [CrossRef]
- Eckardt, N.A. Sequencing the rice genome. Plant Cell 2000, 12, 2011–2018. [Google Scholar] [CrossRef]
- Garris, A.J.; Tai, T.H.; Coburn, J.; Kresovich, S.; McCouch, S. Genetic structure and diversity in Oryza sativa L. Genetics 2005, 169, 1631–1638. [Google Scholar] [CrossRef]
- Song, S.; Tian, D.; Zhang, Z.; Hu, S.; Yu, J. Rice genomics: Over the past two decades and into the future. Genom. Proteom. Bioinform. 2018, 16, 397–404. [Google Scholar] [CrossRef]
- Du, H.; Yu, Y.; Ma, Y.; Gao, Q.; Cao, Y.; Chen, Z.; Ma, B.; Qi, M.; Li, Y.; Zhao, X.; et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 2017, 8, 15324. [Google Scholar] [CrossRef]
- Sharma, T.R.; Devanna, B.N.; Kiran, K.; Singh, P.K.; Arora, K.; Jain, P.; Tiwari, I.M.; Dubey, H.; Saklani, B.; Kumari, M.; et al. Status and Prospects of Next generation Sequencing Technologies in Crop Plants. Curr. Issues Mol. Biol. 2018, 27, 1–36. [Google Scholar] [CrossRef]
- GigaScience. The 3000 Rice Genomes Project. GigaScience 2014, 3, 7. [Google Scholar]
- Sun, C.; Hu, Z.; Zheng, T.; Lu, K.; Zhao, Y.; Wang, W.; Shi, J.; Wang, C.; Lu, J.; Zhang, D.; et al. RPAN: Rice pan-genome browser for ∼3000 rice genomes. Nucleic Acids Res. 2017, 45, 597–605. [Google Scholar] [CrossRef]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef]
- Devanna, N.B.; Vijayan, J.; Sharma, T.R. The blast resistance gene Pi54of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PLoS ONE 2014, 9, e104840. [Google Scholar] [CrossRef]
- Thao, N.P.; Chen, L.; Nakashima, A.; Hara, S.I.; Umemura, K.; Takahashi, A.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 2007, 19, 4035–4045. [Google Scholar] [CrossRef]
- Li, W.; Chern, M.; Yin, J.; Wang, J.; Chen, X. Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 2019, 50, 114–120. [Google Scholar] [CrossRef]
- Faivre-Rampant, O.; Thomas, J.; Allègre, M.; Morel, J.B.; Tharreau, D.; Nottéghem, J.L.; Lebrun, M.H.; Schaffrath, U.; Piffanelli, P. Characterization of the model system rice–Magnaporthe for the study of nonhost resistance in cereals. N. Phytol. 2008, 180, 899–910. [Google Scholar] [CrossRef]
- Dai, Y.; Jia, Y.; Correll, J.; Wang, X.; Wang, Y. Diversification and evolution of the avirulence gene AVR-Pita1 in field isolates of Magnaporthe oryzae. Fungal Genet. Biol. 2010, 47, 973–980. [Google Scholar] [CrossRef]
- Liu, G.; Lu, G.; Zeng, L.; Wang, G.L. Two broad-spectrum blast resistance genes, Pi9(t) and Pi2(t), are physically linked on rice chromosome 6. Mol. Genet. Genom. 2002, 267, 472–480. [Google Scholar] [CrossRef]
- Sharma, T.R.; Madhav, M.S.; Singh, B.K.; Shanker, P.; Jana, T.K.; Dalal, V.; Pandit, A.; Singh, A.; Gaikwad, K.; Upreti, H.C.; et al. High-resolution mapping, cloning and molecular characterization of the Pi-k h gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genom. 2005, 274, 569–578. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef]
- Su, J.; Wang, W.; Han, J.; Chen, S.; Wang, C.; Zeng, L.; Feng, A.; Yang, J.; Zhou, B.; Zhu, X. Functional divergence of duplicated genes results in a novel blast resistance gene Pi50 at the Pi2/9 locus. Theor. Appl. Genet. 2015, 128, 2213–2225. [Google Scholar] [CrossRef]
- Chaipanya, C.; Telebanco-Yanoria, M.J.; Quime, B.; Longya, A.; Korinsak, S.; Korinsak, S.; Toojinda, T.; Vanavichit, A.; Jantasuriyarat, C.; Zhou, B. Dissection of broad-spectrum resistance of the Thai rice variety Jao Hom Nin conferred by two resistance genes against rice blast. Rice 2017, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Liu, S.; Xu, P.; Deng, W.; Li, X.; Tharreau, D.; Li, J.; Zhou, J.; Wang, Q.; Tao, D.; et al. Fine mapping of Pi57(t) conferring broad spectrum resistance against Magnaporthe oryzae in introgression line IL-E1454 derived from Oryza longistaminata. PLoS ONE 2017, 12, e0186201. [Google Scholar] [CrossRef]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Jia, Y.; Minkenberg, B.; Wheatley, M.; Fan, J.; Jia, M.H.; Famoso, A.; Edwards, J.D.; Wamishe, Y.; et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, L.; Zhang, X.; Zhang, Q.; Jia, Y.; Wang, G.; Li, S.; Tian, D.; Li, W.H.; Yang, S. Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc. Natl. Acad. Sci. USA 2019, 116, 18479–18487. [Google Scholar] [CrossRef]
- Kiyosawa, S. Inheritance of a particular sensitivity of the rice variety, Sekiguchi Asahi, to pathogens and chemicals, and linkage relationship with blast resistance genes. Bull. Nat. Inst. Agric. Sci. 1970, 21, 61–72. [Google Scholar]
- Wang, J.; Qu, B.; Dou, S.; Li, L.; Yin, D.; Pang, Z.; Zhou, Z.; Tian, M.; Liu, G.; Xie, Q.; et al. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 2015, 15, 49. [Google Scholar] [CrossRef]
- Liu, H.; Dong, S.; Gu, F.; Liu, W.; Yang, G.; Huang, M.; Xiao, W.; Liu, Y.; Guo, T.; Wang, H.; et al. NBS-LRR protein Pik-H4 interacts with OsBIHD1 to balance rice blast resistance and growth by coordinating ethylene-brassinosteroid pathway. Front. Plant Sci. 2017, 8, 127. [Google Scholar] [CrossRef][Green Version]
- Kang, S.G.; Lee, K.E.; Singh, M.; Kumar, P.; Matin, M.N. Rice lesion mimic mutants (LMM): The current understanding of genetic mutations in the failure of ROS scavenging during lesion formation. Plants 2021, 10, 1598. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Yoshida, H.; Ashikawa, I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 2006, 113, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Sallaud, C.; Lorieux, M.; Roumen, E.; Tharreau, D.; Berruyer, R.; Svestasrani, P.; Garsmeur, O.; Ghesquiere, A.; Notteghem, J.-L. dentification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy. Theor. Appl. Genet. 2003, 106, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, S. Durability of resistance to rice blast disease. JIRCAS Work. Rep. 2007, 53, 1–10. [Google Scholar]
- Barman, S.R.; Gowda, M.; Venu, R.C.; Chattoo, B.B. Identification of a major blast resistance gene in the rice cultivar ‘Tetep’. Plant Breed. 2004, 123, 300–302. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Koizumi, S.; La, T.N.; Zenbayashi, K.S.; Ashizawa, T.; Yasuda, N.; Imazaki, I.; Miyasaka, A. Pi35 (t), a new gene conferring partial resistance to leaf blast in the rice cultivar Hokkai 188. Theor. Appl. Genet. 2006, 113, 697–704. [Google Scholar] [CrossRef]
- Lin, F.; Chen, S.; Que, Z.; Wang, L.; Liu, X.; Pan, Q. The blast resistance gene Pi37 encodes a nucleotide binding site–leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics 2007, 177, 1871–1880. [Google Scholar] [CrossRef]
- Imbe, T.; Matsumoto, S. Inheritance of resistance of rice varieties to the blast fungus strains virulent to the variety ‘‘Reiho’’. Jpn. J. Breed. 1985, 35, 332–339. [Google Scholar] [CrossRef]
- Chen, X.W.; Li, S.G.; Xu, J.C.; Zhai, W.X.; Ling, Z.Z.; Ma, B.T.; Wang, Y.P.; Wang, W.M.; Cao, G.; Ma, Y.Q.; et al. Identification of two blast resistance genes in a rice variety, Digu. J. Phytopathol. 2004, 152, 77–85. [Google Scholar] [CrossRef]
- Zhu, X.; Yang, Q.; Yang, J.; Lei, C.; Wang, J.; Ling, Z. Differentiation ability of monogenic lines to Magnaporthe grisea in indica rice. Acta Phytopathol Sin 2004, 34, 361–368. [Google Scholar]
- Tabien, R.; Li, Z.; Paterson, A.; Marchetti, M.; Stansel, J.; Pinson, S. Mapping QTLs for field resistance to the rice blast pathogen and evaluating their individual and combined utility in improved varieties. Theor. Appl. Genet. 2002, 105, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.L.; Huang, D.Y.; Li, W.; Wang, J.L.; Liu, Z.L.; Wang, X.T.; Shi, K.; Cheng, Z.J.; Zhang, X.; Ling, Z.Z.; et al. Molecular mapping of a blast resistance gene in an indica rice cultivar Yanxian No. 1. Rice Genet. Newsl. 2005, 22, 76–77. [Google Scholar]
- Wang, Z.X.; Yano, M.; Yamanouchi, U.; Iwamoto, M.; Monna, L.; Hayasaka, H.; Katayose, Y.; Sasaki, T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999, 19, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.S.; Tanksley, S.D. Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol. Gen. Genet. 1993, 241, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wang, L.; Ikehashi, H.; Tanisaka, T. Identification of a new blast resistance gene in the indica rice cultivar Kasalath using Japanese differential cultivars and isozyme markers. Phytopathology 1996, 86, 1071–1075. [Google Scholar] [CrossRef]
- Utani, D.W.; Moeljopawiro, S.; Aswidinnoor, H.; Setiawan, A.; Hanarida, I. Blast resistance genes in wild rice Oryza rufipogon and rice cultivar IR64 [online]. Indones. J. Agric. 2008, 2, 71–76. [Google Scholar]
- Tabien, R.E.; Pinson, S.R.M.; Marchetti, M.A.; Li, Z.; Park, W.D.; Paterson, A.H.; Stansel, J.W. Blast resistance genes from Teqing and Lemont. In Rice Genetics III: (In 2 Parts); World Scientific: Singapore, 1996; pp. 451–455. [Google Scholar]
- Devi, S.J.S.R.; Singh, K.; Umakanth, B.; Vishalakshi, B.; Rao, K.V.S.; Suneel, B.; Sharma, S.K.; Kadambari, G.K.M.; Prasad, M.S.; Senguttvel, P.; et al. Identification and characterization of a large effect QTL from Oryza glumaepatula revealed Pi68(t) as putative candidate gene for rice blast resistance. Rice 2020, 13, 1–13. [Google Scholar] [CrossRef]
- Pan, Q.H.; Tanisaka, T.; Ikehashi, H. Studies on the genetics and breeding of blast resistance in rice VII. Gene analysis for the blast resistance of Indian native cultivar, Aus 373. Breed Sci. 1997, 47 (Suppl. 1), 35. [Google Scholar]
- Ahn, S.N.; Kim, Y.K.; Hong, H.C.; Choi, H.C.; Moon, H.P.; Han, S.S.; Mccouch, S.R. Mapping of genes conferring resistance to Korean isolates of rice blast fungus using DNA markers. Korean J. Breed. 1997, 29, 416–423. [Google Scholar]
- Goto, I. Genetic Studieson Resistance of Rice Plant to Blast Fungus (VII) Blast Resistance Genes of Kuroka. Jpn. J. Phytopathol. 1988, 54, 460–465. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Q.; Lin, F.; Hua, L.; Wang, C.; Wang, L.; Pan, Q. Identification and fine mapping of Pi39 (t), a major gene conferring the broad-spectrum resistance to Magnaporthe oryzae. Mol. Genet. Genom. 2007, 278, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Causse, M.A.; Fulton, T.M.; Cho, Y.G.; Ahn, S.N.; Chunwongse, J.; Wu, K.; Xiao, J.; Yu, Z.; Ronald, P.C.; Harrington, S.E. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 1994, 138, 1251–1274. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, N.I.; Bonman, J.M.; Mackill, D.J.; Nelson, R.J.; Chattoo, B.B. Identification of RAPD markers linked to a major blast resistance gene in rice. Mol. Breed. 1995, 1, 341–348. [Google Scholar] [CrossRef]
- Wu, J.L.; Fan, Y.Y.; Li, D.B.; Zheng, K.L.; Leung, H.; Zhuang, J.Y. Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates. Theor. Appl. Genet. 2005, 111, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Jeung, J.U.; Kim, B.R.; Cho, Y.C.; Han, S.S.; Moon, H.P.; Lee, Y.T.; Jena, K.K. A novel gene, Pi40 (t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor. Appl. Genet. 2007, 115, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.N.; Kim, Y.K.; Han, S.S.; Choi, H.C.; Moon, H.P.; McCouch, S.R. Molecular mapping of a gene for resistance to a Korean isolate of rice blast. Rice Genet. Newsl. 1996, 13, 74–76. [Google Scholar]
- Qu, S.; Liu, G.; Zhou, B.; Bellizzi, M.; Zeng, L.; Dai, L.; Han, B.; Wang, G.L. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 2006, 172, 1901–1914. [Google Scholar] [CrossRef]
- Mackill, D.J.; Bonman, J.M. Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 1992, 82, 746–749. [Google Scholar] [CrossRef]
- Tabien, R.E.; Li, Z.; Paterson, A.H.; Marchetti, M.A.; Stansel, J.W.; Pinson, S.R.M.; Park, W.D. Mapping of four major rice blast resistance genes from’Lemont’and’Teqing’and evaluation of their combinatorial effect for field resistance. Theor. Appl. Genet. 2000, 101, 1215–1225. [Google Scholar] [CrossRef]
- Pan, Q.H.; Tanisaka, T.; Ikehashi, H. Studies on the genetics and breeding of blast resistance in rice VI. Gene analysis for the blast resistance of two Yunnan native cultivars GA20 and GA25. Breed. Sci. 1996, 46, 70. [Google Scholar]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G.; et al. AB-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhu, X.; Shen, Y.; He, Z. Genetic characterization and fine mapping of the blast resistance locus Pigm (t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor. Appl. Genet. 2006, 113, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Kiyosawa, S. Some considerations on linkage relationships between Pii and Piz in the blast resistance of rice. Rice Genet. Newsl. 1997, 14, 57–59. [Google Scholar]
- Kwon, S.W.; Cho, Y.C.; Kim, Y.G.; Suh, J.P.; Jeung, J.U.; Roh, J.H.; Lee, S.K.; Jeon, J.S.; Yang, S.J.; Lee, Y.T. Development of near-isogenic Japonica rice lines with enhanced resistance to Magnaporthe grisea. Mol. Cells 2008, 25, 407–416. [Google Scholar]
- Ise, K. Linkage analysis of some blast resistance gene in rice, Oryza sativa L. Jpn. J. Breed. 1991, 42 (Suppl. 2), 388–389. [Google Scholar]
- Chauhan, R.; Farman, M.A.R.K.; Zhang, H.B.; Leong, S. Genetic and physical mapping of a rice blast resistance locus, Pi-CO39 (t), that corresponds to the avirulence gene AVR1-CO39 of Magnaporthe grisea. Mol. Genet. Genom. 2002, 267, 603–612. [Google Scholar] [CrossRef]
- Wang, G.L.; Mackill, D.; Bonman, J.; McCouch, S.R.; Champoux, M.C.; Nelson, R.J. RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 1994, 136, 1421–1434. [Google Scholar] [CrossRef]
- Zenbayashi, K.; Ashizawa, T.; Tani, T.; Koizumi, S. Mapping of the QTL. Theor. Appl. Genet. 2002, 104, 547–552. [Google Scholar] [CrossRef]
- Gowda, M.; Roy-Barman, S.; Chattoo, B.B. Molecular mapping of a novel blast resistance gene Pi38 in rice using SSLP and AFLP markers. Plant Breed. 2006, 125, 596–599. [Google Scholar] [CrossRef]
- Fujii, K.; Hayano-Saito, Y.; Shumiya, A.; Inoue, M. Genetical mapping based on the RFLP analysis for the panicle blast resistance derived from a rice parental line St. No. 1. Breed. Sci. 1995, 45 (Suppl. 1), 209. [Google Scholar]
- Fujii, K.; Hayano-Saito, Y.; Saito, K.; Sugiura, N.; Hayashi, N.; Tsuji, T.; Izawa, T.; Iwasaki, M. Identification of a RFLP marker tightly linked to the panicle blast resistance gene Pb1 in rice. Breed. Sci. 2000, 50, 183–188. [Google Scholar] [CrossRef]
- Chen, D.H.; Inukai, T.; Mackill, D.J.; Ronald, P.C.; Nelson, R.J. Molecular mapping of the blast resistance gene, Pi44 (t), in a line derived from a durably resistant rice cultivar. Theor. Appl. Genet. 1999, 98, 1046–1053. [Google Scholar] [CrossRef]
- Hittalmani, S.; Parco, A.; Mew, T.V.; Zeigler, R.S.; Huang, N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 2000, 100, 1121–1128. [Google Scholar] [CrossRef]
- Li, L.Y.; Wang, L.; Jing, J.X.; Li, Z.Q.; Lin, F.; Huang, L.F.; Pan, Q.H. The Pik m gene, conferring stable resistance to isolates of Magnaporthe oryzae, was finely mapped in a crossover-cold region on rice chromosome 11. Mol. Breed. 2007, 20, 179–188. [Google Scholar] [CrossRef]
- Hayasaka, H.; Miyao, A.; Yano, M.; Matsunaga, K.; Sasaki, T. RFLP mapping of a rice blast resistance gene Pi-k. Breed Sci. 1996, 46 (Suppl. 2), 68. [Google Scholar]
- Fjellstrom, R.; Conaway-Bormans, C.A.; McClung, A.M.; Marchetti, M.A.; Shank, A.R.; Park, W.D. Development of DNA markers suitable for marker assisted selection of three Pi genes conferring resistance to multiple Pyricularia grisea pathotypes. Crop Sci. 2004, 44, 1790–1798. [Google Scholar] [CrossRef]
- Goto, I. Genetic studies on the resistance of rice plant to the blast fungus 1. Inheritance of resistance in crosses Sensho× H-79 and Imochi-shirazu× H-79. Jpn. J. Phytopathol. 1970, 36, 304–312. [Google Scholar] [CrossRef]
- Shinoda, H. Studies on the varietal resistance of rice to blast. 6. Linkage relationship of blast resistance genes. Annu. Rev. Chugoku Natl. Agric. Exp. Stn. 1971, 20, 1–25. [Google Scholar]
- Goto, I. Genetic Studies on Resistance of Rice Plant to Blast Fungus II Difference in resistance to the blast disease between Fukunishiki and its parental cultivar, Zenith. Jpn. J. Phytopathol. 1976, 42, 253–260. [Google Scholar] [CrossRef]
- Zhuang, J.-Y.; Ma, W.-B.; Wu, J.-L.; Chai, R.-Y.; Lu, J.; Fan, Y.-Y.; Jin, M.-Z.; Leung, H.; Zheng, K.-L. Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica 2002, 128, 363–370. [Google Scholar] [CrossRef]
- Wu, K.S.; Martinez, C.; Lentini, Z.; Tohme, J.; Chumley, F.G.; Scolnik, P.A.; Valent, B. Cloning a blast resistance gene by chromosome walking. In Rice Genetics III: (In 2 Parts); World Scientific: Singapore, 1996; pp. 669–674. [Google Scholar]
- McCouch, S.R.; Nelson, R.J.; Tohme, J.; Zeigler, R.S. Mapping of blast resistance genes in rice. In Rice Blast Disease; CAB International: Wallingford, UK, 1994; pp. 167–186. [Google Scholar]
- Inukai, T.; Nelson, R.J.; Zeigler, R.S.; Sarkarung, S.; Mackill, D.J.; Bonman, J.M.; Takamure, I.; Kinoshita, T. Genetic analysis of blast resistance in tropical rice cultivars using near-isogenic lines. In Rice Genetics III: (In 2 Parts); World Scientific: Singapore, 1996; pp. 447–450. [Google Scholar]
- Nakamura, S.; Asakawa, S.; Ohmido, N.; Fukui, K.; Shimizu, N.; Kawasaki, S. Construction of an 800-kb contig in the near-centromeric region of the rice blast resistance gene Pi-ta 2 using a highly representative rice BAC library. Mol. Gen. Genet. 1997, 254, 611–620. [Google Scholar] [CrossRef]
- Iwata, N. Registration of new gene symbols. Rice Genet. Newsl. 1996, 13, 12–18. [Google Scholar]
- Bryan, G.T.; Wu, K.S.; Farrall, L.; Jia, Y.; Hershey, H.P.; McAdams, S.A.; Faulk, K.N.; Donaldson, G.K.; Tarchini, R.; Valent, B. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2033–2045. [Google Scholar] [CrossRef]
- Liu, W.G.; Jin, S.J.; Zhu, X.Y.; Feng, W.A.N.G.; Li, J.H.; Liu, Z.R.; Liao, Y.L.; Zhu, M.S.; Huang, H.J.; Liu, Y.B. Improving blast resistance of a thermo-sensitive genic male sterile rice line GD-8S by molecular marker-assisted selection. Rice Sci. 2008, 15, 179–185. [Google Scholar] [CrossRef]
- Kaji, R.; Ogawa, T. RFLP mapping of blast resistance gene Pi-km in rice. Breed Sci. 1996, 46 (Suppl. 2), 70. [Google Scholar]
- Rai, A.K.; Kumar, S.P.; Gupta, S.K.; Gautam, N.; Singh, N.K.; Sharma, T.R. Functional complementation of rice blast resistance gene Pi-k h (Pi54) conferring resistance to diverse strains of Magnaporthe oryzae. J. Plant Biochem. Biotechnol. 2011, 20, 55–65. [Google Scholar] [CrossRef]
- Kumari, M.; Rai, A.K.; Devanna, B.N.; Singh, P.K.; Kapoor, R.; Rajashekara, H.; Prakash, G.; Sharma, V. Co-transformation mediated stacking of blast resistance genes Pi54 and Pi54rh in rice provides broad spectrum resistance against Magnaporthe oryzae. Plant Cell Rep. 2017, 36, 1747–1755. [Google Scholar] [CrossRef]
- Kumari, M.; Rai, A.K.; Devanna, B.N.; Singh, P.K.; Kapoor, R.; Rajashekara, H.; Prakash, G.; Sharma, V.; Sharma, T.R. Stacking of blast resistance orthologue genes in susceptible indica rice line improves resistance against Magnaporthe oryzae. 3 Biotech 2018, 8, 37. [Google Scholar] [CrossRef]
- Arora, K.; Rai, A.K.; Devanna, B.N.; Kumari, B.; Sharma, T.R. Functional validation of the Pi54 gene by knocking down its expression in a blast-resistant rice line using RNA interference and its effects on other traits. Funct. Plant Biol. 2018, 45, 1241–1250. [Google Scholar] [CrossRef]
- Singh, J.; Gupta, S.K.; Devanna, B.N.; Singh, S.; Upadhyay, A.; Sharma, T.R. Blast resistance gene Pi54 over-expressed in rice to understand its cellular and sub-cellular localization and response to different pathogens. Sci. Rep. 2020, 10, 5243. [Google Scholar] [CrossRef]
- Kumari, A.; Das, A.; Devanna, B.N.; Thakur, S.; Singh, P.K.; Singh, N.K.; Sharma, T.R. Mining of rice blast resistance gene Pi54 shows effect of single nucleotide polymorphisms on phenotypic expression of the alleles. Eur. J. Plant Pathol. 2013, 137, 55–65. [Google Scholar] [CrossRef]
- Das, A.; Soubam, D.; Singh, P.K.; Thakur, S.; Singh, N.K.; Sharma, T.R. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genom. 2012, 12, 215–228. [Google Scholar] [CrossRef]
- Sarkar, C.; Saklani, B.K.; Singh, P.K.; Asthana, R.K.; Sharma, T.R. Variation in the LRR region of Pi54 protein alters its interaction with the AvrPi54 protein revealed by in silico analysis. PLoS ONE 2019, 14, e0224088. [Google Scholar] [CrossRef]
- Chen, L.; Hamada, S.; Fujiwara, M.; Zhu, T.; Thao, N.P.; Wong, H.L.; Krishna, P.; Ueda, T.; Kaku, H.; Shibuya, N.; et al. The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 2010, 7, 185–196. [Google Scholar] [CrossRef]
- Nakashima, A.; Chen, L.; Thao, N.P.; Fujiwara, M.; Wong, H.L.; Kuwano, M.; Umemura, K.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 2008, 20, 2265–2279. [Google Scholar] [CrossRef]
- Lieberherr, D.; Thao, N.P.; Nakashima, A.; Umemura, K.; Kawasaki, T.; Shimamoto, K. A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice. Plant Physiol. 2005, 138, 1644–1652. [Google Scholar] [CrossRef]
- Arora, K.; Rai, A.K.; Devanna, B.N.; Dubey, H.; Narula, A.; Sharma, T.R. Deciphering the role of microRNAs during Pi54 gene mediated Magnaporthe oryzae resistance response in rice. Physiol. Mol. Biol. Plants 2021, 27, 633–647. [Google Scholar] [CrossRef]
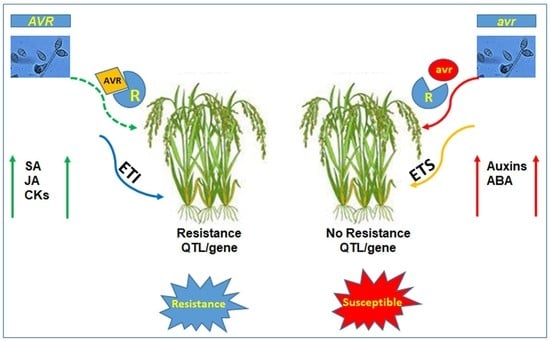
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Thomma, B.P.; Nürnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.; Jones, J.D. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Gupta, S.K.; Rai, A.K.; Kanwar, S.S.; Chand, D.; Singh, N.K.; Sharma, T.R. The single functional blast resistance gene Pi54 activates a complex defence mechanism in rice. J. Exp. Bot. 2012, 63, 757–772. [Google Scholar] [CrossRef]
- Seo, S.; Mitsuhara, I.; Feng, J.; Iwai, T.; Hasegawa, M.; Ohashi, Y. Cyanide, a coproduct of plant hormone ethylene biosynthesis, contributes to the resistance of rice to blast fungus. Plant Physiol. 2011, 155, 502–514. [Google Scholar] [CrossRef]
- Norvienyeku, J.; Lin, L.; Waheed, A.; Chen, X.; Bao, J.; Aliyu, S.R.; Lin, L.; Shabbir, A.; Batool, W.; Zhong, Z.; et al. Bayogenin 3-O-cellobioside confers non-cultivar-specific defence against the rice blast fungus Pyricularia oryzae. Plant Biotechnol. J. 2021, 19, 589. [Google Scholar] [CrossRef]
- Dillon, V.M.; Overton, J.; Grayer, R.J.; Harborne, J.B. Differences in phytoalexin response among rice cultivars of different resistance to blast. Phytochemistry 1997, 44, 599–603. [Google Scholar] [CrossRef]
- Koga, J.; Ogawa, N.; Yamauchi, T.; Kikuchi, M.; Ogasawara, N.; Shimura, M. Functional moiety for the antifungal activity of phytocassane E, a diterpene phytoalexin from rice. Phytochemistry 1997, 44, 249–253. [Google Scholar] [CrossRef]
- Zhan, C.; Lei, L.; Liu, Z.; Zhou, S.; Yang, C.; Zhu, X.; Guo, H.; Zhang, F.; Peng, M.; Zhang, M.; et al. Selection of a subspecies-specific diterpene gene cluster implicated in rice disease resistance. Nat. Plants 2020, 6, 1447–1754. [Google Scholar] [CrossRef]
- Yang, C.; Li, W.; Cao, J.; Meng, F.; Yu, Y.; Huang, J.; Jiang, L.; Liu, M.; Zhang, Z.; Chen, X.; et al. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 2017, 89, 338–353. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Silverman, P.; Seskar, M.; Kanter, D.; Schweizer, P.; Metraux, J.P.; Raskin, I. Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 1995, 108, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bao, Y.; Shan, D.; Wang, Z.; Song, X.; Wang, Z.; Wang, J.; He, L.; Wu, L.; Zhang, Z.; et al. Magnaporthe oryzae induces the expression of a MicroRNA to suppress the immune response in rice. Plant Physiol. 2018, 177, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Patkar, R.N.; Naqvi, N.I. Fungal manipulation of hormone-regulated plant defense. PLoS Pathog. 2017, 13, e1006334. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.J.; Liu, X.L.; Liu, X.Q.; Zhang, H.; Yu, Y.J.; Liang, Z.W. Stunted growth caused by blast disease in rice seedlings is associated with changes in phytohormone signaling pathways. Front. Plant Sci. 2017, 8, 1558. [Google Scholar] [CrossRef]
- Kyndt, T.; Zemene, H.Y.; Haeck, A.; Singh, R.; De Vleesschauwer, D.; Denil, S.; De Meyer, T.; Höfte, M.; Demeestere, K.; Gheysen, G. Below-ground attack by the root knot nematode Meloidogyne graminicola predisposes rice to blast disease. Mol. Plant-Microbe Interact. 2017, 30, 255–266. [Google Scholar] [CrossRef]
- Jiang, C.J.; Shimono, M.; Sugano, S.; Kojima, M.; Liu, X.; Inoue, H.; Sakakibara, H. and Takatsuji, H.; Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol. Plant-Microbe Interact. 2013, 26, 287–296. [Google Scholar] [CrossRef]
- Zhang, S.; Deng, Y.Z.; Zhang, L.H. Phytohormones: The chemical language in Magnaporthe oryzae-rice pathosystem. Mycology 2018, 9, 233–237. [Google Scholar] [CrossRef]
- Yazawa, K.; Jiang, C.J.; Kojima, M.; Sakakibara, H.; Takatsuji, H. Reduction of abscisic acid levels or inhibition of abscisic acid signaling in rice during the early phase of Magnaporthe oryzae infection decreases its susceptibility to the fungus. Physiol. Mol. Plant Pathol. 2012, 78, 1–7. [Google Scholar] [CrossRef]
- Sharma, T.R.; Das, A.; Thakur, S.; Devanna, B.N.; Singh, P.K.; Jain, P.; Vijayan, J.; Kumar, S. Oscillating transcriptome during rice-Magnaporthe interaction. Curr. Issues Mol. Biol. 2016, 19, 99–120. [Google Scholar]
- Jain, P.; Dubey, H.; Singh, P.K.; Solanke, A.U.; Singh, A.K.; Sharma, T.R. Deciphering signalling network in broad spectrum Near Isogenic Lines of rice resistant to Magnaporthe oryzae. Sci. Rep. 2019, 9, 16939. [Google Scholar] [CrossRef]
- Jain, P.; Singh, P.K.; Kapoor, R.; Khanna, A.; Solanke, A.U.; Krishnan, S.G.; Singh, A.K.; Sharma, V.; Sharma, T.R. Understanding host-pathogen interactions with expression profiling of NILs carrying rice-blast resistance Pi9 gene. Front. Plant Sci. 2017, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Arlorio, M.; Ludwig, A.; Boller, T.; Bonfante, P. Inhibition of fungal growth by plant chitinases andβ-1,3-glucanases. Protoplasma 1992, 171, 34–43. [Google Scholar] [CrossRef]
- Bennett, R.N.; Wallsgrove, R.M. Secondary metabolites in plant defence mechanisms. N. Phytol. 1994, 127, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kwon, S.J.; Wu, J.; Choi, J.; Lee, Y.H.; Agrawal, G.K.; Tamogami, S.; Rakwal, R.; Park, S.R.; Kim, B.G.; et al. Transcriptome analysis of early responsive genes in rice during Magnaporthe oryzae infection. Plant Pathol. J. 2014, 30, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Wally, O.; Punja, Z.K. Enhanced disease resistance in transgenic carrot (Daucus carota L.) plants over-expressing a rice cationic peroxidase. Planta 2010, 232, 1229–1239. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, J.; Li, Y.; Yuan, Z.; He, H.; Yang, H.; Qu, H.; Ma, C.; Qu, S. Transcriptome analysis highlights defense and signaling pathways mediated by rice pi21 gene with partial resistance to Magnaporthe oryzae. Front. Plant Sci. 2016, 7, 1834. [Google Scholar] [CrossRef]
- Delteil, A.; Gobbato, E.; Cayrol, B.; Estevan, J.; Michel-Romiti, C.; Dievart, A.; Kroj, T.; Morel, J.-B. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016, 16, 17. [Google Scholar] [CrossRef]
- Albrecht, C.; Boutrot, F.; Segonzac, C.; Schwessinger, B.; Gimenez-Ibanez, S.; Chinchilla, D.; Rathjen, J.P.; de Vries, S.C.; Zipfel, C. Brassinosteroids inhibit pathogen-associated molecular pattern–triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 2012, 109, 303–308. [Google Scholar] [CrossRef]
- Liu, W.; Liu, J.; Triplett, L.; Leach, J.E.; Wang, G.-L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014, 52, 213–241. [Google Scholar] [CrossRef]
- Chujo, T.; Takai, R.; Akimoto-Tomiyama, C.; Ando, S.; Minami, E.; Nagamura, Y.; Kaku, H.; Shibuya, N.; Yasuda, M.; Nakashita, H.; et al. Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta—Gene Struct. Expr. 2007, 1769, 497–505. [Google Scholar] [CrossRef]
- Vijayan, J.; Jain, S.; Jain, N.; Devanna, B.N.; Rathour, R.; Variar, M.; Prashanthi, S.K.; Singh, A.K.; Singh, U.D.; Singh, N.K.; et al. Identification of differentially expressed genes in rice during its early phases of interaction with Magnaporthe oryzae. Indian J. Genet. 2013, 73, 233–243. [Google Scholar] [CrossRef]
- Peng, Y.; Bartley, L.E.; Canlas, P.; Ronald, P.C. OsWRKY IIa transcription factors modulate rice innate immunity. Rice 2010, 3, 36–42. [Google Scholar] [CrossRef]
- Bagnaresi, P.; Biselli, C.; Orrù, L.; Urso, S.; Crispino, L.; Abbruscato, P.; Piffanelli, P.; Lupotto, E.; Cattivelli, L.; Valè, G. Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS ONE 2012, 12, e51609. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Ou, B.; Li, J.; Zhao, Y.; Guo, D.; Zhu, Y.; Chen, Z.; Gu, H.; Li, C.; Qin, G.; et al. Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance. PLoS ONE 2013, 8, e59720. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Qi, M.; Sheng, G.; Yang, Y. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant-Microbe Interact. 2006, 19, 1127–1137. [Google Scholar] [CrossRef]
- Yamada, S.; Kano, A.; Tamaoki, D.; Miyamoto, A.; Shishido, H.; Miyoshi, S.; Taniguchi, S.; Akimitsu, K.; Gomi, K. Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 2012, 53, 2060–2072. [Google Scholar] [CrossRef]
- Umemura, K.; Ogawa, N.; Shimura, M.; Koga, J.; Usami, H.; Kono, T. Possible role of phytocassane, rice phytoalexin, in disease resistance of rice against the blast fungus Magnaporthe grisea. Biosci. Biotechnol. Biochem. 2003, 67, 899–902. [Google Scholar] [CrossRef]
- Okada, K. The biosynthesis of isoprenoids and the mechanisms regulating it in plants. Biosci. Biotechnol. Biochem. 2011, 75, 1219–1225. [Google Scholar] [CrossRef]
- Jwa, N.-S.; Agrawal, G.K.; Tamogami, S.; Yonekura, M.; Han, O.; Iwahashi, H.; Rakwal, R. Role of defense/stress-related marker genes, proteins and secondary metabolites in defining rice self-defense mechanisms. Plant Physiol. Biochem. 2006, 44, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Mitsuhara, I.; Seo, S.; Imai, T.; Koga, J.; Okada, K.; Yamane, H.; Ohashi, Y. Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol. Plant-Microbe Interact. 2010, 23, 1000–1011. [Google Scholar] [CrossRef]
- Yamane, H. Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Biosci. Biotechnol. Biochem. 2013, 77, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef]
- Yao, S.; Yang, Z.; Yang, R.; Huang, Y.; Guo, G.; Kong, X.; Lan, Y.; Zhou, T.; Wang, H.; Wang, W.; et al. Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol. Plant 2019, 12, 1114–1122. [Google Scholar] [CrossRef]
- Baldrich, P.; Campo, S.; Wu, M.T.; Liu, T.T.; Hsing, Y.I.C.; Segundo, B.S. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 2015, 12, 847–863. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, M.; Tang, M.; Dong, B.; Wu, D.; Zhang, Z.; Zhou, B. Repression of microRNA biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice. Plant Sci. 2015, 237, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jeyakumar, J.M.J.; Feng, Q.; Zhao, Z.-X.; Fan, J.; Khaskheli, M.I.; Wang, W.-M. The roles of rice microRNAs in rice-Magnaporthe oryzae interaction. Phytopathol. Res. 2019, 1, 33. [Google Scholar] [CrossRef]
- Singh, P.K.; Ray, S.; Thakur, S.; Rathour, R.; Sharma, V.; Sharma, T.R. Co-evolutionary interactions between host resistance and pathogen avirulence genes in rice-Magnaporthe oryzae pathosystem. Fungal Genet. Biol. 2018, 115, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Sharma, T.R.; Das, A.; Thakur, S.; Jalali, B.L. Recent Understanding on Structure, Function and Evolution of Plant Disease Resistance Genes. Proc. Indian Natl. Sci. Acad. 2014, 80, 83–93. [Google Scholar] [CrossRef]
- Sharma, T.R.; Das, A.; Kumar, S.P.; Lodha, M.L. Resistance Gene Analogues as a Tool for Rapid Identification and Cloning of Disease Resistance genes in Plants. J. Plant Biochem. Biotechnol. 2009, 18, 1–11. [Google Scholar] [CrossRef]
- Pruitt, R.N.; Schwessinger, B.; Joe, A.; Thomas, N.; Liu, F.; Albert, M.; Robinson, M.R.; Chan, L.J.G.; Luu, D.D.; Chen, H.; et al. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci. Adv. 2015, 1, e1500245. [Google Scholar] [CrossRef]
- Van der Biezen, E.A.; Jones, J.D.G. Plant disease- resistance proteins and the gene-for-gene concept. Trends Plant Sci. 1998, 23, 454–456. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D.G. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Ashikawa, I.; Hayashi, N.; Yamane, H.; Kanamori, H.; Wu, J.; Matsumoto, T.; Ono, K.; Yano, M. Two adjacent nucleotide-binding site–leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 2008, 180, 2267–2276. [Google Scholar] [CrossRef]
- Lee, S.K.; Song, M.Y.; Seo, Y.S.; Kim, H.K.; Ko, S.; Cao, P.J.; Suh, J.P.; Yi, G.; Roh, J.H.; Lee, S.; et al. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil–nucleotide-binding–leucine-rich repeat genes. Genetics 2009, 181, 1627–1638. [Google Scholar] [CrossRef]
- Yuan, B.; Zhai, C.; Wang, W.; Zeng, X.; Xu, X.; Hu, H.; Lin, F.; Wang, L.; Pan, Q. The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor. Appl. Genet. 2011, 122, 1017–1028. [Google Scholar] [CrossRef]
- Zhai, C.; Zhang, Y.; Yao, N.; Lin, F.; Liu, Z.; Dong, Z.; Wang, L.; Pan, Q. Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PLoS ONE 2014, 9, e98067. [Google Scholar] [CrossRef]
- Paulus, J.K.; van der Hoorn, R.A.L. Tricked or trapped—Two decoy mechanisms in host–pathogen interactions. PLoS Pathol. 2018, 14, e1006761. [Google Scholar] [CrossRef]
- Kim, S.H.; Qi, D.; Ashfield, T.; Helm, M.; Innes, R.W. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 2016, 351, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Kroj, T.; Chanclud, E.; Michel-Romiti, C.; Grand, X.; Morel, J.B. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. N. Phytol. 2016, 210, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, D.; de Guillen, K.; Cesari, S.; Chalvon, V.; Gracy, J.; Padilla, A.; Kroj, T. Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 2017, 29, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Kearsey, M.J.; Pooni, H.S. The Genetical Analysis of Quantitative Traits; Stanley Thornes Publishers Ltd.: Kingston upon Thames, UK, 1998. [Google Scholar]
- Lopez-Gerena, J. Mapping QTL Controlling Durable Resistance to Rice Blast in the Cultivar Oryzica Llanos 5. Ph.D. Thesis, Plant Pathology College of Agriculture, Universidad del Valle, Cali, Colombia, Kansas State University, Manhattan, KS, USA, 2006. [Google Scholar]
- Bonman, J.M.; Mackill, D. Durable resistance to rice blast disease. Oryza 1988, 25, 103–110. [Google Scholar]
- Zenbayashi-Sawata, K.; Ashizawa, T.; Koizumi, S. Pi34-AVRPi34: A new gene-for-gene interaction for partial resistance in rice to blast caused by Magnaporthe grisea. J. Genet. Plant Pathol. 2005, 71, 395–401. [Google Scholar] [CrossRef]
- Yunoki, T.; Ezuka, A.; Morinaka, Y. Sakurai, Studies on the varietal resistance to rice blast. 4. Variation of field resistance due to fungus strains. Bull. Chugoku Agric. Exp. Stn. Ser. A 1970, 6, 21–41. [Google Scholar]
- Fukuoka, S.; Okuno, K. QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice. Theor. Appl. Genet. 2001, 103, 185–190. [Google Scholar] [CrossRef]
- Latif, M.; Yusop, M.R.; Rahman, M.M.; Talukdar, M.B. Microsatellite and minisatellite markers based DNA fingerprinting and genetic diversity of blast and ufra resistant genotypes. Comptes Rendus Biol. 2011, 334, 282–289. [Google Scholar] [CrossRef]
- Kou, Y.; Wang, S. Broad-spectrum and durability: Understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 2010, 13, 181–185. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Zhang, A.; Ren, Y.; Wu, F.; Wang, G.; Xu, Y.; Lei, C.; Zhu, S.; Pan, T.; et al. A cyclic nucleotide--gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 2019, 29, 820–831. [Google Scholar] [CrossRef]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T.; et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010, 64, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shi, Y.; Liu, W.; Chai, R.; Fu, Y.; Zhuang, J.; Wu, J. A Pid3 allele from rice cultivar Gumei 2 confers resistance to Magnaporthe oryzae. J. Genet. Genom. 2011, 38, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Lei, C.; Xu, X.; Hao, K.; Wang, J.; Cheng, Z.; Ma, X.; Zhou, K.; Zhang, X.; Guo, X.; et al. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol. Plant Microbe Interact. 2015, 28, 558–568. [Google Scholar] [CrossRef]
- Cao, N.; Chen, Y.; Ji, Z.; Zeng, Y.; Yang, C.; Liang, Y. Recent progress in molecular mechanism of rice blast resistance. Chin. J. Rice Sci. 2019, 33, 489–498. [Google Scholar]
- Li, Y.; Wu, C.; Xing, Y.; Chen, H.; He, Y. Dynamic QTL analysis for rice blast resistance under natural infection conditions. Aust. J. Crop Sci. 2008, 2, 73–82. [Google Scholar]
- Takahashi, A.; Hayashi, N.; Miyao, A.; Hirochika, H. Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging. BMC Plant Biol. 2010, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Yoshida, H. Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 2009, 57, 413–425. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.; Ishikawa, K.; Kosami, K.I.; Uno, K.; Nagawa, S.; Tan, L.; Du, J.; Shimamoto, K.; Kawano, Y. Resistance protein Pit interacts with the GEF OsSPK1 to activate OsRac1 and trigger rice immunity. Proc. Natl. Acad. Sci. 2018, 115, E11551–E11560. [Google Scholar] [CrossRef]
- Xu, X.; Hayashi, N.; Wang, C.-T.; Fukuoka, S.; Kawasaki, S.; Takatsuji, H.; Jiang, C.-J. Rice blast resistance gene Pikahei-1(t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol. Breed. 2014, 34, 691–700. [Google Scholar] [CrossRef]
- Liu, M.H.; Kang, H.; Xu, Y.; Peng, Y.; Wang, D.; Gao, L.; Wang, X.; Ning, Y.; Wu, J.; Liu, W.; et al. Genome-wide association study identifies an NLR gene that confers partial resistance to Magnaporthe oryzae in rice. Plant Biotechnol. J. 2020, 18, 1376–1383. [Google Scholar] [CrossRef]
- Zhou, B.; Qu, S.; Liu, G.; Dolan, M.; Sakai, H.; Lu, G.; Bellizzi, M.; Wang, G.L. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant-Microbe Interact. 2006, 19, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Yan, B.; Shou, J.; Tang, J.; Wang, X.; Zhai, K.; Liu, J.; Li, Q.; Luo, M.; Deng, Y.; et al. A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Philos. Trans. R. Soc. B 2019, 374, 20180308. [Google Scholar] [CrossRef] [PubMed]
- Zhai, K.; Deng, Y.; Liang, D.; Tang, J.; Liu, J.; Yan, B.; Yin, X.; Lin, H.; Chen, F.; Yang, D.; et al. RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol. Cell 2019, 74, 996–1009. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Tao, Y.; Chen, X.; Zou, Y.; Lei, C.; Wang, J.; Li, X.; Zhao, X.; Zhang, M.; Lu, Z.; et al. Identification of a new rice blast resistance gene, Pid3, by genome-wide comparison of paired nucleotide-binding site-leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 2009, 182, 1303–1311. [Google Scholar] [CrossRef]
- Zhou, Z.; Pang, Z.; Zhao, S.; Zhang, L.; Lv, Q.; Yin, D.; Li, D.; Liu, X.; Zhao, X.; Li, X.; et al. Importance of OsRac1 and RAI1 in signalling of nucleotide-binding site leucine-rich repeat protein-mediated resistance to rice blast disease. New Phytol. 2019, 223, 828–838. [Google Scholar] [CrossRef]
- Lv, Q.; Xu, X.; Shang, J.; Jiang, G.; Pang, Z.; Zhou, Z.; Wang, J.; Liu, Y.; Li, T.; Li, X.; et al. Functional analysis of Pid3-A4, an ortholog of rice blast resistance gene Pid3 revealed by allele mining in common wild rice. Phytopathology 2013, 103, 594–599. [Google Scholar] [CrossRef]
- Liu, X.; Lin, F.; Wang, L.; Pan, Q. The in silico map-based cloning of Pi36, a rice coiled-coil–nucleotide-binding site–leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics 2007, 176, 2541–2549. [Google Scholar] [CrossRef]
- Takagi, H.; Uemura, A.; Yaegashi, H.; Tamiru, M.; Abe, A.; Mitsuoka, C.; Utsushi, H.; Natsume, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap-Gap: Whole-genome resequencing of mutant F 2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 2013, 200, 276–283. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, B.; Zhu, X.; Yang, J.; Bordeos, A.; Wang, G.; Leach, J.E.; Leung, H. Fine-mapping and molecular marker development for Pi56 (t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor. Appl. Genet. 2013, 126, 985–998. [Google Scholar] [CrossRef]
- Inoue, H.; Hayashi, N.; Matsushita, A.; Xinqiong, L.; Nakayama, A.; Sugano, S.; Jiang, C.J.; Takatsuji, H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein–protein interaction. Proc. Natl. Acad. Sci. USA 2013, 110, 9577–9582. [Google Scholar] [CrossRef]
- Zhai, C.; Lin, F.; Dong, Z.; He, X.; Yuan, B.; Zeng, X.; Wang, L.; Pan, Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011, 189, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Peng, P.; Tian, J.; He, Y.; Zhang, L.; Liu, Z.; Yin, D.; Zhang, Z. Pike, a rice blast resistance allele consisting of two adjacent NBS–LRR genes, was identified as a novel allele at the Pik locus. Mol. Breed. 2015, 35, 1–15. [Google Scholar] [CrossRef]
- Hua, L.; Wu, J.; Chen, C.; Wu, W.; He, X.; Lin, F.; Wang, L.; Ashikawa, I.; Matsumoto, T.; Wang, L.; et al. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theoretical and Applied Genetics 2012, 125, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.R.; Rai, A.K.; Gupta, S.K.; Singh, N.K. Broad-spectrum blast resistance gene pi-k h cloned from rice line Tetep designated as Pi54. J. Plant Biochem. Biotechnol. 2010, 19, 87–89. [Google Scholar] [CrossRef]
- Chen, L.; Shiotani, K.; Togashi, T.; Miki, D.; Aoyama, M.; Wong, H.L.; Kawasaki, T.; Shimamoto, K. Analysis of the Rac/Rop small GTPase family in rice: Expression, subcellular localization and role in disease resistance. Plant Cell Physiol. 2010, 51, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Ribot, C.; Cesari, S.; Abidi, I.; Chalvon, V.; Bournaud, C.; Vallet, J.; Lebrun, M.H.; Morel, J.B.; Kroj, T. The M agnaporthe oryzae effector AVR 1–CO 39 is translocated into rice cells independently of a fungal-derived machinery. Plant J. 2013, 74, 1–12. [Google Scholar] [CrossRef]
- Wang, L.; Ma, Z.; Kang, H.; Gu, S.; Mukhina, Z.; Wang, C.; Wang, H.; Bai, Y.; Sui, G.; Zheng, W.; et al. Cloning and functional analysis of the novel rice blast resistance gene Pi65 in japonica rice. Theor. Appl. Genet. 2022, 135, 173–183. [Google Scholar] [CrossRef]
- Vasudevan, K.; Gruissem, W.; Bhullar, N.K. Identification of novel alleles of the rice blast resistance gene Pi54. Sci. Rep. 2015, 5, 15678. [Google Scholar] [CrossRef]
- Leung, H.; Raghavan, C.; Zhou, B.; Oliva, R.; Choi, I.R.; Lacorte, V.; Jubay, M.L.; Cruz, C.V.; Gregorio, G.; Singh, R.K.; et al. Allele mining and enhanced genetic recombination for rice breeding. Rice 2015, 8, 34. [Google Scholar] [CrossRef]
- Till, B.J.; Reynolds, S.H.; Greene, E.A.; Codomo, C.A.; Enns, L.C.; Johnson, J.E.; Burtner, C.; Odden, A.R.; Young, K.; Taylor, N.E.; et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003, 13, 524–530. [Google Scholar] [CrossRef]
- Kumar, G.R.; Sakthivel, K.; Sundaram, R.M.; Neeraja, C.N.; Balachandran, S.M.; Rani, N.S.; Viraktamath, B.C.; Madhav, M.S. Allele mining in crops: Prospects and potentials. Biotechnol. Adv. 2010, 18, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Rawal, H.C.; Mithra, S.V.A.; Arora, K.; Kumar, V.; Goel, N.; Mishra, D.C.; Chaturvedi, K.K.; Rai, A.; Devi, S.V.; Sharma, T.R.; et al. Genome-wide analysis in wild and cultivated Oryza species reveals abundance of NBS genes in progenitors of cultivated rice. Plant Mol. Biol. Report. 2018, 36, 373–386. [Google Scholar] [CrossRef]
- Yang, M.-Z.; Cheng, Z.-Q.; Chen, S.-N.; Qian, J.; Xu, L.-L.; Huang, X.-Q. A rice blast resistance genetic resource from wild rice in Yunnan, China, J. Plant Physiol. Mol. Biol. 2007, 33, 589–595. [Google Scholar]
- Geng, X.S.; Yang, M.Z.; Huang, X.Q.; Cheng, Z.Q.; Fu, J.; Sun, T.; Li, J. Cloning and analyzing of rice blast resistance gene Pi-ta+ allele from Jinghong erect type of common wild rice (Oryza rufipogon Griff) in Yunnan. Yi Chuan 2008, 30, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Hwang, S.; Chiang, Y.; Lin, T. Molecular evolution of the Pi-ta gene resistant to rice blast in wild rice (Oryza rufipogon). Genetics 2008, 179, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jia, Y.; Shu, Q.Y.; Wu, D. Haplotype diversity at the Pi-ta locus in cultivated rice and its wild relatives. Phytopathology 2008, 98, 1305–1311. [Google Scholar] [CrossRef][Green Version]
- Lee, S.; Costanzo, S.; Jia, Y.; Olsen, K.; Caicedo, A.L. Evolutionary dynamics of the genomic region around the blast resistance gene Pi-ta in AA genome Oryza species. Genetics 2009, 183, 1315–1325. [Google Scholar] [CrossRef]
- Imam, J.; Mandal, N.P.; Variar, M.; Shukla, P. Allele Mining and Selective Patterns of Pi9 Gene in a Set of Rice Landraces from India. Front. Plant Sci. 2016, 7, 1846. [Google Scholar] [CrossRef]
- Thakur, S.; Gupta, Y.K.; Singh, P.K.; Rathour, R.; Variar, M.; Prashanthi, S.K.; Singh, U.D.; Chand, D.; Rana, J.C.; Sharma, T.R. Molecular diversity in rice blast resistance gene Pi-ta makes it highly effective against dynamic population of Magnaporthe oryzae. Funct. Integr. Genom. 2013, 13, 309–322. [Google Scholar] [CrossRef]
- Liu, J.; Hu, Y.; Ning, Y.; Jiang, N.; Wu, J.; Jeon, J.-S.; Xiao, Y.; Liu, X.; Dai, L.; Wang, G.-L. Genetic variation and evolution of the Pi9 blast resistance locus in the AA genome Oryza species. J. Plant Biol. 2011, 54, 294–302. [Google Scholar] [CrossRef]
- Thakur, S.; Singh, P.K.; Rathour, R.; Variar, M.; Prashanthi, S.K.; Singh, A.K.; Singh, U.D.; Chand, D.; Singh, N.K.; Sharma, T.R. Positive selection pressure on rice blast resistance allele Piz-t makes it divergent in Indian land races. J. Plant Interact. 2013, 8, 34–44. [Google Scholar] [CrossRef][Green Version]
- Thakur, S.; Singh, P.K.; Das, A.; Rathour, R.; Variar, M.; Prashanthi, S.K.; Singh, A.K.; Singh, U.D.; Chand, D.; Singh, N.K.; et al. Extensive sequence variation in rice blast resistance gene Pi54 makes it broad spectrum in nature. Front. Plant Sci. 2015, 6, 345. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lv, Q.; Huang, Z.; Xu, X.; Tang, L.; Liu, H.; Wang, C.; Zhou, Z.; Xin, Y.; Xing, J.; Peng, Z.; et al. Allelic variation of the rice blast resistance gene Pid3 in cultivated rice worldwide. Sci. Rep. 2017, 7, 10362. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lei, F.; Wang, Q.; He, W.; Yuan, B.; Yuan, W. Identification of Novel Alleles of the Rice Blast-Resistance Gene Pi9 through Sequence-Based Allele Mining. Rice 2020, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Scardaci, S.C.; Webster, R.K.; Greer, C.A.; Hill, J.E.; William, J.F.; Mutters, R.G.; Brandon, D.M.; McKenzie, K.S.; Oster, J.J. Rice blast: A new disease in California. Agron. Fact Sheet Ser. 1997, 2, 2–5. [Google Scholar]
- Puri, K.D.; Shrestha, S.M.; Khhatri Chhetri, G.B.; Joshi, K.D. Leaf and neck blast resistance reaction in Tropical rice lines under greenhouse condition. Euphytica 2009, 165, 523–532. [Google Scholar] [CrossRef]
- Kumar, V.; Jain, P.; Venkadesan, S.; Karkute, S.; Bhati, J.; Abdin, M.; Sevanthi, A.; Mishra, D.; Chaturvedi, K.; Rai, A.; et al. Understanding rice-Magnaporthe oryzae interaction in resistant and susceptible cultivars of rice under panicle blast infection using a time-course transcriptome analysis. Genes 2021, 12, 301. [Google Scholar] [CrossRef]
- Bonman, J.M. Durable resistance to rice blast disease-environmental influences. In Breeding for Disease Resistance; Springer: Dordrecht, The Netherlands, 1992; pp. 115–123. [Google Scholar]
- Sirithunya, P.; Tragoonrung, S.; Vanavichit, A.; Pa-In, N.; Vongsaprom, C.; Toojinda, T. Quantitative Trait loci associated with leaf and neck blast resistance in recombinant inbred line population of rice (Oryza sativa). DNA Res. 2002, 9, 79–88. [Google Scholar] [CrossRef][Green Version]
- Noenplab, A.; Vanavichit, A.; Toojinda, T.; Sirithunya, P.; Tragoonrung, S.; Sriprakhon, S.; Vongsaprom, C. QTL Mapping for leaf and neck blast resistance in Khao Dawk Mali 105 and Jao Hom Nin recombinant inbred lines. Sci. Asia 2006, 32, 133–142. [Google Scholar] [CrossRef]
- Kalia, S.; Rathour, R. Current status on mapping of genes for resistance to leaf- and neck-blast disease in rice. 3Biotech 2019, 9, 209. [Google Scholar] [CrossRef]
- Ishihara, T.; Hayano-Saito, Y.; Oide, S.; Ebana, K.; La, N.T.; Hayashi, K.; Ashizawa, T.; Suzuki, F.; Koizumi, S. Quantitative trait locus analysis of resistance to panicle blast in the rice cultivar Miyazakimochi. Rice 2014, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Fang, N.; Wang, R.; He, W.; Yin, C.; Guan, C.; Chen, H.; Huang, J.; Wang, J.; Bao, Y.; Zhang, H. QTL mapping of panicle blast resistance in japonica landrace Heikezijing and its application in rice breeding. Mol. Breed. 2016, 36, 171–179. [Google Scholar] [CrossRef]
- Biswal, A.K.; Shamim, M.; Cruzado, K.; Soriano, G.; Ghatak, A.; Toleco, M.; Vikram, P. Role of biotechnology in rice production. In Rice Production Worldwide; Springer: Cham, Switzerland, 2017; pp. 487–547. [Google Scholar]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Ghasemzadeh, A.; Ravanfar, S.A.; Latif, M.A. Molecular progress on the mapping and cloning of functional genes for blast disease in rice (Oryza sativa L.): Current status and future considerations. Crit. Rev. Biotechnol. 2016, 36, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Nizolli, V.O.; Pegoraro, C.; de Oliveira, A.C. Rice blast: Strategies and challenges for improving genetic resistance. Crop Breed. Appl. Biotechnol. 2021, 30, 21. [Google Scholar] [CrossRef]
- Wongsaprom, C.; Sirithunya, P.; Vanavichit, A.; Pantuwan, G.; Jongdee, B.; Sidhiwong, N.; Lanceras-Siangliw, J.; Toojinda, T. Two introgressed quantitative trait loci confer a broad-spectrum resistance to blast disease in the genetic background of the cultivar RD6 a Thai glutinous jasmine rice. Field Crops Res. 2010, 119, 245–251. [Google Scholar] [CrossRef]
- Sreewongchai, T.; Toojinda, T.; Thanintorn, N.; Kosawang, C.; Vanavichit, A.; Tharreau, D.; Sirithunya, P. Development of elite indica rice lines with wide spectrum of resistance to Thai blast isolates by pyramiding multiple resistance QTLs. Plant Breed. 2010, 129, 176–180. [Google Scholar] [CrossRef]
- Manivong, P.; Korinsak, S.; Korinsak, S.; Siangliw, J.L.; Vanavichit, A.; Toojinda, T. Marker-assisted selection to improve submergence tolerance, blast resistance and strong fragrance in glutinous rice. Genom. Genet. 2014, 7, 110–122. [Google Scholar]
- Fukuoka, S.; Saka, N.; Mizukami, Y.; Koga, H.; Yamanouchi, U.; Yoshioka, Y.; Hayashi, N.; Ebana, K.; Mizobuchi, R.; Yano, M. Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 2015, 5, 7773. [Google Scholar] [CrossRef]
- Suwannual, T.; Chankaew, S.; Monkham, T.; Saksirirat, W.; Sanitchon, J. Pyramiding of four blast resistance QTLs into Thai rice cultivar RD6 through marker-assisted selection. Czech J. Genet. Plant Breed. 2017, 53, 1–8. [Google Scholar] [CrossRef]
- Kiyosawa, S. Breakdown of blast resistance in relation to general strategies of resistance gene deployment to prolong effectiveness of resistance in plants. Plant Dis. Epidemiol. 1989, 2, 251–283. [Google Scholar]
- Khanna, A.; Sharma, V.; Ellur, R.K.; Shikari, A.; Krishnan, S.G.; Singh, U.D.; Prakash, G.; Sharma, T.R.; Rathour, R.; Variar, M.; et al. Development and evaluation of near-isogenic lines for major blast resistance gene(s) in Basmati rice. Theor. Appl. Genet. 2015, 128, 1243–1259. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Mou, T.; Yu, H.; Zhou, F. Molecular breeding of thermo-sensitive genic male sterile (TGMS) lines of rice for blast resistance using Pi2 gene. Rice 2015, 8, 1–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tian, D.; Chen, Z.; Chen, Z.; Zhou, Y.; Wang, Z.; Wang, F.; Chen, S. Allele-specific marker-based assessment revealed that the rice blast resistance genes Pi2 and Pi9 have not been widely deployed in Chinese indica rice cultivars. Rice 2016, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Yunyu, W.; Aihong, L. Strategy for use of rice blast resistance genes in rice molecular breeding. Rice Sci. 2020, 27, 263–277. [Google Scholar] [CrossRef]
- Xing, X.; Liu, X.L.; Chen, H.L.; Yang, F.Y.; Li, Y.C.; Liao, H.; You, L.; Liu, J.L.; Dai, L.Y.; Wang, G.L. Improving blast resistance of rice restorer R288 by molecular marker-assisted selection of Pi9 Gene. Crop Res. 2016, 30, 487–491. [Google Scholar]
- Huang, Y.L.; Yan, Z.; Wang, H.; Shen, G.L.; Zhang, C.H. Directed improvement of rice blast resistance of sterile line Q211S with molecular marker-assisted selection. Chin. Agric. Sci. Bull. 2018, 34, 135–140. [Google Scholar]
- Beser, N.; Del Valle, M.M.; Kim, S.M.; Vinarao, R.B.; Surek, H.; Jena, K.K. Marker-assisted introgression of a broad-spectrum resistance gene, Pi40 improved blast resistance of two elite rice (Oryza sativa L.) cultivars of Turkey. Mol. Plant Breed. 2016, 7. [Google Scholar]
- Kumar, S.V.; Srinivas Prasad, M.; Rambabu, R.; Madhavi, K.R.; Bhaskar, B.; Abhilash Kumar, V.; Sundaram, R.M.; Krishna Satya, A.; Sheshu Madhav, M.; Prakasam, V. Marker-assisted introgression of Pi-1 gene conferring resistance to rice blast pathogen pyricularia oryzae in the background of Samba Mahsuri. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2133–2146. [Google Scholar] [CrossRef]
- Kumar, S.V.; Rambabu, R.; Bhaskar, B.; Madhavi, K.R.; Srikanth, S.; Prakasam, V.; Sundaram, R.M.; Madhav, M.S.; Rao, L.S.; Prasad, M.S. Introgression of durable blast resistance gene Pi-54 into indica rice cv. samba mahsuri, through Marker Assisted Backcross Breeding. Electron. J. Plant Breed. 2018, 9, 705–715. [Google Scholar] [CrossRef]
- Khan, G.H.; Shikari, A.B.; Vaishnavi, R.; Najeeb, S.; Padder, B.A.; Bhat, Z.A.; Parray, G.A.; Bhat, M.A.; Kumar, R.; Singh, N.K. Marker-assisted introgression of three dominant blast resistance genes into an aromatic rice cultivar Mushk Budji. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Arunakumari, K.; Durgarani, C.V.; Satturu, V.; Sarikonda, K.R.; Chittoor, P.D.R.; Vutukuri, B.; Laha, G.S.; Nelli, A.P.K.; Gattu, S.; Jamal, M.; et al. Marker-assisted pyramiding of genes conferring resistance against bacterial blight and blast diseases into Indian rice variety MTU1010. Rice Sci. 2016, 23, 306–316. [Google Scholar] [CrossRef]
- Xiao, W.M.; Luo, L.X.; Hui, W.A.N.G.; Tao, G.U.O.; Liu, Y.Z.; Zhou, J.Y.; Zhu, X.Y.; Yang, Q.Y.; Chen, Z.Q. Pyramiding of Pi46 and Pita to improve blast resistance and to evaluate the resistance effect of the two R genes. J. Integr. Agric. 2016, 15, 2290–2298. [Google Scholar] [CrossRef]
- Liu, W.; Wang, F.; Jin, S.; Zhu, X.; Li, J.; Liu, Z.; Liao, Y.; Zhu, M.; Huang, H.; Fu, F.; et al. Improvement of rice blast resistance in TGMS line by pyramiding of Pi1 and Pi2 through molecular marker-assisted selection. Acta Agron. Sin. 2008, 34, 1128–1136. [Google Scholar] [CrossRef]
- Tanweer, F.A.; Rafii, M.Y.; Sijam, K.; Rahim, H.A.; Ahmed, F.; Ashkani, S.; Latif, M.A. Introgression of blast resistance genes (putative Pi-b and Pi-kh) into elite rice cultivar MR219 through marker-assisted selection. Front. Plant Sci. 2015, 6, 1002. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Pan, C.; Chen, X.; Yu, L.; Zhang, X.; Li, Y.; Xiao, N.; Gong, H.; Sheng, S.; Pan, X.; et al. Resistance spectrum difference between two broad-spectrum blast resistance genes, Pigm and Pi2, and their interaction effect on Pi1. Acta Agron. Sin. 2013, 39, 1927–1934. [Google Scholar] [CrossRef]
- Ellur, R.K.; Khanna, A.; Yadav, A.; Pathania, S.; Rajashekara, H.; Singh, V.K.; Krishnan, S.G.; Bhowmick, P.K.; Nagarajan, M.; Vinod, K.K.; et al. Improvement of Basmati rice varieties for resistance to blast and bacterial blight diseases using marker assisted backcross breeding. Plant Sci. 2016, 242, 330–341. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.; Singh, S.P.; Ellur, R.K.; Choudhary, V.; Sarkel, S.; Singh, D.; Krishnan, S.G.; Nagarajan, M.; Vinod, K.K.; et al. Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker assisted backcross breeding. Field Crops Res. 2012, 128, 8–16. [Google Scholar] [CrossRef]
- Xiao, N.; Wu, Y.; Pan, C.; Yu, L.; Chen, Y.; Liu, G.; Li, Y.; Zhang, X.; Wang, Z.; Dai, Z.; et al. Improving of rice blast resistances in japonica by pyramiding major R genes. Front. Plant Sci. 2017, 7, 1918. [Google Scholar] [CrossRef]
- Patroti, P.; Vishalakshi, B.; Umakanth, B.; Suresh, J.; Senguttuvel, P.; Madhav, M.S. Marker-assisted pyramiding of major blast resistance genes in Swarna-Sub1, an elite rice variety (Oryza sativa L.). Euphytica 2019, 215, 1–11. [Google Scholar] [CrossRef]
- Jiang, H.; Feng, Y.; Bao, L.; Li, X.; Gao, G.; Zhang, Q.; Xiao, J.; Xu, C.; He, Y. Improving blast resistance of Jin 23B and its hybrid rice by marker-assisted gene pyramiding. Mol. Breed. 2012, 30, 1679–1688. [Google Scholar] [CrossRef]
- Khanna, A.; Sharma, V.; Ellur, R.K.; Shikari, A.B.; Krishnan, S.G.; Singh, U.D.; Prakash, G.; Sharma, T.R.; Rathour, R.; Variar, M.; et al. Marker assisted pyramiding of major blast resistance genes Pi9 and Pita in the genetic background of an elite Basmati rice variety, Pusa Basmati 1. Indian J. Genet. 2015, 75, 417–425. [Google Scholar] [CrossRef]
- Usatov, A.V.; Kostylev, P.I.; Azarin, K.V.; Markin, N.V.; Makarenko, M.S.; Khachumov, V.A.; Bibov, M.Y. Introgression of the rice blast resistance genes Pi1, Pi2 and Pi33 into Russian rice varieties by marker-assisted selection. Indian J. Genet. Plant Breed. 2016, 76. [Google Scholar] [CrossRef][Green Version]
- Divya, B.; Robin, S.; Rabindran, R.; Senthil, S.; Raveendran, M.; Joel, A.J. Marker assisted backcross breeding approach to improve blast resistance in Indian rice (Oryza sativa) variety ADT43. Euphytica 2014, 200, 61–77. [Google Scholar] [CrossRef]
- Ratna Madhavi, K.; Rambabu, R.; Abhilash Kumar, V.; Vijay Kumar, S.; Aruna, J.; Ramesh, S.; Sundaram, R.M.; Laha, G.S.; Sheshu Madhav, M.; Prasad, M.S. Marker assisted introgression of blast (Pi-2 and Pi-54) genes in to the genetic background of elite, bacterial blight resistant indica rice variety, Improved Samba Mahsuri. Euphytica 2016, 212, 331–342. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, N.; Yu, L.; Pan, C.; Li, Y.; Zhang, X.; Liu, G.; Dai, Z.; Pan, X.; Li, A. Combination patterns of major R genes determine the level of resistance to the M. oryzae in rice (Oryza sativa L.). PLoS ONE 2015, 10, e0126130. [Google Scholar] [CrossRef]
- Jiang, H.; Li, Z.; Liu, J.; Shen, Z.; Gao, G.; Zhang, Q.; He, Y. Development and evaluation of improved lines with broad-spectrum resistance to rice blast using nine resistance genes. Rice 2019, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Balachiranjeevi, C.H.; Bhaskar, N.S.; Abhilash, V.; Akanksha, S.; Viraktamath, B.C.; Madhav, M.S.; Hariprasad, A.S.; Laha, G.S.; Prasad, M.S.; Balachandran, S.M.; et al. Marker-assisted introgression of bacterial blight and blast resistance into DRR17B, an elite, fine-grain type maintainer line of rice. Mol. Breed. 2015, 35, 1–12. [Google Scholar] [CrossRef]
- Jamaloddin, M.; Durga Rani, C.V.; Swathi, G.; Anuradha, C.; Vanisri, S.; Rajan, C.P.D.; Krishnam Raju, S.; Bhuvaneshwari, V.; Jagadeeswar, R.; Laha, G.S.; et al. Marker Assisted Gene Pyramiding (MAGP) for bacterial blight and blast resistance into mega rice variety “Tellahamsa”. PLoS ONE 2020, 15, e0234088. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.; Singh, S.P.; Ellur, R.K.; Singh, D.; Gopala Krishnan, S.; Bhowmick, P.K.; Nagarajan, M.; Vinod, K.K.; Singh, U.D.; et al. Marker-assisted simultaneous but stepwise backcross breeding for pyramiding blast resistance genes Piz5 and Pi54 into an elite Basmati rice restorer line ‘PRR 78’. Plant Breed. 2013, 132, 486–495. [Google Scholar]
- Hari, Y.; Srinivasarao, K.; Viraktamath, B.C.; Hari Prasad, A.S.; Laha, G.S.; Ahmed, M.I.; Natarajkumar, P.; Sujatha, K.; Srinivas Prasad, M.; Pandey, M.; et al. Marker-assisted introgression of bacterial blight and blast resistance into IR 58025B, an elite maintainer line of rice. Plant Breed. 2013, 132, 586–594. [Google Scholar] [CrossRef]
- Singh, A.; Singh, V.K.; Singh, S.P.; Ellur, R.K.; Singh, D.; Bhowmick, K.; Gopala Krishnan, S.; Nagarajan, M.; Vinod, K.K.; Mohapatra, T.; et al. Marker aided improvement of Pusa1460 an elite Basmati rice for resistance to Blast diseases. AoB Plants 2012, 2012, pls029. [Google Scholar]
- Divya, B.; Biswas, A.; Robin, S.; Rabindran, R.; Joel, A. Gene interactions and genetics of blast resistance and yield attributes in rice (Oryza sativa L.). J. Genet. 2014, 93, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Dantre, R.K.; Bhaskar, B.; Kumar, S.V.; Madhavi, K.R.; Prasad, M.S. Performance of Gene Introgressed Lines against Blast Disease under Different Agro Climatic Locations of Chhattisgarh and Telangana. Int. J. Pure Appl. Biosci. 2018, 6, 1472–1477. [Google Scholar] [CrossRef]
- Ravindrababu, V.; Srinivas Prasad, M. Enhancement of leaf blast resistance in rice cultivar ‘Swarna’ by marker assisted backcross breeding. Int. J. Dev. Res. 2016, 08, 9194–9198. [Google Scholar]
- Rekha, G.; Abhilash Kumar, V.; Viraktamath, B.C.; Pranathi, K.; Kousik, M.B.V.N.; Laxmi Prasanna, B.; Backiyalakshmi, C.; Sinha, P.; Ravindra, R.K.; Bhaskar, S.; et al. Improvement of blast resistance of the popular high-yielding, medium slender-grain type, bacterial blight resistant rice variety, Improved Samba Mahsuri by marker-assisted breeding. J. Plant Biochem. Biotechnol. 2018, 27, 463–472. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, N.; Chen, Y.; Yu, L.; Pan, C.; Li, Y.; Zhang, X.; Huang, N.; Ji, H.; Dai, Z.; et al. Comprehensive evaluation of resistance effects of pyramiding lines with different broad-spectrum resistance genes against Magnaporthe oryzae in rice (Oryza sativa L.). Rice 2019, 12, 1–13. [Google Scholar] [CrossRef]
- Ramalingam, J.; Palanisamy, S.; Alagarasan, G.; Renganathan, V.G.; Ramanathan, A.; Saraswathi, R. Improvement of stable restorer lines for blast resistance through functional marker in rice (Oryza sativa L.). Genes 2020, 11, 1266. [Google Scholar] [CrossRef]
- Ramkumar, G.; Madhav, M.S.; Rama Devi, S.J.S.; Umakanth, B.; Pandey, M.K.; Prasad, M.S.; Sundaram, R.M.; Viraktamath, B.C.; Ravindra Babu, V. Identification and validation of novel alleles of rice blast resistant gene Pi54, and analysis of their nucleotide diversity in landraces and wild Oryza species. Euphytica 2016, 209, 725–737. [Google Scholar] [CrossRef]
- Shikari, A.B.; Rajashekara, H.; Khanna, A.; Krishnan, S.G.; Rathour, R.; Singh, U.D.; Sharma, T.R.; Prabhu, K.V.; Singh, A.K. Identification and validation of rice blast resistance genes in Indian rice germplasm. Indian J. Genet. Plant Breed. 2014, 74, 286–299. [Google Scholar] [CrossRef]
- Zhang, L.; Nakagomi, Y.; Endo, T.; Teranishi, M.; Hidema, J.; Sato, S.; Higashitani, A. Divergent evolution of rice blast resistance Pi54 locus in the genus Oryza. Rice 2018, 11, 1–13. [Google Scholar] [CrossRef]
- Wan, B.; Liu, K.; Zhao, S.; Zhu, J.; Liu, Y.; Zhang, G.; Zhu, G. Distribution of rice blast resistance genes Pi-ta, Pi-b, Pigm and Pi54 in backbone parent and their relationships with neck blast resistance. Southwest China J. Agric. Sci. 2020, 33, 1–6. [Google Scholar]
- Azizi, P.; Rafii, M.Y.; Abdullah, S.N.; Hanafi, M.M.; Maziah, M.; Sahebi, M.; Ashkani, S.; Taheri, S.; Jahromi, M.F. Over-expression of the Pikh gene with a CaMV 35S promoter leads to improved blast disease (Magnaporthe oryzae) tolerance in rice. Front. Plant Sci. 2016, 7, 773. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peng, M.; Lin, X.; Xiang, X.; Ren, H.; Fan, X.; Chen, K. Characterization and evaluation of transgenic rice pyramided with the Pi Genes Pib, Pi25 and Pi54. Rice 2021, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Voytas, D.F. Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 2015, 33, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.B.; Aboulhouda, S.; Hysolli, E.; Smith, C.J.; Wang, S.; Castanon, O.; Church, G.M. The future of multiplexed eukaryotic genome engineering. ACS Chem. Biol. 2018, 13, 313–325. [Google Scholar] [CrossRef]
- Devanna, B.N.; Molla, K.A.; Parameswaran, C.; Katara, J.L.; Kumar, A.; Panda, R.S.; Kishor, J.; Bandita, S.; Cayalvizhi, B.; Samantaray, S. CRISPR/Cas mediated genome-editing for rice improvement. ORYZA-Int. J. Rice 2021, 58, 89–102. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Mukhtar, M.S.; Mansoor, S. Genome editing: Targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Liu, D.; Chen, X.; Liu, J.; Ye, J.; Guo, Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 2012, 63, 3899–3911. [Google Scholar] [CrossRef]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.; Wang, S.; Lei, C.; Cheng, Z. Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 2018, 69, 1051–1064. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Shen, L.; Hu, P.; Liu, Q.; Zhu, X.; Qian, Q.; Wang, K.; Wang, Y. Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing. J. Integr. Plant Biol. 2019, 61, 1201–1205. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Yan, F.; Kuang, Y.; Li, N.; Zhang, D.; Zhou, X.; Lin, H.; Zhou, H. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. Plant 2018, 11, 623–626. [Google Scholar] [CrossRef] [PubMed]

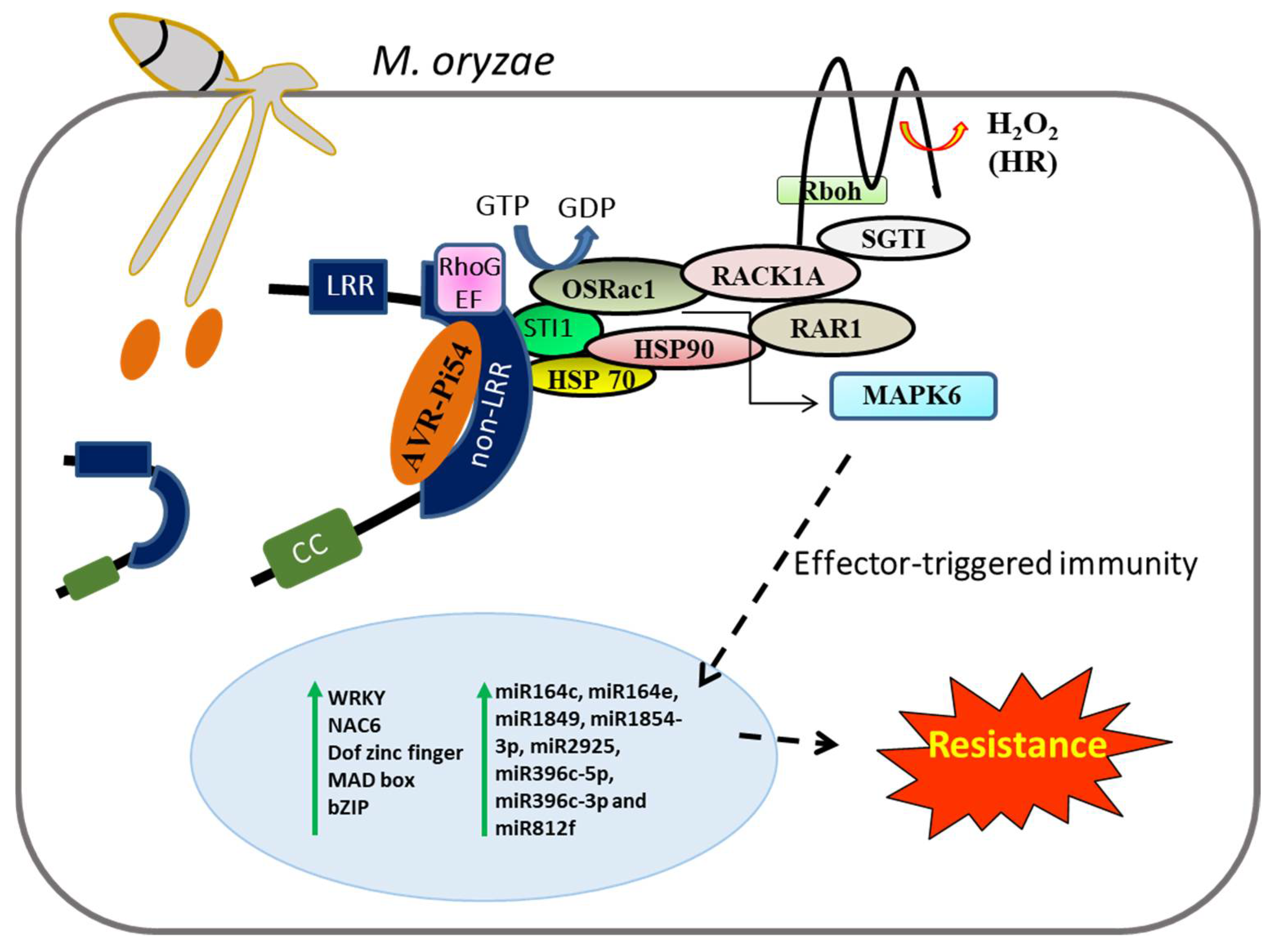


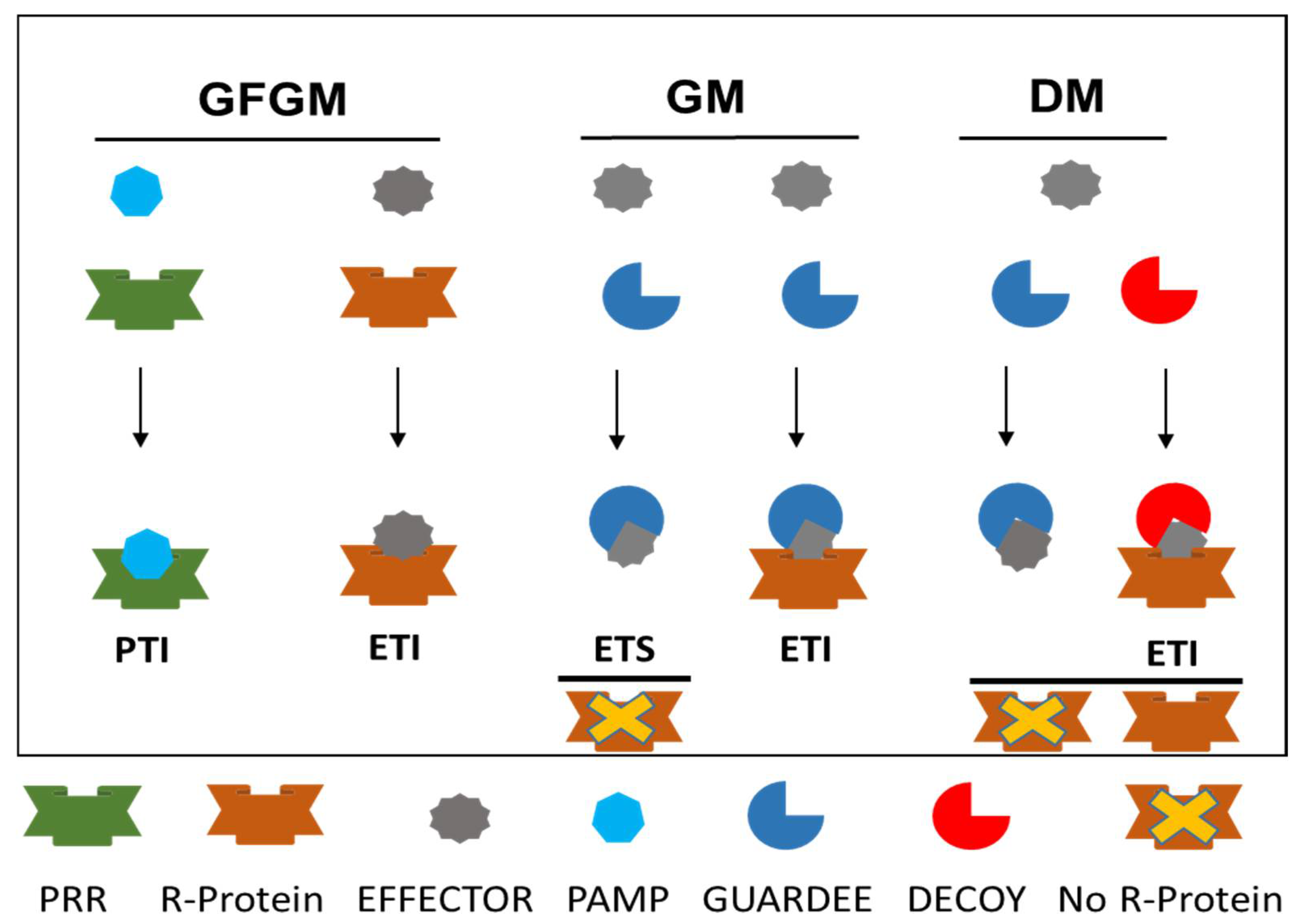

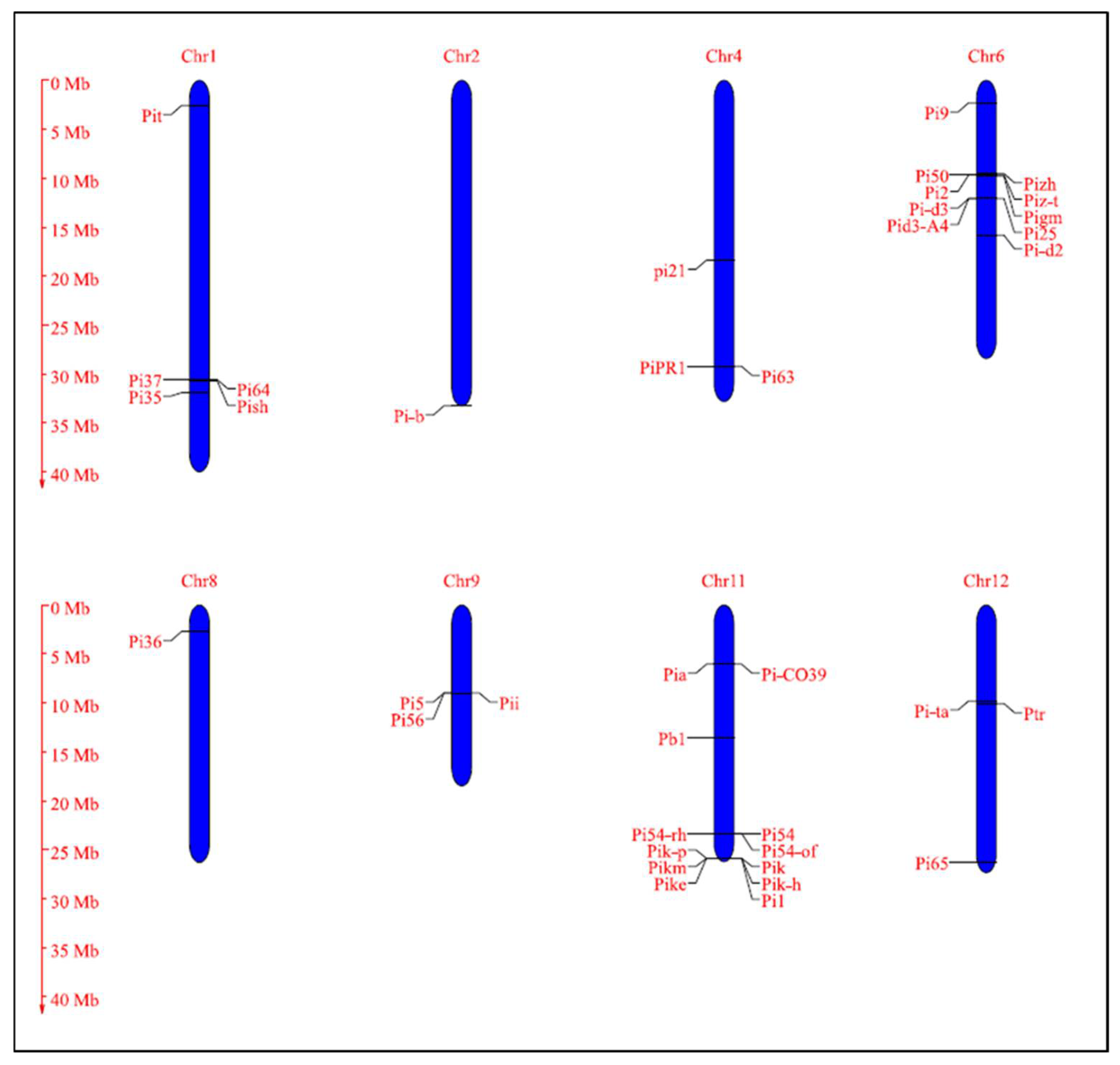
| Strain | Genome Size (Mb) | N 50 Value | No. of Genes | % of Repeats in Genome | Reference |
|---|---|---|---|---|---|
| 70-15 | 38.8 | 1.6 Mb | 11,109 | 9.7 | [11] |
| Ina168 | 38.0 | 28.4 Kb | NA | NA | [16] |
| P131 | 37.95 | 12.3 Kb | 12,714 | 3.15 | [13] |
| Y34 | 38.87 | 11.6 Kb | 12,862 | 3.41 | [13] |
| FJ81278 | 37.3 | 151.7 Kb | 10,453 | 2.73 | [14] |
| HN19311 | 37.1 | 147.4 Kb | 10,256 | 2.83 | [14] |
| 98-06 | 42.1 | 88.6 Kb | 14,019 | 9.3 | [17] |
| B157 | 41 | 92.4 Kb | 12,535 | 10.4 | [15] |
| MG01 | 43 | 54.6 Kb | 13,135 | 10.39 | [15] |
| GFSI1-7-2 | 39.1 | 88.3 Kb | 12,468 | NA | [16] |
| Br48 | 40.7 | 97.5 Kb | 12,671 | NA | [16] |
| Br58 | 40.2 | 91.1 Kb | 12,626 | NA | [16] |
| Z2-1 | 39.5 | 64.1 Kb | 12,383 | NA | [16] |
| Dig41 | 41.3 | 24.3 Kb | 11,457 | NA | [16] |
| RML-29 | 42.2 | 10.4 Kb | 12,746 | 11.78 | [12] |
| RP-2421 | 44.85 | 35.35 Kb | 12,957 | 12.28 | [12] |
| 2539 | 38.08 | 107 Kb | 12,116 | NA | [18] |
| RMg_Dl | 42.42 | 524.2 Kb | 10,555 | NA | [19] |
| FR13 | 46.45 | 5.39 Mb | 14,322 | 13.23 | [20] |
| US71 | 45.61 | 2.81 Mb | 14,348 | 13.23 | [20] |
| CD156 | 43.39 | 5.53 Mb | 14,304 | 6.21 | [20] |
| BR32 | 41.85 | 5.09 Mb | NA | 6.26 | [20] |
| QJ08-2006 | 38.41 | 127.4 Kb | 10,432 | 2.28 | [21] |
| QJ10-10 | 38.28 | 105.1 Kb | 10,418 | 2.22 | [21] |
| QJ10-3001 | 38.40 | 133.1 Kb | 10,401 | 2.28 | [21] |
| RMg-Dl | 34.82 | 45.894 Kb | 12,747 | NA | [22] |
| 70-15 | 40.90 | NA | 12,991 | 11.1 | [23] |
| FR13 | 42.40 | 0.104 Mb | 14,384 | 1.56 | [23] |
| GY11 | 39.00 | 0.226 Mb | 14,781 | 1.00 | [23] |
| PH14 | 40.00 | 0.757 Mb | 13,816 | 1.16 | [23] |
| TH12 | 40.10 | 0.716 Mb | 14,026 | 1.46 | [23] |
| TH16 | 39.10 | 0.939 Mb | 13,571 | 1.60 | [23] |
| US71 | 41.20 | 0.814 Mb | 13,803 | 2.40 | [23] |
| BR32 | 41.90 | 1.760 Mb | 14,336 | 2.00 | [23] |
| CD156 | 42.70 | 1.066 Mb | 14,067 | 1.47 | [23] |
| BR29 | 40.90 | 0.955 Mb | 12,283 | 1.60 | [23] |
| V86010 | 38.9 | 93.4 Kb | 11,857 | 5.1 | [24] |
| 76_3 | 38.35 | 0.159 Mb | NA | NA | [25] |
| 82_0835 | 40.07 | 0.136 Mb | NA | NA | [25] |
| 90_4_1 | 39.92 | 0.151 Mb | NA | NA | [25] |
| BF17 | 39.72 | 0.138 Mb | NA | NA | [25] |
| BF32 | 40.18 | 0.120 Mb | NA | NA | [25] |
| BF48 | 40.01 | 0.144 Mb | NA | NA | [25] |
| BF5 | 40.96 | 0.122 Mb | NA | NA | [25] |
| BN0293 | 38.14 | 0.178 Mb | NA | NA | [25] |
| EG308 | 41.56 | 0.149 Mb | NA | NA | [25] |
| Glhn3 | 39.39 | 0.134 Mb | NA | NA | [25] |
| Glhn4 | 39.52 | 0.134 Mb | NA | NA | [25] |
| JUM1 | 40.50 | 0.127 Mb | NA | NA | [25] |
| KE002 | 40.25 | 0.147 Mb | NA | NA | [25] |
| KE016 | 40.27 | 0.156 Mb | NA | NA | [25] |
| KE017 | 40.10 | 0.141 Mb | NA | NA | [25] |
| KE019 | 39.37 | 0.176 Mb | NA | NA | [25] |
| KE021 | 40.07 | 0.152 Mb | NA | NA | [25] |
| KE029 | 41.03 | 0.154 Mb | NA | NA | [25] |
| KE041 | 39.00 | 0.146 Mb | NA | NA | [25] |
| KE210 | 38.85 | 15.86 Kb | NA | NA | [25] |
| KE255 | 39.83 | 0.119 Mb | NA | NA | [25] |
| KE332 | 41.00 | 35.0 Kb | NA | NA | [25] |
| KE415 | 39.45 | 33.91 Kb | NA | NA | [25] |
| KE443 | 40.88 | 36.41 Kb | NA | NA | [25] |
| KE473 | 40.61 | 37.27 Kb | NA | NA | [25] |
| KE491 | 40.05 | 34.50 Kb | NA | NA | [25] |
| NG0110 | 39.60 | 0.115 Mb | NA | NA | [25] |
| NG0135 | 39.85 | 0.109 Mb | NA | NA | [25] |
| NG0153 | 39.80 | 0.130 Mb | NA | NA | [25] |
| NGO104 | 38.94 | 0.128 Mb | NA | NA | [25] |
| TG004 | 39.96 | 0.125 Mb | NA | NA | [25] |
| TH3 | 37.30 | 22.62 Kb | NA | NA | [25] |
| TZ090 | 38.95 | 0.127 Mb | NA | NA | [25] |
| UG08 | 39.18 | 0.122 Mb | NA | NA | [25] |
| V0104 | 40.34 | 0.126 Mb | NA | NA | [25] |
| V0108 | 39.90 | 0.153 Mb | NA | NA | [25] |
| V0113 | 39.75 | 0.151 Mb | NA | NA | [25] |
| Avr Gene | Protein Size (aa) | Chromosome | Effector Type * | Cognate R Gene | Reference |
|---|---|---|---|---|---|
| PWL1 | 147 | 2 | Glycine-rich | Unknown | [29] |
| PWL2 | 145 | 2 | Glycine-rich | Unknown | [30] |
| AVR1-CO39 | 89 | 1 | ToxB like | Pi-CO39 | [31] |
| AVR-Pita | 224 | 3 | Zinc metalloprotease | Pi-ta | [32] |
| ACE1 | 4035 | 1 | PKS/NRPS | Pi33 (not cloned) | [33] |
| AVR-Pia | 85 | 5 or 7 ** | ToxB like | Pia | [16,34] |
| AVR-Pii | 70 | 7 | Unknown | Pii | [16] |
| AVR-Pik/km/kp; (AVR-Pikh) | 113 (5 alleles) | 1 | ToxB like | Pik/Pik-m/Pik-p, Pik-h | [16,35] |
| AvrPiz-t | 108 | 7 | ToxB like | Piz-t | [36] |
| AVR-Pi9 | 91 | 7 | Six cysteine | Pi9 | [37] |
| AVRPib | 75 | 3 | Unknown | Pib | [38] |
| AVR-Pi54 | 153 | 4 | ToxB like | Pi54, Pi54rh, Pi54of | [39] |
| MoHTR1 | Unknown | Unknown | zinc-finger TF | Unknown | [28] |
| MoHTR2 | Unknown | Unknown | zinc-finger TF | Unknown | [28] |
| Target Gene | Recipient Parent | Chromosome | Marker Used | Reference |
|---|---|---|---|---|
| Pi2 | C815S | 6 | RM527 | [315] |
| Pigm | Kongyu 131, Longjing 26, Kenjiandao 6 | 6 | M80362 | [316] |
| Pigm | KT27S | 6 | G8900 | [316] |
| Pi9 | E32 | 6 | Ins2-3 | [317] |
| Pi9 | R288 | 6 | Clon2-1 | [318] |
| Pi9 | Q211S | 6 | Nbs21 | [319] |
| Pi40 | Osmancik-97, Halilbey | 6 | 9871.T7E2b | [320] |
| Pi1 | BPT5204 | 11 | RM224 | [321] |
| Pi54 | BPT5204 | 11 | Pi-54MAS | [322] |
| Pi54 | R1, R2 | 11 | RM224 | [323] |
| Pi54 | MTU1010 | 11 | Pi54MAS/RM206 | [324] |
| Pi46/Pi-ta | Hanghui 179 | 11, 12 | RM224, YL155/YL87//YL155/87 | [325] |
| Pi1/Pi54/Pi-ta | Mushk Budji | 11, 12, 12 | Pi54MAS, RM224, YL155/YL87//YL155/87 | [323] |
| Pib/Pik | K6415 | 2, 11 | NSb, K6415 | [326] |
| Pib/Pi54 | MR219 | 2, 11 | RM208, RM206 | [107] |
| Pi1/Pi2 | GD-7S | 6, 11 | RM144, AP22 | [327] |
| Pi1/Pi2 | Pusa RH-10 | 6, 11 | RM5926, AP5659-5 | [326] |
| Pi1/Pi2, Pi1/Pigm | GZ63S, 97S, R084, R609 | 6, 11 | RM224, ZJ58.7, AP22 | [328] |
| Pi2/Pi54 | PB1121 | 6, 11 | AP5659-5, RM206 | [329] |
| Pi2/Pi54 | PRR78 | 6, 11 | AP5930, RM206 | [330] |
| Piz-t/Pi54, Pi9/Pi54 | 07GY31 | 6, 11 | Z4794, Pikh-1 | [331] |
| Pi2/Pi1/Pi54 | Swarna-Sub1 | 6, 11 | Pi54MAS, RM224, AP5659-5 | [332] |
| Pi1/Pi2/D12 | Jin 23B | 6, 11, 12 | RM144/RM224, PI2-4/HC28, RM277/RM309 | [333] |
| Pi9/Pi-ta | Pusa Basmati 1 | 6, 12 | AP5659-5, YL155/YL87//YL155/87 | [334] |
| Pi1/Pi2/Pi33 | Kuboyar | 6, 8, 11 | RM224, RM527, RM310 | [335] |
| Pi1/Pi2/Pi33 | ADT43 | 6, 8, 11 | RM224, RM527, RM25 | [336] |
| Pi2/Pi54 | Sambha Mahsuri | 6,11 | Pi54MAS, AP5659-5 | [337] |
| Country | Approach | Applications | Cultivar Developed | Reference |
|---|---|---|---|---|
| China; Beijing | MAS-Gene pyramiding | Pi9, Pizt, and Pi54 for blast resistance | NILs | [331] |
| China; Yangzhou | MAS-Gene pyramiding | Combination of major R genes including Pi54 for blast resistance | NILs | [338] |
| China; Wuhan | MAS-Gene pyramiding | Pi54, Pi37, Pit, Pid3, Pigm, Pi36, Pi5, Pikm, and Pb1 for blast resistance | Improved Y58S, GuangZhan63S (GZ63), C815S and HD9802S | [339] |
| India (ICAR-IIRR) | MAS-Gene pyramiding | Pi54 blast and Xa21, xa13 blight resistance | MTU1010 | [324] |
| India (ICAR-IARI) | MAS-Gene pyramiding | Pi2, Pi54 blast and xa13, Xa21 blight resistance | PB1121-NILs and PB6-NILs | [329] |
| India (ICAR-IIRR) | MAS-Gene pyramiding | Pi54 blast and Xa2 blight resistance | DRR17B | [340] |
| India (ICAR-IIRR) | MAS-Gene pyramiding | Pi54 and Pi2 blast resistance | Improved Samba Mahsuri | [337] |
| India (ICAR-IARI) | MAS-Gene pyramiding | Pi54, Pi1, Pita, Pi2, and Pi9 | PB1 NILs | [314] |
| India (PJTSAU, Hyderabad) | MAS-Gene pyramiding | Pi54 and Pi1 for blast resistance | Tellahamsa | [341] |
| ICAR-IARI | MAS-Gene pyramiding | Piz5 and Pi54 blast resistance | Basmati restorer PRR78 | [342] |
| ICAR-IIRR | MAS-Gene pyramiding | Pi54 blast and Xa2 blight resistance | IR58025B | [343] |
| ICAR-IARI | MAS-Gene pyramiding | Pi54 blast, xa13, Xa21 blight and QTL qSBR11-1 ShB resistance | Improved Pusa Basmati 1 | [344] |
| ICAR-IIRR | MAS | Pi1, Pi2, Pi33, and Pi54 for blast resistance | ADT 43 NIL | [345] |
| ICAR-IIRR | MAS | Pi1, Pi2, and Pi54 | 16 introgressed lines | [346] |
| Universiti Putra, Malaysia | MABB | Pi54 (Pi-kh) and Pi-b | MR219 | [327] |
| ICAR-IIRR | MABB | Pi54 blast resistance | Swarna | [347] |
| ICAR-IIRR | MABB | Pi54 introgression for blast resistance | Samba Mahsuri | [322] |
| ICAR-IIRR | MABB | Pi2, Pi54, Xa21, xa13, and xa5 | Improved Samba Mahsuri | [348] |
| UAS & Tech, Kashmir | MABB | Pi54, Pi1, and Pita | Mushk Budji | [323] |
| ICAR-IIRR | MAS-Gene pyramiding | PizPi1, Pi2, and Pi54 | Swarna-Sub1 | [332] |
| China: Yangzhou | MAS-Gene pyramiding | Pi1, Pi33, and Pi54, Piz | 15-pyramided lines | [349] |
| TNAU | MAS | Pi54 introgression for blast resistance | Restorer lines | [350] |
| ICAR-IIRR | MAS-Gene pyramiding | Pi54, Pi1, Xa21, and xa13 | Tellahamsa | [341] |
| ICAR-IARI | Allele mining | Pi54 allele mining land races and wild rice | - | [351] |
| Switzerland (ETH Zurich) | Allele mining | Pi54 mining from 885 Indian rice genotype | - | [279] |
| ICAR-NRCPB | Allele mining | Pi54 mining from 92 rice lines | - | [293] |
| ICAR-IARI | Allele mining | Pi54 mining from 100 rice germplasm | - | [352] |
| ICAR-NRCPB | Allele mining | Pi54 mining from land races and wild rice | - | [170] |
| Tohoku University, Japan | Allele mining | Pi54 evolution in the Oryza genus | - | [353] |
| China; Yancheng | Allele mining | Field resistance for blast Pi-ta, Pigm, and Pi54 for blast disease | Rice accessions | [354] |
| Malaysia | Over-expression | Constitutive expression of Pi54 homologue from rice line PH9 | Transgenic- MR219 | [355] |
| ICAR-NRCPB | Over-expression | Pi54 orthologue from O. officinalis | Transgenic TP309 | [89] |
| ICAR-NRCPB | Over-expression | Pi54 orthologue from O. rhizomatis | Transgenic TP309 | [171] |
| ICAR-NIPB | Over-expression | Pi54 | TP309 | [169] |
| China; Chengdu | Over-expression | Pib, Pi25, and Pi54 | Kasalath, Zhenghan 10 | [356] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devanna, B.N.; Jain, P.; Solanke, A.U.; Das, A.; Thakur, S.; Singh, P.K.; Kumari, M.; Dubey, H.; Jaswal, R.; Pawar, D.; et al. Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions. J. Fungi 2022, 8, 584. https://doi.org/10.3390/jof8060584
Devanna BN, Jain P, Solanke AU, Das A, Thakur S, Singh PK, Kumari M, Dubey H, Jaswal R, Pawar D, et al. Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions. Journal of Fungi. 2022; 8(6):584. https://doi.org/10.3390/jof8060584
Chicago/Turabian StyleDevanna, Basavantraya N., Priyanka Jain, Amolkumar U. Solanke, Alok Das, Shallu Thakur, Pankaj K. Singh, Mandeep Kumari, Himanshu Dubey, Rajdeep Jaswal, Deepak Pawar, and et al. 2022. "Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions" Journal of Fungi 8, no. 6: 584. https://doi.org/10.3390/jof8060584
APA StyleDevanna, B. N., Jain, P., Solanke, A. U., Das, A., Thakur, S., Singh, P. K., Kumari, M., Dubey, H., Jaswal, R., Pawar, D., Kapoor, R., Singh, J., Arora, K., Saklani, B. K., AnilKumar, C., Maganti, S. M., Sonah, H., Deshmukh, R., Rathour, R., & Sharma, T. R. (2022). Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions. Journal of Fungi, 8(6), 584. https://doi.org/10.3390/jof8060584